High-Resolution 3D Crop Reconstruction and Automatic Analysis of Phenotyping Index Using Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Crop and Experimental Environment
2.2. Image Sensor
2.3. Measurement Method and Application Software
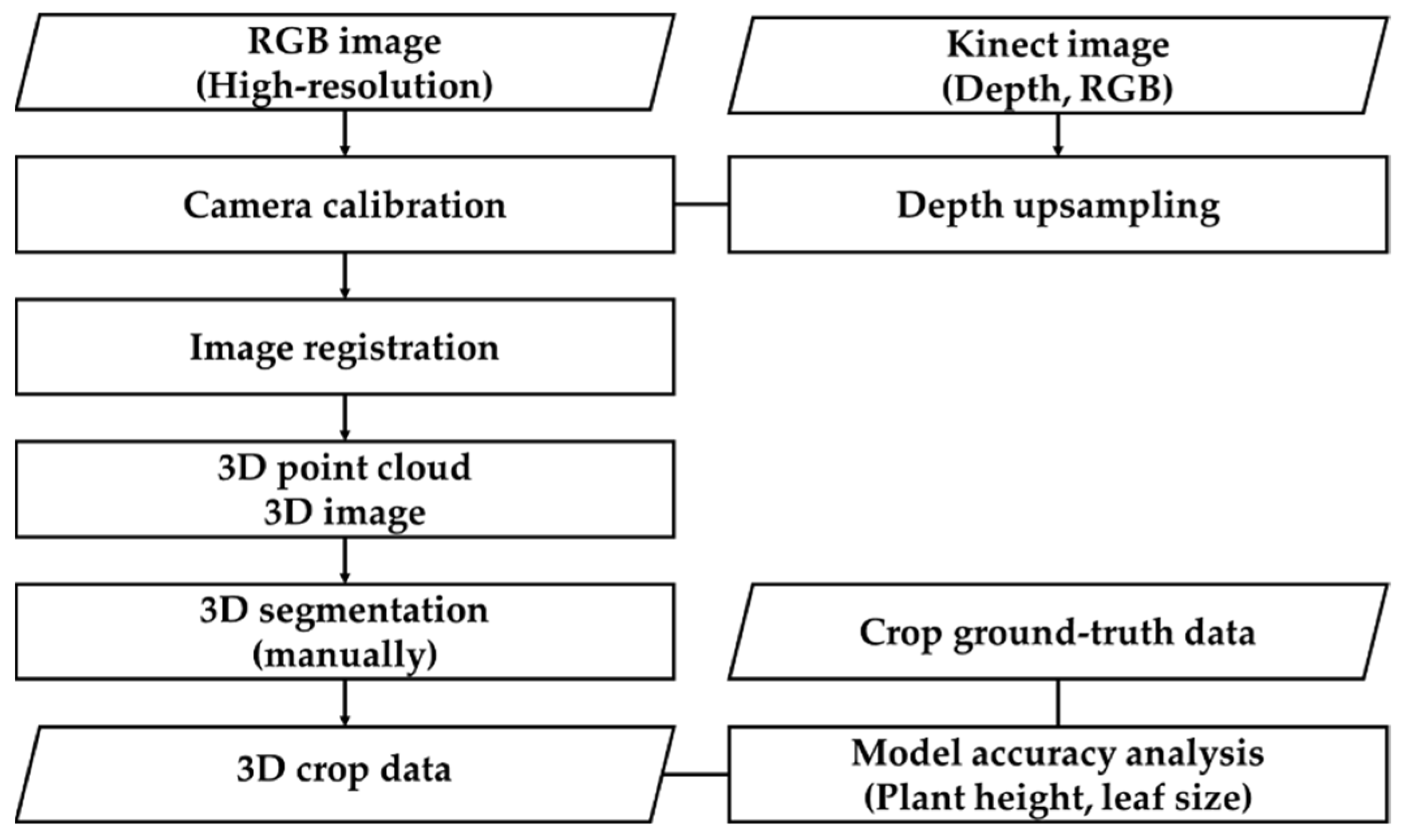
2.4. Algorithm for High-Resolution 3D Reconstruction
2.5. Machine Learning-Based Crop Extraction and