Machine Learning for Plant Breeding and Biotechnology
Abstract
:1. Introduction
2. Traditional Plant Breeding
2.1. Assessment and Classification of Genetic Diversity
2.2. Yield Component Analysis and Indirect Selection (Prediction)
2.3. Yield Stability and Genotype × Environment Interaction
2.4. Biotic and Abiotic Stress Assessment
2.5. Classical Mating Designs and Hybrid Breeding Programs
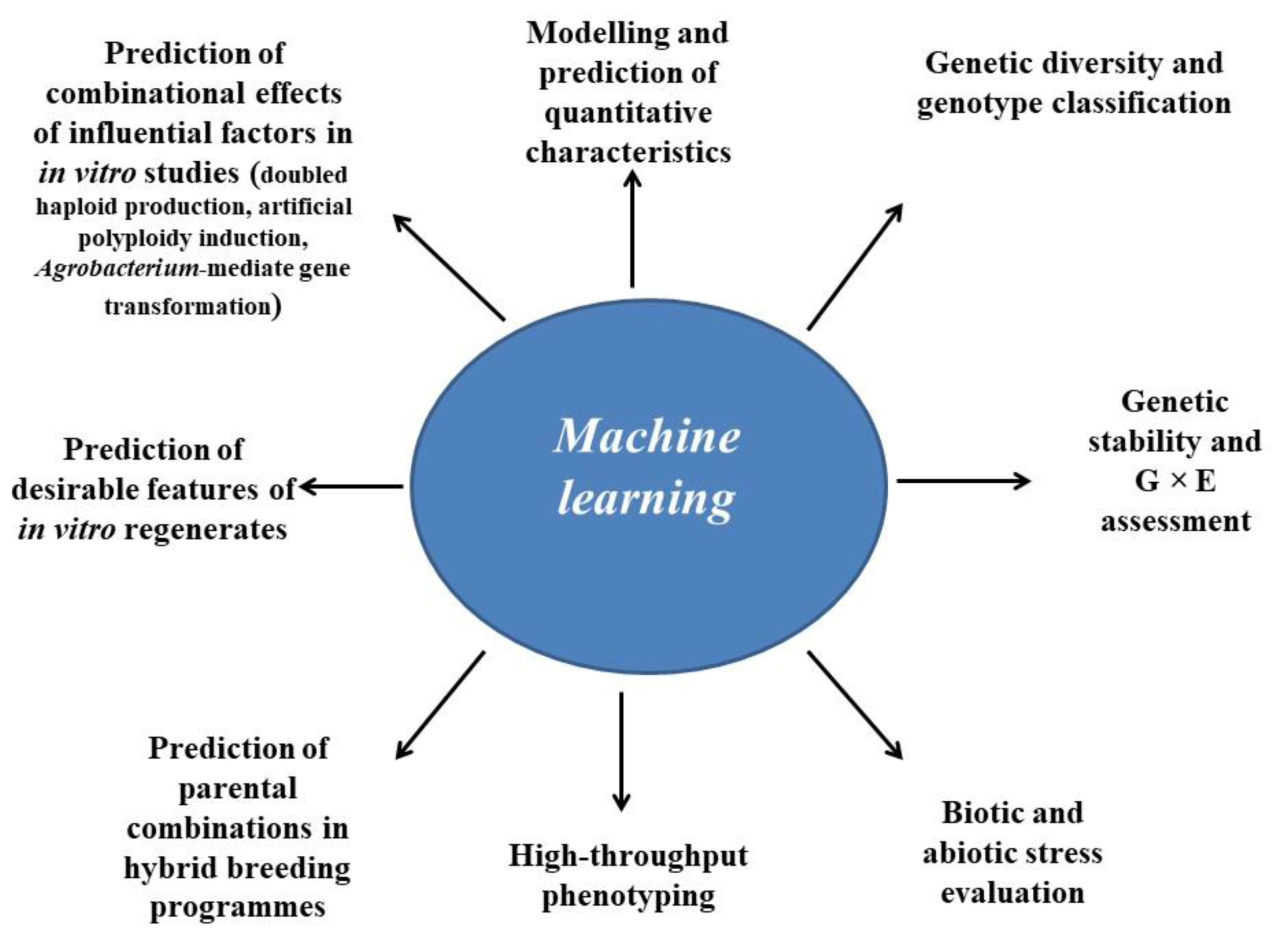
3. Applications of Machine Learning in In Vitro-Based Plant Biotechnology
4. Coupled Machine Learning-Image Processing for High-Throughput Phenotyping and Precision Agriculture
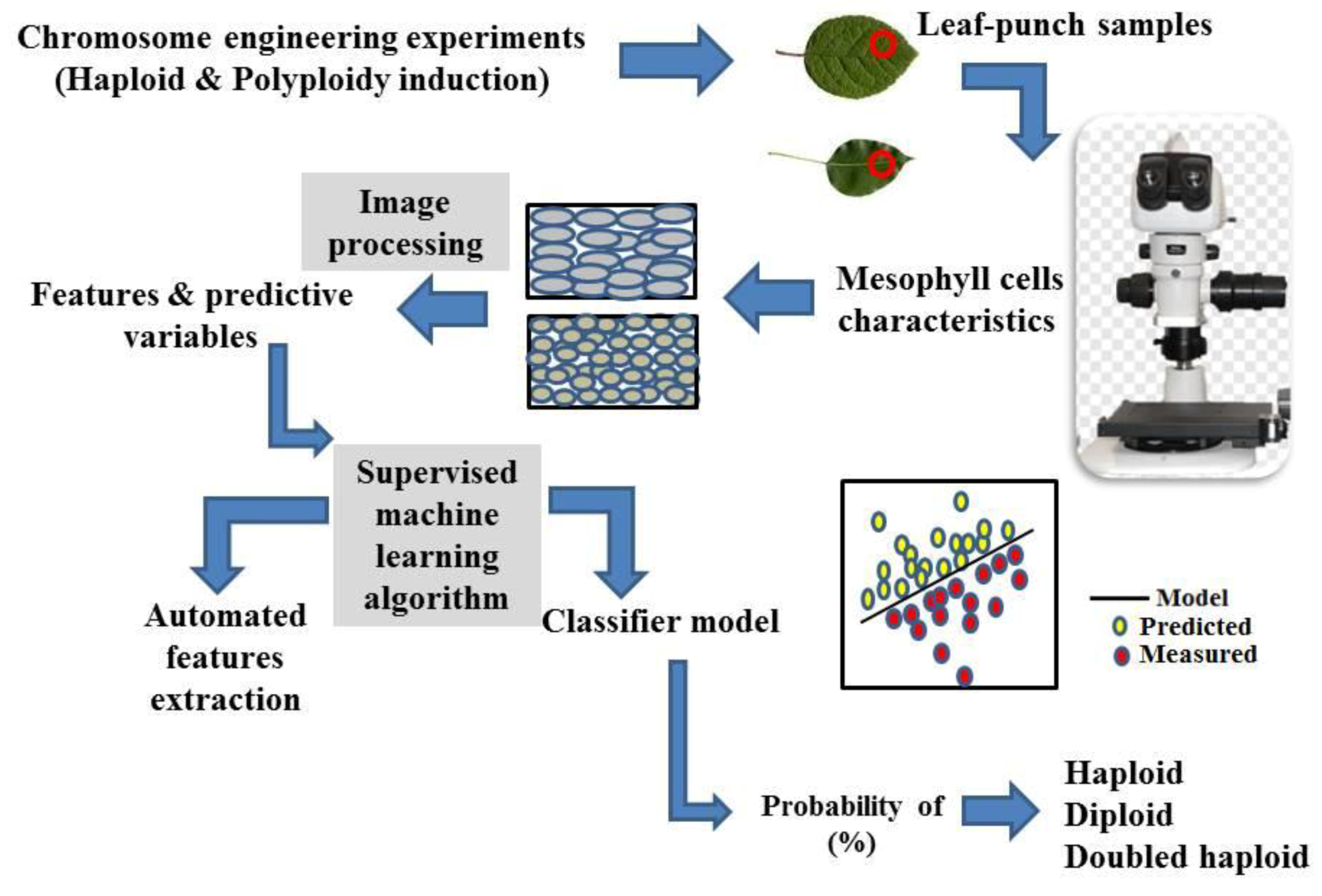
5. A Proposed Idea for Plant Ploidy Level Determination through Image Processing-Machine Learning
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ali, S.; Shafique, O.; Mahmood, T.; Hanif, M.A.; Ahmed, I.; Khan, B.A. A Review about Perspectives of Nanotechnology in Agriculture. Pakistan J. Agric. Res. 2018, 31, 116–121. [Google Scholar] [CrossRef]
- Niazian, M.; Sadat-Noori, S.A.; Abdipour, M. Modeling the seed yield of Ajowan (Trachyspermum ammi L.) using artificial neural network and multiple linear regression models. Ind. Crops Prod. 2018, 117, 224–234. [Google Scholar] [CrossRef]
- Hesami, M.; Naderi, R.; Tohidfar, M.; Yoosefzadeh-Najafabadi, M. Application of Adaptive Neuro-Fuzzy Inference System-Non-dominated Sorting Genetic Algorithm-II (ANFIS-NSGAII) for Modeling and Optimizing Somatic Embryogenesis of Chrysanthemum. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Chegini, G.R.; Khazaei, J.; Ghobadian, B.; Goudarzi, A.M. Prediction of process and product parameters in an orange juice spray dryer using artificial neural networks. J. Food Eng. 2008, 84, 534–543. [Google Scholar] [CrossRef]
- Zheng, H.; Li, W.; Jiang, J.; Liu, Y.; Cheng, T.; Tian, Y.; Zhu, Y.; Cao, W.; Zhang, Y.; Yao, X.A. Comparative Assessment of Different Modeling Algorithms for Estimating Leaf Nitrogen Content in Winter Wheat Using Multispectral Images from an Unmanned Aerial Vehicle. Remote Sens. 2018, 10, 2026. [Google Scholar] [CrossRef] [Green Version]
- Hesami, M.; Naderi, R.; Yoosefzadeh-Najafabadi, M.; Rahmati, M. Data-driven modeling in plant tissue culture. J. Appl. Environ. Biol. Sci 2017, 7, 37–44. [Google Scholar]
- Salehi, M.; Farhadi, S.; Moieni, A.; Safaie, N.; Ahmadi, H. Mathematical Modeling of Growth and Paclitaxel Biosynthesis in Corylus avellana Cell Culture Responding to Fungal Elicitors Using Multilayer Perceptron-Genetic Algorithm. Front. Plant Sci. 2020, 11, 1148. [Google Scholar] [CrossRef]
- Asefpour Vakilian, K. Machine learning improves our knowledge about miRNA functions towards plant abiotic stresses. Sci. Rep. 2020, 10, 3041. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Cimen, E.; Singh, N.; Buckler, E. Deep learning for plant genomics and crop improvement. Curr. Opin. Plant Biol. 2020, 54, 34–41. [Google Scholar] [CrossRef]
- Hu, H.; Scheben, A.; Edwards, D. Advances in Integrating Genomics and Bioinformatics in the Plant Breeding Pipeline. Agriculture 2018, 8, 75. [Google Scholar] [CrossRef] [Green Version]
- Orozco-Arias, S.; Isaza, G.; Guyot, R. Retrotransposons in Plant Genomes: Structure, Identification, and Classification through Bioinformatics and Machine Learning. Int. J. Mol. Sci. 2019, 20, 3837. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, R. Predicting average regional yield and production of wheat in the Argentine Pampas by an artificial neural network approach. Eur. J. Agron. 2009, 30, 70–77. [Google Scholar] [CrossRef]
- Azevedo, A.M.; Andrade Júnior, V.C.D.; Pedrosa, C.E.; Oliveira, C.M.D.; Dornas, M.F.S.; Cruz, C.D.; Valadares, N.R. Application of artificial neural networks in indirect selection: A case study on the breeding of lettuce. Bragantia 2015, 74, 387–393. [Google Scholar] [CrossRef]
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Shahroudy, A.; Shuai, B.; Liu, T.; Wang, X.; Wang, G.; Cai, J.; et al. Recent advances in convolutional neural networks. Pattern Recognit. 2018, 77, 354–377. [Google Scholar] [CrossRef] [Green Version]
- Hesami, M.; Naderi, R.; Tohidfar, M.; Yoosefzadeh-Najafabadi, M. Development of support vector machine-based model and comparative analysis with artificial neural network for modeling the plant tissue culture procedures: Effect of plant growth regulators on somatic embryogenesis of chrysanthemum, as a case study. Plant Methods 2020, 16, 112. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.M.; Darvishzadeh, R.; Skidmore, A.; Gara, T.W.; Heurich, M. Machine learning methods’ performance in radiative transfer model inversion to retrieve plant traits from Sentinel-2 data of a mixed mountain forest. Int. J. Digit. Earth 2020, 1–15. [Google Scholar] [CrossRef]
- Gold, K.M.; Townsend, P.A.; Herrmann, I.; Gevens, A.J. Investigating potato late blight physiological differences across potato cultivars with spectroscopy and machine learning. Plant Sci. 2020, 295, 110316. [Google Scholar] [CrossRef]
- Barth, R.; IJsselmuiden, J.; Hemming, J.; Van Henten, E.J. Synthetic bootstrapping of convolutional neural networks for semantic plant part segmentation. Comput. Electron. Agric. 2019, 161, 291–304. [Google Scholar] [CrossRef]