Segmentation Algorithm
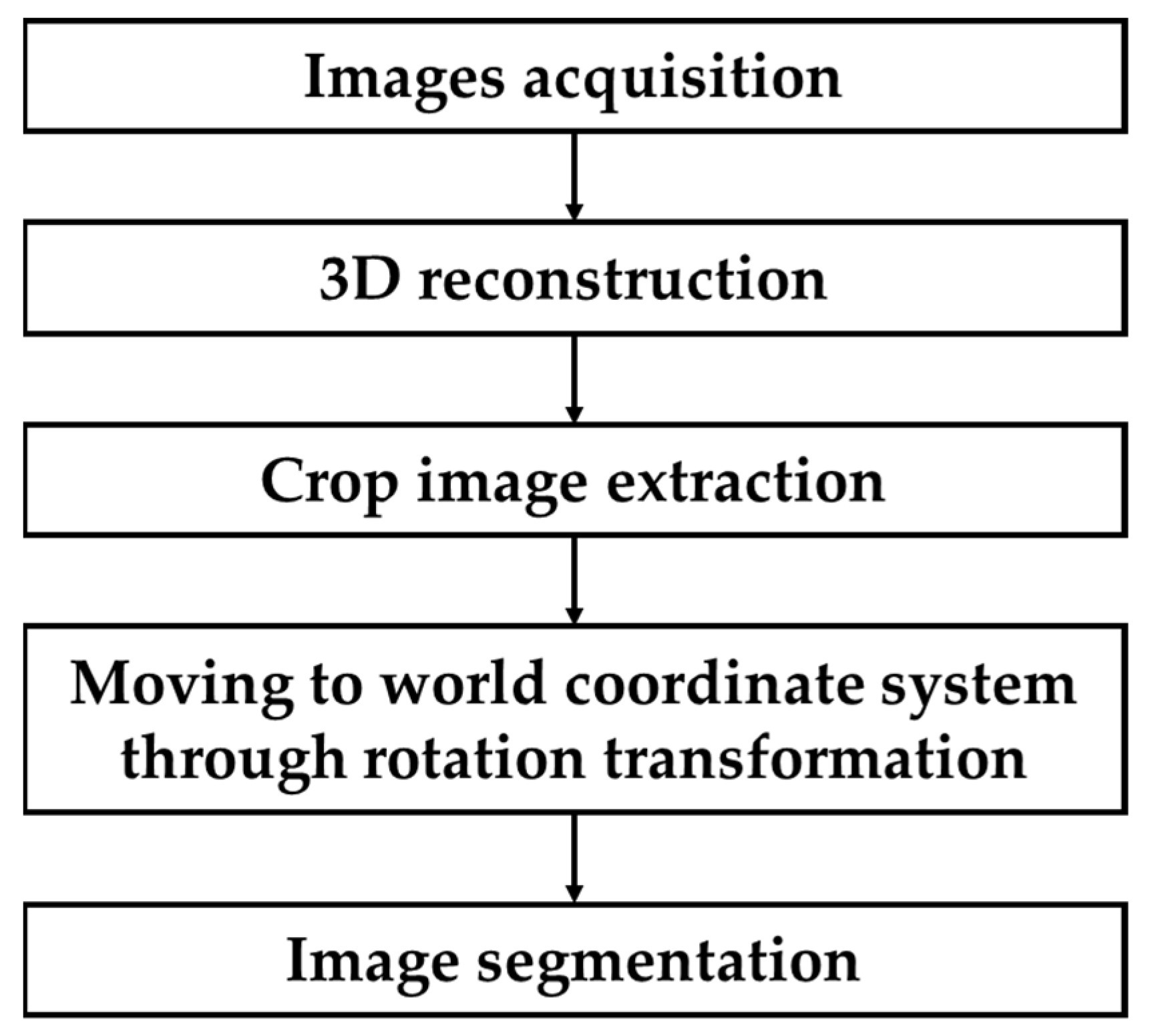
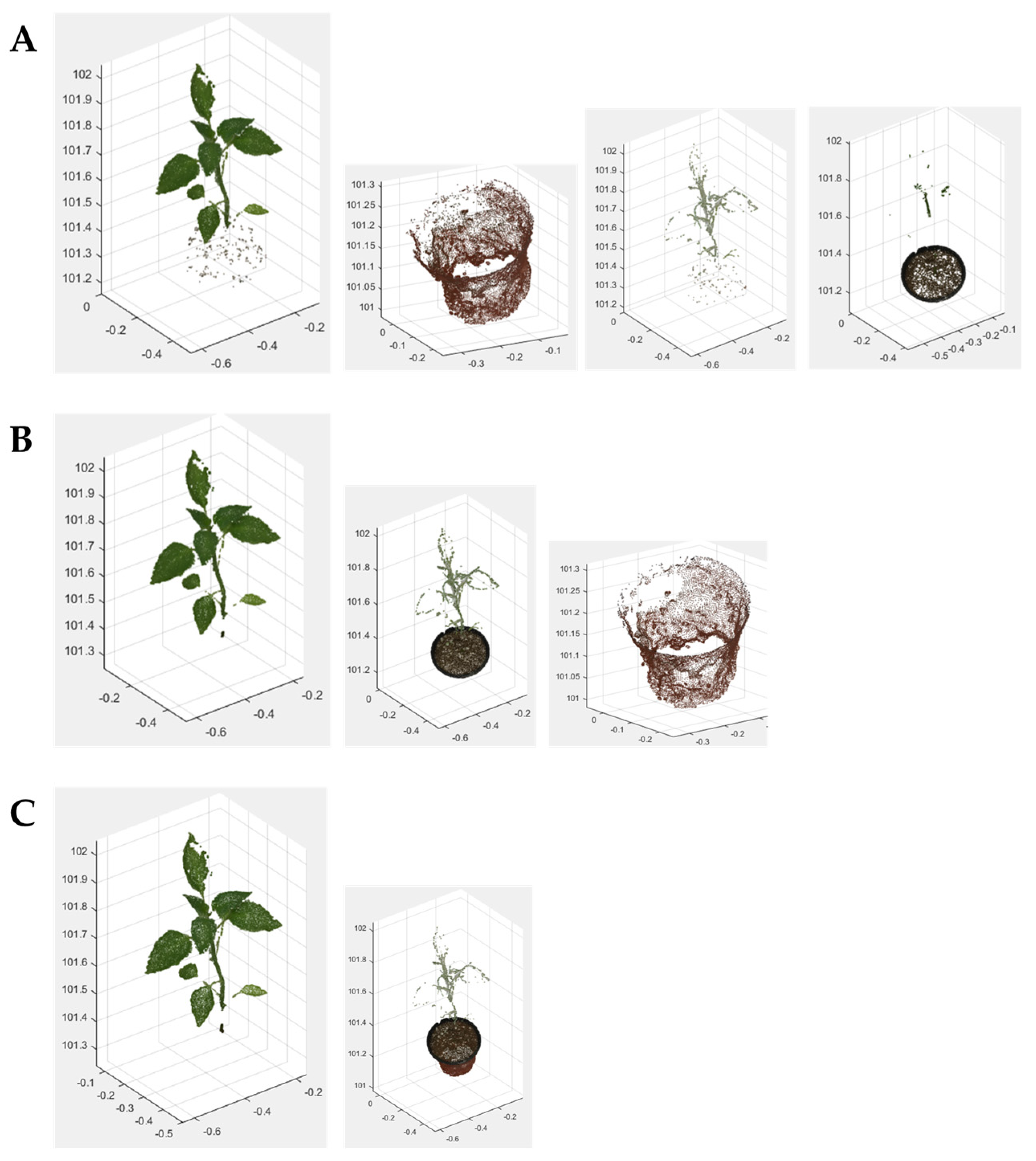
2.5.1. 3D Crop Extraction