- An, J.; Li, W.; Li, M.; Cui, S.; Yue, H. Identification and Classification of Maize Drought Stress Using Deep Convolutional Neural Network. Symmetry 2019, 11, 256. [Google Scholar] [CrossRef] [Green Version]
- Geetharamani, G.; Arun Pandian, J. Identification of plant leaf diseases using a nine-layer deep convolutional neural network. Comput. Electr. Eng. 2019, 76, 323–338. [Google Scholar]
- Kattenborn, T.; Eichel, J.; Wiser, S.; Burrows, L.; Fassnacht, F.E.; Schmidtlein, S. Convolutional Neural Networks accurately predict cover fractions of plant species and communities in Unmanned Aerial Vehicle imagery. Remote Sens. Ecol. Conserv. 2020. [Google Scholar] [CrossRef] [Green Version]
- Iniyan, S.; Jebakumar, R.; Mangalraj, P.; Mohit, M.; Nanda, A. Plant Disease Identification and Detection Using Support Vector Machines and Artificial Neural Networks. In Artificial Intelligence and Evolutionary Computations in Engineering Systems; Advances in Intelligent Systems and Computing; Dash, S., Lakshmi, C., Das, S., Panigrahi, B., Eds.; Springer: Singapore, 2020; pp. 15–27. [Google Scholar]
- Wang, Y.; Li, T.; Jin, G.; Wei, Y.; Li, L.; Kalkhajeh, Y.K.; Ning, J.; Zhang, Z. Qualitative and quantitative diagnosis of nitrogen nutrition of tea plants under field condition using hyperspectral imaging coupled with chemometrics. J. Sci. Food Agric. 2020, 100, 161–167. [Google Scholar] [CrossRef]
- Niazian, M.; Sadat-Noori, S.A.; Abdipour, M. Artificial neural network and multiple regression analysis models to predict essential oil content of ajowan (Carum copticum L.). J. Appl. Res. Med. Aromat. Plants 2018, 9, 124–131. [Google Scholar] [CrossRef]
- Niazian, M.; Sadat-Noori, S.A.; Abdipour, M.; Tohidfar, M.; Mortazavian, S.M.M. Image Processing and Artificial Neural Network-Based Models to Measure and Predict Physical Properties of Embryogenic Callus and Number of Somatic Embryos in Ajowan (Trachyspermum ammi (L.) Sprague). Vitr. Cell. Dev. Biol. Plant 2018, 54, 54–68. [Google Scholar] [CrossRef]
- Wei, M.C.F.; Maldaner, L.F.; Ottoni, P.M.N.; Molin, J.P. Carrot Yield Mapping: A Precision Agriculture Approach Based on Machine Learning. Al 2020, 1, 229–241. [Google Scholar]
- Hesami, M.; Naderi, R.; Tohidfar, M. Modeling and Optimizing in vitro Sterilization of Chrysanthemum via Multilayer Perceptron-Non-dominated Sorting Genetic Algorithm-II (MLP-NSGAII). Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, K.; Gong, L.; Huang, Y.; Liu, C.; Pan, J. Deep Learning-Based Segmentation and Quantification of Cucumber Powdery Mildew Using Convolutional Neural Network. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Arab, M.M.; Yadollahi, A.; Shojaeiyan, A.; Ahmadi, H. Artificial Neural Network Genetic Algorithm as Powerful Tool to Predict and Optimize In vitro Proliferation Mineral Medium for G × N15 Rootstock. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Arab, M.M.; Yadollahi, A.; Ahmadi, H.; Eftekhari, M.; Maleki, M. Mathematical Modeling and Optimizing of in Vitro Hormonal Combination for G × N15 Vegetative Rootstock Proliferation Using Artificial Neural Network-Genetic Algorithm (ANN-GA). Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Arab, M.M.; Yadollahi, A.; Eftekhari, M.; Ahmadi, H.; Akbari, M.; Khorami, S.S. Modeling and Optimizing a New Culture Medium for In Vitro Rooting of G×N15 Prunus Rootstock using Artificial Neural Network-Genetic Algorithm. Sci. Rep. 2018, 8, 9977. [Google Scholar] [CrossRef]
- Costa, M.O.; Capel, L.S.; Maldonado, C.; Mora, F.; Mangolin, C.A.; Machado, M.D. High genetic differentiation of grapevine rootstock varieties determined by molecular markers and artificial neural networks. Acta Sci. Agron. 2019, 42, e43475. [Google Scholar] [CrossRef] [Green Version]
- Altuntaş, Y.; Cömert, Z.; Kocamaz, A.F. Identification of haploid and diploid maize seeds using convolutional neural networks and a transfer learning approach. Comput. Electron. Agric. 2019, 163, 104874. [Google Scholar] [CrossRef]
- Darwish, A.; Ezzat, D.; Hassanien, A.E. An optimized model based on convolutional neural networks and orthogonal learning particle swarm optimization algorithm for plant diseases diagnosis. Swarm Evol. Comput. 2020, 52, 100616. [Google Scholar] [CrossRef]
- Mishra, S.; Sachan, R.; Rajpal, D. Deep Convolutional Neural Network based Detection System for Real-time Corn Plant Disease Recognition. Procedia Comput. Sci. 2020, 167, 2003–2010. [Google Scholar] [CrossRef]
- Feng, X.; Zhan, Y.; Wang, Q.; Yang, X.; Yu, C.; Wang, H.; Tang, Z.; Jiang, D.; Peng, C.; He, Y. Hyperspectral imaging combined with machine learning as a tool to obtain high-throughput plant salt-stress phenotyping. Plant J. 2020, 101, 1448–1461. [Google Scholar] [CrossRef]
- Coulibaly, S.; Kamsu-Foguem, B.; Kamissoko, D.; Traore, D. Deep neural networks with transfer learning in millet crop images. Comput. Ind. 2019, 108, 115–120. [Google Scholar] [CrossRef] [Green Version]
- Azizi, A.; Abbaspour-Gilandeh, Y.; Nooshyar, M.; Afkari-Sayah, A. Identifying Potato Varieties Using Machine Vision and Artificial Neural Networks. Int. J. Food Prop. 2016, 19, 618–635. [Google Scholar] [CrossRef]