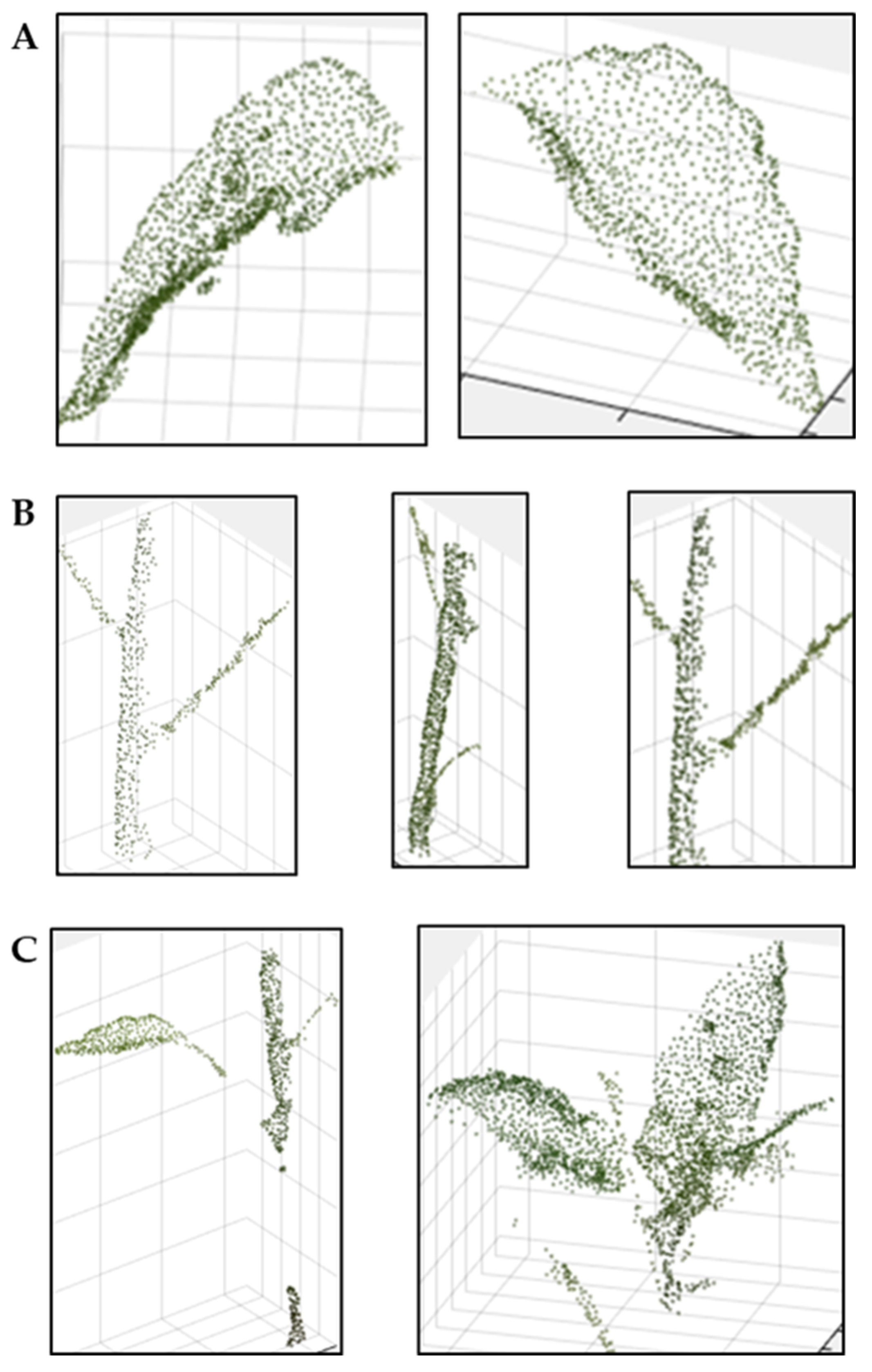
2.5.2. 3D Crop Segmentation and Automatic Analysis
3. Results
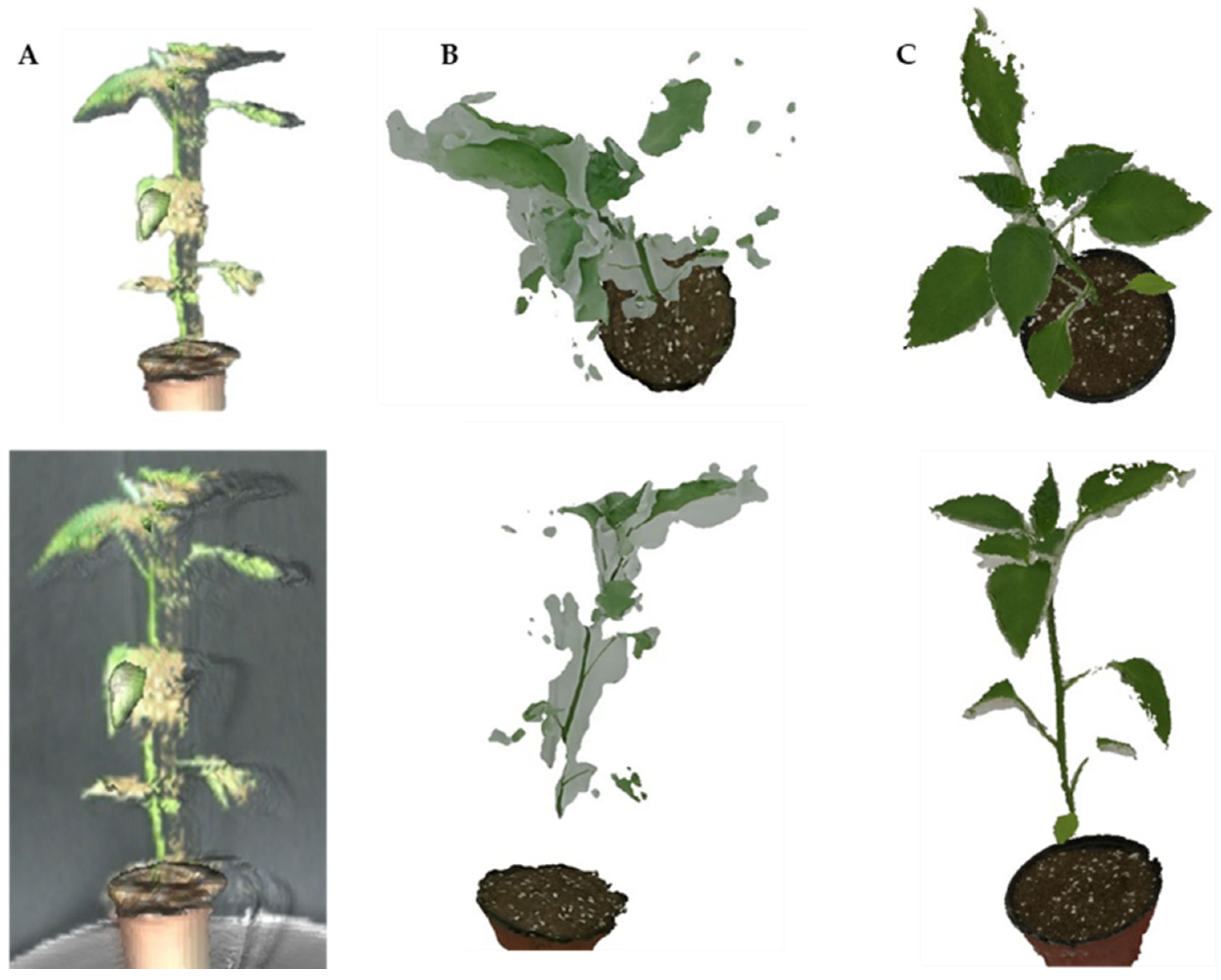