- Niedbała, G.; Piekutowska, M.; Weres, J.; Korzeniewicz, R.; Witaszek, K.; Adamski, M.; Pilarski, K.; Czechowska-Kosacka, A.; Krysztofiak-Kaniewska, A. Application of Artificial Neural Networks for Yield Modeling of Winter Rapeseed Based on Combined Quantitative and Qualitative Data. Agronomy 2019, 9, 781. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Zhao, B.; Yang, C.; Shi, Y.; Liao, Q.; Zhou, G.; Wang, C.; Xie, T.; Jiang, Z.; Zhang, D.; et al. Rapeseed Stand Count Estimation at Leaf Development Stages With UAV Imagery and Convolutional Neural Networks. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Niedbała, G. Application of artificial neural networks for multi-criteria yield prediction of winter rapeseed. Sustainability 2019, 11, 533. [Google Scholar]
- Niedbała, G. Simple model based on artificial neural network for early prediction and simulation winter rapeseed yield. J. Integr. Agric. 2019, 18, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Wang, R.; Xie, C.; Liu, L.; Zhang, J.; Li, R.; Wang, F.; Zhou, M.; Liu, W. A Recognition Method for Rice Plant Diseases and Pests Video Detection Based on Deep Convolutional Neural Network. Sensors 2020, 20, 578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, C.R.; Arko, P.S.; Ali, M.E.; Iqbal Khan, M.A.; Apon, S.H.; Nowrin, F.; Wasif, A. Identification and recognition of rice diseases and pests using convolutional neural networks. Biosyst. Eng. 2020, 194, 112–120. [Google Scholar] [CrossRef] [Green Version]
- Abdipour, M.; Younessi-Hmazekhanlu, M.; Ramazani, S.H.R.; Omidi, A. Hassan Artificial neural networks and multiple linear regression as potential methods for modeling seed yield of safflower (Carthamus tinctorius L.). Ind. Crops Prod. 2019, 127, 185–194. [Google Scholar] [CrossRef]
- Abdipour, M.; Ramazani, S.H.R.; Younessi-Hmazekhanlu, M.; Niazian, M. Modeling Oil Content of Sesame (Sesamum indicum L.) Using Artificial Neural Network and Multiple Linear Regression Approaches. J. Am. Oil Chem. Soc. 2018, 95, 283–297. [Google Scholar] [CrossRef]
- Parsaeian, M.; Shahabi, M.; Hassanpour, H. Estimating Oil and Protein Content of Sesame Seeds Using Image Processing and Artificial Neural Network. J. Am. Oil Chem. Soc. 2020, 97, 691–702. [Google Scholar] [CrossRef]
- Uzal, L.C.; Grinblat, G.L.; Namías, R.; Larese, M.G.; Bianchi, J.S.; Morandi, E.N.; Granitto, P.M. Seed-per-pod estimation for plant breeding using deep learning. Comput. Electron. Agric. 2018, 150, 196–204. [Google Scholar] [CrossRef]
- Sakoda, K.; Watanabe, T.; Sukemura, S.; Kobayashi, S.; Nagasaki, Y.; Tanaka, Y.; Shiraiwa, T. Genetic Diversity in Stomatal Density among Soybeans Elucidated Using High-throughput Technique Based on an Algorithm for Object Detection. Sci. Rep. 2019, 9, 7610. [Google Scholar] [CrossRef]
- Niazian, M.; Shariatpanahi, M.E.; Abdipour, M.; Oroojloo, M. Modeling callus induction and regeneration in an anther culture of tomato (Lycopersicon esculentum L.) using image processing and artificial neural network method. Protoplasma 2019, 256, 1317–1332. [Google Scholar] [CrossRef]
- Verma, S.; Chug, A.; Singh, A.P. Application of convolutional neural networks for evaluation of disease severity in tomato plant. J. Discret. Math. Sci. Cryptogr. 2020, 23, 273–282. [Google Scholar] [CrossRef]
- Ravari, S.Z.; Dehghani, H.; Naghavi, H. Assessment of salinity indices to identify Iranian wheat varieties using an artificial neural network. Ann. Appl. Biol. 2016, 168, 185–194. [Google Scholar] [CrossRef]
- Niedbała, G.; Kozłowski, R.J. Application of Artificial Neural Networks for Multi-Criteria Yield Prediction of Winter Wheat. J. Agric. Sci. Technol. 2019, 21, 51–61. [Google Scholar]
- Niedbała, G.; Nowakowski, K.; Rudowicz-Nawrocka, J.; Piekutowska, M.; Weres, J.; Tomczak, R.J.; Tyksiński, T.; Pinto, A.Á. Multicriteria prediction and simulation of winter wheat yield using extended qualitative and quantitative data based on artificial neural networks. Appl. Sci. 2019, 9, 2773. [Google Scholar] [CrossRef] [Green Version]
- Sadeghi-Tehran, P.; Virlet, N.; Ampe, E.M.; Reyns, P.; Hawkesford, M.J. DeepCount: In-Field Automatic Quantification of Wheat Spikes Using Simple Linear Iterative Clustering and Deep Convolutional Neural Networks. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Haider, S.A.; Naqvi, S.R.; Akram, T.; Umar, G.A.; Shahzad, A.; Sial, M.R.; Khaliq, S.; Kamran, M. LSTM Neural Network Based Forecasting Model for Wheat Production in Pakistan. Agronomy 2019, 9, 72. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; Qiu, Z.; Song, J.; Li, J.; Cheng, Q.; Zhai, J.; Ma, C. A deep convolutional neural network approach for predicting phenotypes from genotypes. Planta 2018, 248, 1307–1318. [Google Scholar] [CrossRef]
- Hesami, M.; Condori-Apfata, J.A.; Valderrama Valencia, M.; Mohammadi, M. Application of Artificial Neural Network for Modeling and Studying In Vitro Genotype-Independent Shoot Regeneration in Wheat. Appl. Sci. 2020, 10, 5370. [Google Scholar] [CrossRef]
- Niedbała, G.; Kurasiak-Popowska, D.; Stuper-Szablewska, K.; Nawracała, J. Application of Artificial Neural Networks to Analyze the Concentration of Ferulic Acid, Deoxynivalenol, and Nivalenol in Winter Wheat Grain. Agriculture 2020, 10, 127. [Google Scholar] [CrossRef] [Green Version]
- Ray, A.; Halder, T.; Jena, S.; Sahoo, A.; Ghosh, B.; Mohanty, S.; Mahapatra, N.; Nayak, S. Application of artificial neural network (ANN) model for prediction and optimization of coronarin D content in Hedychium coronarium. Ind. Crops Prod. 2020, 146, 112186. [Google Scholar] [CrossRef]
- Srivastava, A.; Gupta, S.; Shanker, K.; Gupta, N.; Gupta, A.K.; Lal, R.K. Genetic diversity in Indian poppy (P. somniferum L.) germplasm using multivariate and SCoT marker analyses. Ind. Crops Prod. 2020, 144, 112050. [Google Scholar] [CrossRef]
- Niazian, M.; Sadat Noori, S.A.; Tohidfar, M.; Mortazavian, S.M.M. Essential Oil Yield and Agro-morphological Traits in Some Iranian Ecotypes of Ajowan (Carum copticum L.). J. Essent. Oil Bear. Plants 2017, 20, 1151–1156. [Google Scholar] [CrossRef]
- Schulman, A.H. Molecular markers to assess genetic diversity. Euphytica 2007, 158, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Boonsrangsom, T. Genetic diversity of ‘Wan Chak Motluk’ (Curcuma comosa Roxb.) in Thailand using morphological characteristics and random amplification of polymorphic DNA (RAPD) markers. South Afr. J. Bot. 2020, 130, 224–230. [Google Scholar] [CrossRef]
- Pandolfi, C.; Mugnai, S.; Azzarello, E.; Bergamasco, S.; Masi, E.; Mancuso, S. Artificial neural networks as a tool for plant identification: A case study on Vietnamese tea accessions. Euphytica 2009, 166, 411–421. [Google Scholar] [CrossRef]
- Raza, A.; Mehmood, S.S.; Ashraf, F.; Khan, R.S.A. Genetic Diversity Analysis of Brassica Species Using PCR-Based SSR Markers. Gesunde Pflanz. 2019, 71, 1–7. [Google Scholar] [CrossRef]
- Bird, C.; Schweizer, M.; Roberts, A.; Austin, W.E.N.; Knudsen, K.L.; Evans, K.M.; Filipsson, H.L.; Sayer, M.D.J.; Geslin, E.; Darling, K.F. The genetic diversity, morphology, biogeography, and taxonomic designations of Ammonia (Foraminifera) in the Northeast Atlantic. Mar. Micropaleontol. 2020, 155, 101726. [Google Scholar] [CrossRef] [Green Version]
- Poletto, T.; Poletto, I.; Moraes Silva, L.M.; Brião Muniz, M.F.; Silveira Reiniger, L.R.; Richards, N.; Stefenon, V.M. Morphological, chemical and genetic analysis of southern Brazilian pecan (Carya illinoinensis) accessions. Sci. Hortic. Amst. 2020, 261, 108863. [Google Scholar] [CrossRef]
- Saini, G.; Khamparia, A.; Luhach, A.K. Classification of Plants Using Convolutional Neural Network. In Advances in Intelligent Systems and Computing, Proceedings of the First International Conference on Sustainable Technologies for Computational Intelligence; Luhach, A., Kosa, J., Poonia, R., Gao, X., Singh, D., Eds.; Springer: Singapore, 2020; Volume 1045, pp. 551–561. [Google Scholar]
- Yang, H.-W.; Hsu, H.-C.; Yang, C.-K.; Tsai, M.-J.; Kuo, Y.-F. Differentiating between morphologically similar species in genus Cinnamomum (Lauraceae) using deep convolutional neural networks. Comput. Electron. Agric. 2019, 162, 739–748. [Google Scholar] [CrossRef]
- Sant’Anna, I.C.; Tomaz, R.S.; Silva, G.N.; Nascimento, M.; Bhering, L.L.; Cruz, C.D. Superiority of artificial neural networks for a genetic classification procedure. Genet. Mol. Res. 2015, 14, 9898–9906. [Google Scholar] [CrossRef]
- Korani, W.; Clevenger, J.P.; Chu, Y.; Ozias-Akins, P. Machine Learning as an Effective Method for Identifying True Single Nucleotide Polymorphisms in Polyploid Plants. Plant Genome 2019, 12, 1–10. [Google Scholar] [CrossRef] [Green Version]
- González-Camacho, J.M.; de los Campos, G.; Pérez, P.; Gianola, D.; Cairns, J.E.; Mahuku, G.; Babu, R.; Crossa, J. Genome-enabled prediction of genetic values using radial basis function neural networks. Theor. Appl. Genet. 2012, 125, 759–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peixoto, L.A.; Bhering, L.L.; Cruz, C.D. Artificial neural networks reveal efficiency in genetic value prediction. Genet. Mol. Res. 2015, 14, 6796–6807. [Google Scholar] [CrossRef]
- Zingaretti, L.M.; Gezan, S.A.; Ferrão, L.F.V.; Osorio, L.F.; Monfort, A.; Muñoz, P.R.; Whitaker, V.M.; Pérez-Enciso, M. Exploring Deep Learning for Complex Trait Genomic Prediction in Polyploid Outcrossing Species. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef] [Green Version]
- Ghaffari, H.; Tadayon, M.R.; Razmjoo, J.; Bahador, M.; Soureshjani, H.K.; Yuan, T. Impact of Jasmonic Acid on Sugar Yield and Physiological Traits of Sugar Beet in Response to Water Deficit Regimes: Using Stepwise Regression Approach. Russ. J. Plant Physiol. 2020, 67, 482–493. [Google Scholar] [CrossRef]
- Zou, J.; Hu, W.; Li, Y.; He, J.; Zhu, H.; Zhou, Z. Screening of drought resistance indices and evaluation of drought resistance in cotton (Gossypium hirsutum L.). J. Integr. Agric. 2020, 19, 495–508. [Google Scholar] [CrossRef]
- Lv, C.; Huang, Y.; Sun, W.; Yu, L.; Zhu, J. Response of rice yield and yield components to elevated [CO2]: A synthesis of updated data from FACE experiments. Eur. J. Agron. 2020, 112, 125961. [Google Scholar] [CrossRef]