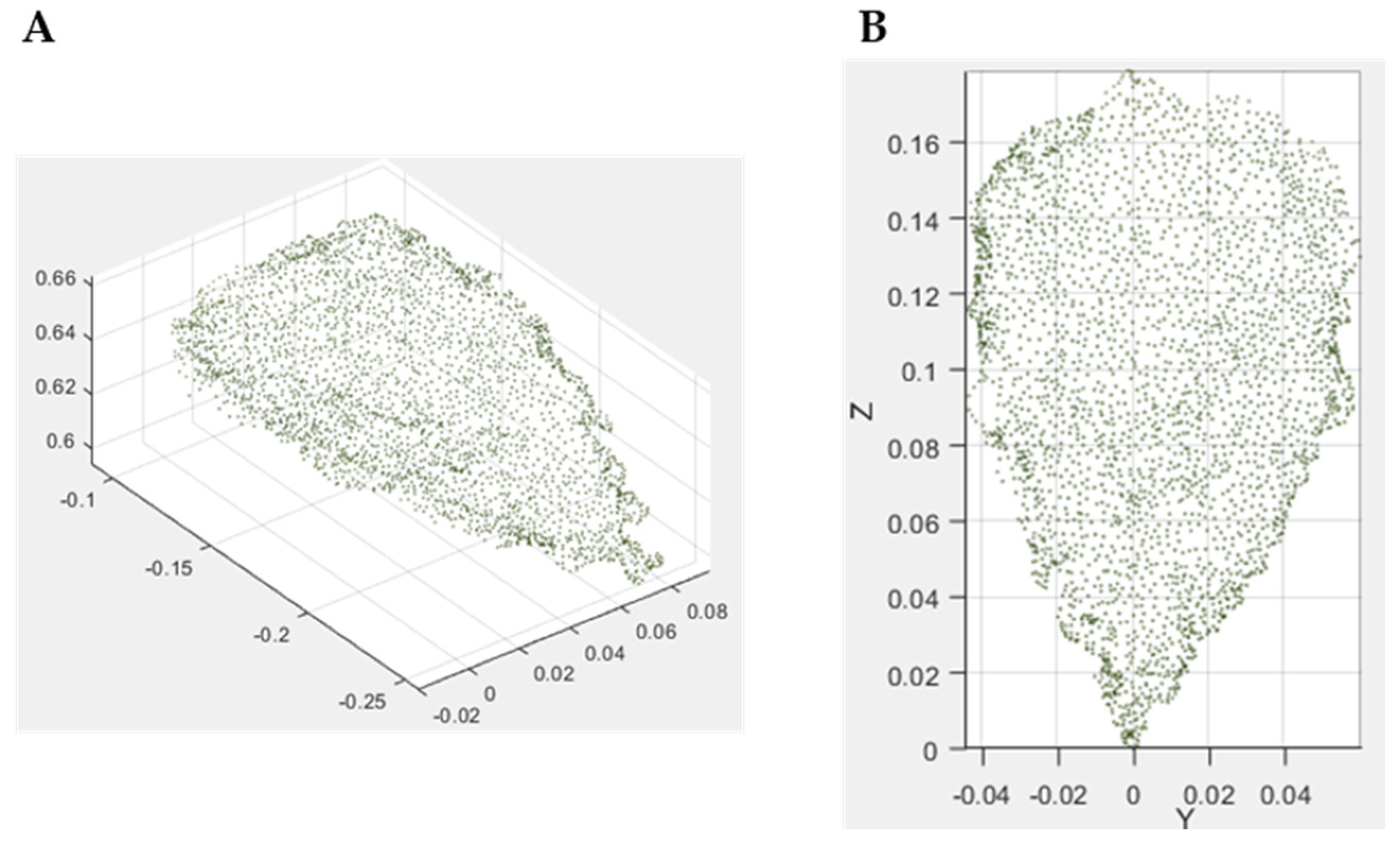
3.1. High-Resolution 3D Reconstruction Results
3.2. Crop Image