- Emamgholizadeh, S.; Parsaeian, M.; Baradaran, M. Seed yield prediction of sesame using artificial neural network. Eur. J. Agron. 2015, 68, 89–96. [Google Scholar] [CrossRef]
- Lee, S.; Jeong, Y.; Son, S.; Lee, B. A Self-Predictable Crop Yield Platform (SCYP) Based on Crop Diseases Using Deep Learning. Sustainability 2019, 11, 3637. [Google Scholar] [CrossRef] [Green Version]
- Ajay, B.C.; Bera, S.K.; Singh, A.L.; Kumar, N.; Gangadhar, K.; Kona, P. Evaluation of Genotype × Environment Interaction and Yield Stability Analysis in Peanut Under Phosphorus Stress Condition Using Stability Parameters of AMMI Model. Agric. Res. 2020, 1–10. [Google Scholar] [CrossRef]
- Finlay, K.; Wilkinson, G. The analysis of adaptation in a plant-breeding programme. Aust. J. Agric. Res. 1963, 14, 742. [Google Scholar] [CrossRef] [Green Version]
- Eberhart, S.A.; Russell, W.A. Stability Parameters for Comparing Varieties 1. Crop Sci. 1966, 6, 36–40. [Google Scholar] [CrossRef] [Green Version]
- Wricke, G. Über eine Methode zur Erfassung der ökologischen Streubreite in Feldversuchen. Z. Pflanzenzuchtg 1962, 47, 92–96. [Google Scholar]
- Shukla, G.K. Some statistical aspects of partitioning genotype-environmental components of variability. Heredity Edinb. 1972, 29, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Francis, C.A.; Prager, M.; Laing, D.R.; Flor, C.A. Genotype × Environment Interactions in Bush Bean Cultivars in Monoculture and Associated with Maize. Crop Sci. 1978, 18, 237–242. [Google Scholar] [CrossRef]
- Lin, C.S.; Binns, M.R. A superiority measure of cultivar performance for cultivar × location data. Can. J. Plant Sci. 1988, 68, 193–198. [Google Scholar] [CrossRef]
- Karimizadeh, R.; Mohammadi, M.; Sabaghni, N.; Mahmoodi, A.A.; Roustami, B.; Seyyedi, F.; Akbari, F. GGE Biplot Analysis of Yield Stability in Multi-environment Trials of Lentil Genotypes under Rainfed Condition. Not. Sci. Biol. 2013, 5, 256–262. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.P.; Das, S.K.; Bhaskarrao, U.M.; Reddy, M.N. Sustainability Index Under Different Management; Annual Report; CRIDA: Hyderabad, India, 1990. [Google Scholar]
- Han, X.; Hu, C.; Chen, Y.; Qiao, Y.; Liu, D.; Fan, J.; Li, S.; Zhang, Z. Crop yield stability and sustainability in a rice-wheat cropping system based on 34-year field experiment. Eur. J. Agron. 2020, 113, 125965. [Google Scholar] [CrossRef]
- Flores, F.; Moreno, M.; Cubero, J. A comparison of univariate and multivariate methods to analyze G×E interaction. Field Crop. Res. 1998, 56, 271–286. [Google Scholar] [CrossRef]
- Hühn, M. Beiträge zur Erfassung der phänotypischen Stabilität. I. Vorschlag einiger auf Ranginformationen beruhender Stabilitätsparameter. EDV Medizin Biol. 1979, 10, 112–117. [Google Scholar]
- Nassar, R.; Hühn, M. Studies on Estimation of Phenotypic Stability: Tests of Significance for Nonparametric Measures of Phenotypic Stability. Biometrics 1987, 43, 45. [Google Scholar] [CrossRef]
- Kang, M.S. A rank-sum method for selecting high-yielding, stable corn genotypes. Cereal Res. Commun. 1988, 16, 113–115. [Google Scholar]
- Ketata, H.; Yan, S.K.; Nachit, M. Relative consistency performance across environments. In Proceedings of the International Symposium on Physiology and Breeding of Winter Cereals for Stressed Mediterranean Environments, Montpellier, France, 3–6 July 1989. [Google Scholar]
- Fox, P.N.; Skovmand, B.; Thompson, B.K.; Braun, H.-J.; Cormier, R. Yield and adaptation of hexaploid spring triticale. Euphytica 1990, 47, 57–64. [Google Scholar] [CrossRef]
- Li, N.; Lin, H.; Wang, T.; Li, Y.; Liu, Y.; Chen, X.; Hu, X. Impact of climate change on cotton growth and yields in Xinjiang, China. Field Crop. Res. 2020, 247, 107590. [Google Scholar] [CrossRef]
- Harfouche, A.L.; Jacobson, D.A.; Kainer, D.; Romero, J.C.; Harfouche, A.H.; Scarascia Mugnozza, G.; Moshelion, M.; Tuskan, G.A.; Keurentjes, J.J.B.; Altman, A. Accelerating Climate Resilient Plant Breeding by Applying Next-Generation Artificial Intelligence. Trends Biotechnol. 2019, 37, 1217–1235. [Google Scholar] [CrossRef]
- Fischer, R.; Maurer, R. Drought resistance in spring wheat cultivars. I. Grain yield responses. Aust. J. Agric. Res. 1978, 29, 897. [Google Scholar] [CrossRef]
- Fischer, R.; Wood, J. Drought resistance in spring wheat cultivars. III.* Yield associations with morpho-physiological traits. Aust. J. Agric. Res. 1979, 30, 1001. [Google Scholar] [CrossRef]
- Rosielle, A.A.; Hamblin, J. Theoretical Aspects of Selection for Yield in Stress and Non-Stress Environment. Crop Sci. 1981, 21, 943–946. [Google Scholar] [CrossRef]
- Bouslama, M.; Schapaugh, W.T. Stress Tolerance in Soybeans. I. Evaluation of Three Screening Techniques for Heat and Drought Tolerance. Crop Sci. 1984, 24, 933–937. [Google Scholar] [CrossRef]
- Fernandez, G.C.J. Effective selection criteria for assessing stress tolerance. In Proceedings of the International Symposium on Adaptation of Vegetables and Other Food Crops in Temperature and Water Stress, Tainan, Taiwan, 13–16 August 1992; Kuo, C.G., Ed.; AVRDC Publication: Tainan, Taiwan, 1992; pp. 257–270. [Google Scholar]
- Gavuzzi, P.; Rizza, F.; Palumbo, M.; Campanile, R.G.; Ricciardi, G.L.; Borghi, B. Evaluation of field and laboratory predictors of drought and heat tolerance in winter cereals. Can. J. Plant Sci. 1997, 77, 523–531. [Google Scholar] [CrossRef]
- Schneider, K.A.; Rosales-Serna, R.; Ibarra-Perez, F.; Cazares-Enriquez, B.; Acosta-Gallegos, J.A.; Ramirez-Vallejo, P.; Wassimi, N.; Kelly, J.D. Improving Common Bean Performance under Drought Stress. Crop Sci. 1997, 37, 43–50. [Google Scholar] [CrossRef]
- Farshadfar, E.; Sutka, J. Screening drought tolerance criteria in maize. Acta Agron. Hungarica 2002, 50, 411–416. [Google Scholar] [CrossRef]
- Niazian, M.; Sadat-Noori, S.A.; Tohidfar, M.; Galuszka, P.; Mortazavian, S.M.M. Agrobacterium-mediated genetic transformation of ajowan (Trachyspermum ammi (L.) Sprague): An important industrial medicinal plant. Ind. Crops Prod. 2019, 132, 29–40. [Google Scholar] [CrossRef]
- Niazian, M.; Shariatpanahi, M.E. In vitro-based doubled haploid production: Recent improvements. Euphytica 2020, 216, 69. [Google Scholar] [CrossRef]
- Marchetti, C.F.; Ugena, L.; Humplík, J.F.; Polák, M.; Ćavar Zeljković, S.; Podlešáková, K.; Fürst, T.; De Diego, N.; Spíchal, L. A Novel Image-Based Screening Method to Study Water-Deficit Response and Recovery of Barley Populations Using Canopy Dynamics Phenotyping and Simple Metabolite Profiling. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Detection of nutrition deficiencies in plants using proximal images and machine learning: A review. Comput. Electron. Agric. 2019, 162, 482–492. [Google Scholar] [CrossRef]
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine Learning for High-Throughput Stress Phenotyping in Plants. Trends Plant Sci. 2016, 21, 110–124. [Google Scholar] [CrossRef] [Green Version]
- Picon, A.; Alvarez-Gila, A.; Seitz, M.; Ortiz-Barredo, A.; Echazarra, J.; Johannes, A. Deep convolutional neural networks for mobile capture device-based crop disease classification in the wild. Comput. Electron. Agric. 2019, 161, 280–290. [Google Scholar] [CrossRef]
- Agarwal, M.; Sinha, A.; Gupta, S.K.; Mishra, D.; Mishra, R. Potato crop disease classification using convolutional neural network. In Smart Systems and IoT: Innovations in Computing; Somani, A.K., Shekhawat, R.S., Mundra, A., Srivastava, S., Verma, V.K., Eds.; Smart Innovation, Systems and Technologies; Springer: Singapore, 2020; Volume 141, ISBN 978-981-13-8405-9. [Google Scholar]
- Anagnostis, A.; Asiminari, G.; Papageorgiou, E.; Bochtis, D. A Convolutional Neural Networks Based Method for Anthracnose Infected Walnut Tree Leaves Identification. Appl. Sci. 2020, 10, 469. [Google Scholar] [CrossRef] [Green Version]
- Khamparia, A.; Saini, G.; Gupta, D.; Khanna, A.; Tiwari, S.; de Albuquerque, V.H.C. Seasonal Crops Disease Prediction and Classification Using Deep Convolutional Encoder Network. Circuits Syst. Signal Process. 2020, 39, 818–836. [Google Scholar] [CrossRef]
- Sibiya, M.; Sumbwanyambe, M. A Computational Procedure for the Recognition and Classification of Maize Leaf Diseases Out of Healthy Leaves Using Convolutional Neural Networks. AgriEngineering 2019, 1, 9. [Google Scholar] [CrossRef] [Green Version]
- Kearsey, M.; Pooni, H. The Genetical Analysis of Quantitative Traits; Stanley Thornes Ltd: Cheltenham, UK, 1998. [Google Scholar]
- Griffing, B. Concept of General and Specific Combining Ability in Relation to Diallel Crossing Systems. Aust. J. Biol. Sci. 1956, 9, 463. [Google Scholar] [CrossRef]
- Hayman, B.I. The Theory and Analysis of Diallel Crosses. II. Genetics 1958, 43, 63–85. [Google Scholar] [PubMed]
- Jinks, J.L. The Analysis of Continuous Variation in a Diallel Cross of Nicotiana Rustica Varieties. Genetics 1954, 39, 767–788. [Google Scholar] [PubMed]
- Kempthorne, O. An Introduction to Genetic Statistics; John Wiley & Sons Inc.: New York, NY, USA, 1957. [Google Scholar]
- Comstock, R.E.; Robinson, H.F. The Components of Genetic Variance in Populations of Biparental Progenies and Their Use in Estimating the Average Degree of Dominance. Biometrics 1948, 4, 254. [Google Scholar] [CrossRef] [PubMed]
- Opsahl, B. The Discrimination of Interactions and Linkage in Continuous Variation. Biometrics 1956, 12, 415. [Google Scholar] [CrossRef]
- Kearsey, M.J.; Jinks, J.L. A general method of detecting additive, dominance and epistatic variation for metrical traits I. Theory. Heredity Edinb. 1968, 23, 403–409. [Google Scholar] [CrossRef] [Green Version]
- Dezfouli, P.M.; Sedghi, M.; Shariatpanahi, M.E.; Niazian, M.; Alizadeh, B. Assessment of general and specific combining abilities in doubled haploid lines of rapeseed (Brassica napus L.). Ind. Crop. Prod. 2019, 141, 111754. [Google Scholar] [CrossRef]
- Khaki, S.; Khalilzadeh, Z.; Wang, L. Predicting yield performance of parents in plant breeding: A neural collaborative filtering approach. PLoS ONE 2020, 15, e0233382. [Google Scholar] [CrossRef]
- Niazian, M. Application of genetics and biotechnology for improving medicinal plants. Planta 2019, 249, 953–973. [Google Scholar] [CrossRef]
- Ayuso, M.; García-Pérez, P.; Ramil-Rego, P.; Gallego, P.P.; Barreal, M.E. In vitro culture of the endangered plant Eryngium viviparum as dual strategy for its ex situ conservation and source of bioactive compounds. Plant Cell Tissue Organ Cult. 2019, 138, 427–435. [Google Scholar] [CrossRef]
- Sugimoto, K.; Temman, H.; Kadokura, S.; Matsunaga, S. To regenerate or not to regenerate: Factors that drive plant regeneration. Curr. Opin. Plant Biol. 2019, 47, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Prasad, V.S.S.; Gupta, S.D. Applications and potentials of artificial neural networks in plant tissue culture. In Plant Tissue Culture Engineering; Gupta, S.D., Ibaraki, Y., Eds.; Focus on Biotechnology; Springer: Dordrecht, The Netherlands, 2008; Volume 6, ISBN 978-1-4020-3594-4. [Google Scholar]
- Niazian, M.; Noori, S.A.S.; Galuszka, P.; Tohidfar, M.; Mortazavian, S.M.M. Genetic stability of regenerated plants via indirect somatic embryogenesis and indirect shoot regeneration of Carum copticum L. Ind. Crops Prod. 2017, 97, 330–337. [Google Scholar] [CrossRef]
- Orłowska, R.; Bednarek, P.T. Precise evaluation of tissue culture-induced variation during optimisation of in vitro regeneration regime in barley. Plant Mol. Biol. 2020, 103, 33–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, G.C.; Garda, M. Plant tissue culture media and practices: An overview. Vitr. Cell. Dev. Biol. Plant 2019, 55, 242–257. [Google Scholar] [CrossRef]
- Alanagh, E.N.; Garoosi, G.; Haddad, R.; Maleki, S.; Landín, M.; Gallego, P.P. Design of tissue culture media for efficient Prunus rootstock micropropagation using artificial intelligence models. Plant Cell Tissue Organ Cult. 2014, 117, 349–359. [Google Scholar] [CrossRef]
- Hesami, M.; Naderi, R.; Tohidfar, M. Modeling and Optimizing Medium Composition for Shoot Regeneration of Chrysanthemum via Radial Basis Function-Non-dominated Sorting Genetic Algorithm-II (RBF-NSGAII). Sci. Rep. 2019, 9, 18237. [Google Scholar] [CrossRef] [Green Version]
- Sadat Noori, S.A.; Norouzi, M.; Karimzadeh, G.; Shirkool, K.; Niazian, M. Effect of colchicine-induced polyploidy on morphological characteristics and essential oil composition of ajowan (Trachyspermum ammi L.). Plant Cell Tissue Organ Cult. 2017, 130, 543–551. [Google Scholar] [CrossRef]
- Castillo, A.M.; Cistué, L.; Vallés, M.P.; Soriano, M. Chromosome Doubling in Monocots. In Advances in Haploid Production in Higher Plants; Springer: Dordrecht, The Netherlands, 2009; pp. 329–338. [Google Scholar]
- Niazian, M.; Sadat Noori, S.A.; Galuszka, P.; Mortazavian, S.M.M. Tissue culture-based Agrobacterium-mediated and in planta transformation methods. Czech J. Genet. Plant Breed. 2017, 53, 133–143. [Google Scholar] [CrossRef] [Green Version]
- Matias, F.I.; Caraza-Harter, M.V.; Endelman, J.B. FIELDimageR: An R package to analyze orthomosaic images from agricultural field trials. Plant Phenome J. 2020, 3, e20005. [Google Scholar] [CrossRef]
- Tsaftaris, S.A.; Minervini, M.; Scharr, H. Machine Learning for Plant Phenotyping Needs Image Processing. Trends Plant Sci. 2016, 21, 989–991. [Google Scholar] [CrossRef] [Green Version]
- Ubbens, J.R.; Stavness, I. Deep Plant Phenomics: A Deep Learning Platform for Complex Plant Phenotyping Tasks. Front. Plant Sci. 2017, 8, 1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; He, Y. Discriminating varieties of tea plant based on Vis/NIR spectral characteristics and using artificial neural networks. Biosyst. Eng. 2008, 99, 313–321. [Google Scholar] [CrossRef]
- Khoshroo, A.; Arefi, A.; Masoumiasl, A.; Jowkar, G.H. Classification of wheat cultivars using image processing and artificial neural networks. Agric. Commun. 2014, 2, 17–22. [Google Scholar]
- Mahlein, A.-K. Plant Disease Detection by Imaging Sensors—Parallels and Specific Demands for Precision Agriculture and Plant Phenotyping. Plant Dis. 2016, 100, 241–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maes, W.H.; Steppe, K. Perspectives for Remote Sensing with Unmanned Aerial Vehicles in Precision Agriculture. Trends Plant Sci. 2019, 24, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Khadka, K.; Earl, H.J.; Raizada, M.N.; Navabi, A. A Physio-Morphological Trait-Based Approach for Breeding Drought Tolerant Wheat. Front. Plant Sci. 2020, 11, 715. [Google Scholar] [CrossRef]
- Singh, A.; Jones, S.; Ganapathysubramanian, B.; Sarkar, S.; Mueller, D.; Sandhu, K.; Nagasubramanian, K. Challenges and Opportunities in Machine-Augmented Plant Stress Phenotyping. Trends Plant Sci. 2020. [Google Scholar] [CrossRef]
- Pan-pan, H.; Wei-xu, L.; Hui-hui, L.; Zeng-xu, X. In vitro induction and identification of autotetraploid of Bletilla striata (Thunb.) Reichb.f. by colchicine treatment. Plant Cell Tissue Organ Cult. 2018, 132, 425–432. [Google Scholar] [CrossRef]
- Ochatt, S.J.; Patat-Ochatt, E.M.; Moessner, A. Ploidy level determination within the context of in vitro breeding. Plant Cell Tissue Organ Cult. 2011, 104, 329–341. [Google Scholar] [CrossRef]
- Ahmadi, B.; Ebrahimzadeh, H. In vitro androgenesis: Spontaneous vs. artificial genome doubling and characterization of regenerants. Plant Cell Rep. 2020, 39, 299–316. [Google Scholar] [CrossRef]
- Santeramo, D.; Howell, J.; Ji, Y.; Yu, W.; Liu, W.; Kelliher, T. DNA content equivalence in haploid and diploid maize leaves. Planta 2020, 251, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blonder, B.; Graae, B.J.; Greer, B.; Haagsma, M.; Helsen, K.; Kapás, R.E.; Pai, H.; Rieksta, J.; Sapena, D.; Still, C.J.; et al. Remote sensing of ploidy level in quaking aspen ( Populus tremuloides Michx.). J. Ecol. 2020, 108, 175–188. [Google Scholar] [CrossRef] [Green Version]

| Leaning Algorithm | Advantages | Disadvantages |
|---|---|---|
| ANNs |
|
|
| CNNs |
|
|
| SVMs |
|
|
| RF |
|
|
| Plant Species | Type of Machine Learning | Techniques | Purpose(s) | Reference |
|---|---|---|---|---|
| Ajowan (Trachyspermum ammi L.) | ANN | MLR | Modeling and predicting of seed yield | [2] |
| ANN | MLR | Modeling and predicting of essential oil content | [24] | |
| ANN | MLR, IP | Predicting physical properties of embryogenic callus and number of somatic embryos | [25] | |
| Arabidopsis thaliana | DT, SVMs, NB | Gaussian kernel | Predict the plant abiotic stresses response through the miRNAs’ concentration | [8] |
| Carrot (Daucus carota) | RF | - | Precision agriculture-yield mapping | [26] |
| Chrysanthemum | ANN | GA | Modeling and optimizing of in vitro sterilization | [27] |
| ANFIS | GA | Modeling and optimizing of somatic embryogenesis | [3] | |
| ANN, SVMs | MLP | Modeling effect of plant growth regulators on somatic embryogenesis | [15] | |
| Cucumber (Cucumis sativus) | CNN | IP | Segmentation and quantification of powdery mildew disease | [28] |
| Garnem (G × N15) Prunus rootstock | ANN | GA | Prediction and optimization of mineral salts of in vitro culture medium | [29] |
| ANN | GA | Modeling and optimizing of in vitro hormonal combination | [30] | |
| ANN | GA | Modeling and optimizing of new in vitro culture medium | [31] | |
| Grapevine rootstock | ANN | Principal coordinate analysis, UPGMA | Genetic diversity assessment through molecular markers (RAPD-SSR) dataset | [32] |
| Maize (Zea mays L.) | CNN | IP | Identification of haploid and diploid maize seeds | [33] |
| CNN | IP | Classification model to identify the infected and healthy leaves | [34] | |
| CNN | IP | Plant diseases recognition | [35] | |
| CNN | IP | Identification and classification of drought stress | [19] | |
| Okra (Abelmoschus esculentus L.) | DNN | IP | High-throughput salt-stress phenotyping | [36] |
| Pearl millet (Pennisetum glaucum) | DNN | IP | Identification of mildew disease | [37] |
| Potato (Solanum tuberosum) | ANN | IP | Identification and discrimination of potato varieties | [38] |
| RF | Classification of Phytophthora infestans infected cultivars | [17] | ||
| Rapeseed (Brassica napus) | ANN | MLP | Seed yield modeling | [39] |
| CNN | IP | Stand count estimation | [40] | |
| ANN | MLP | Multicriteria yield prediction based on meteorological data and mineral fertilization data | [41] | |
| ANN | MLP | Early prediction and simulation of seed yield based on meteorological and mineral fertilization data | [42] | |
| Rice (Oryza sativa) | CNN | Plant diseases and pest recognition | [43,44] | |
| Safflower (Carthamus tinctorius L.) | ANN | MLR | Seed yield modeling | [45] |
| Sesame (Sesamum indicum L.) | ANN | MLR | Oil content modeling | [46] |
| ANN, SVMs | RBF, ERBF, GRNN, M5-Rule, M5-Tree, MLR | Estimation of oil and protein content | [47] | |
| Soybean (Glycine max) | CNN | IP | Estimation of seeds per pod | [48] |
| DNN | IP | Evaluation of stomatal density diversity | [49] | |
| Tomato (Lycopersicon esculentum L.) | ANN | MLR, IP | Modeling of callus induction and regeneration in anther culture | [50] |
| CNN | IP | Evaluation of disease severity | [51] | |
| Wheat (Triticum aestivum L.) | ANN | MLP | Estimation of salinity tolerance | [52] |
| ANN | MLP | Prediction of seed yield based on meteorological data and information on mineral fertilization | [53] | |
| ANN | MLP | Prediction and simulation of seed yield with qualitative and quantitative data sets | [54] | |
| CNN | IP | Quantification of spikes | [55] | |
| DNN | LSTM | Production forecasting | [56] | |
| CNN | - | Genomic selection | [57] | |
| ANN, GRNN | MLP | Modeling in vitro shoot regeneration | [58] | |
| ANN | MLP | Analysis of concentration of ferulic acid, deoxynivalenol, and nivalenol | [59] | |
| White ginger (Hedychium coronarium) | ANN | MLP | Prediction and optimization of coronarin D content | [60] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niazian, M.; Niedbała, G. Machine Learning for Plant Breeding and Biotechnology. Agriculture 2020, 10, 436. https://doi.org/10.3390/agriculture10100436
Niazian M, Niedbała G. Machine Learning for Plant Breeding and Biotechnology. Agriculture. 2020; 10(10):436. https://doi.org/10.3390/agriculture10100436
Chicago/Turabian StyleNiazian, Mohsen, and Gniewko Niedbała. 2020. "Machine Learning for Plant Breeding and Biotechnology" Agriculture 10, no. 10: 436. https://doi.org/10.3390/agriculture10100436
APA StyleNiazian, M., & Niedbała, G. (2020). Machine Learning for Plant Breeding and Biotechnology. Agriculture, 10(10), 436. https://doi.org/10.3390/agriculture10100436