Extraction and Segmentation Results
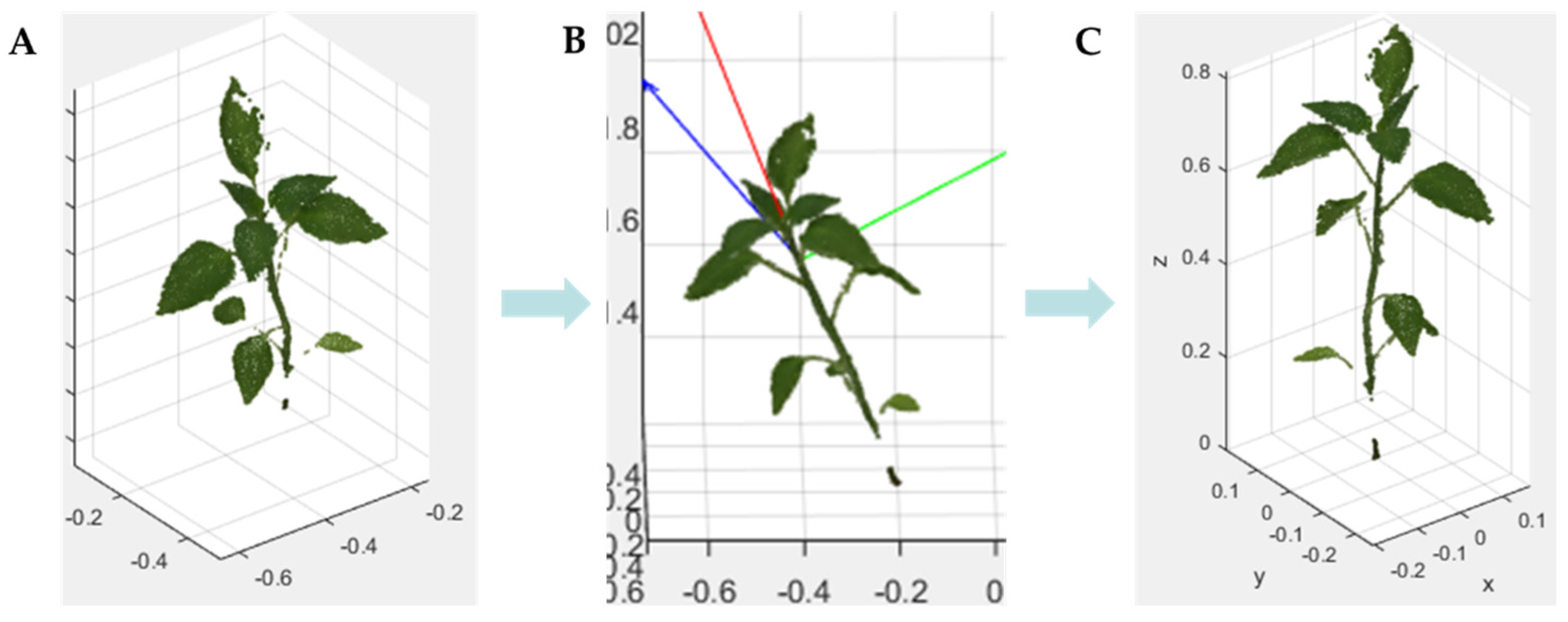
3.2.1. 3D Crop Extraction and Coordinate Transformation
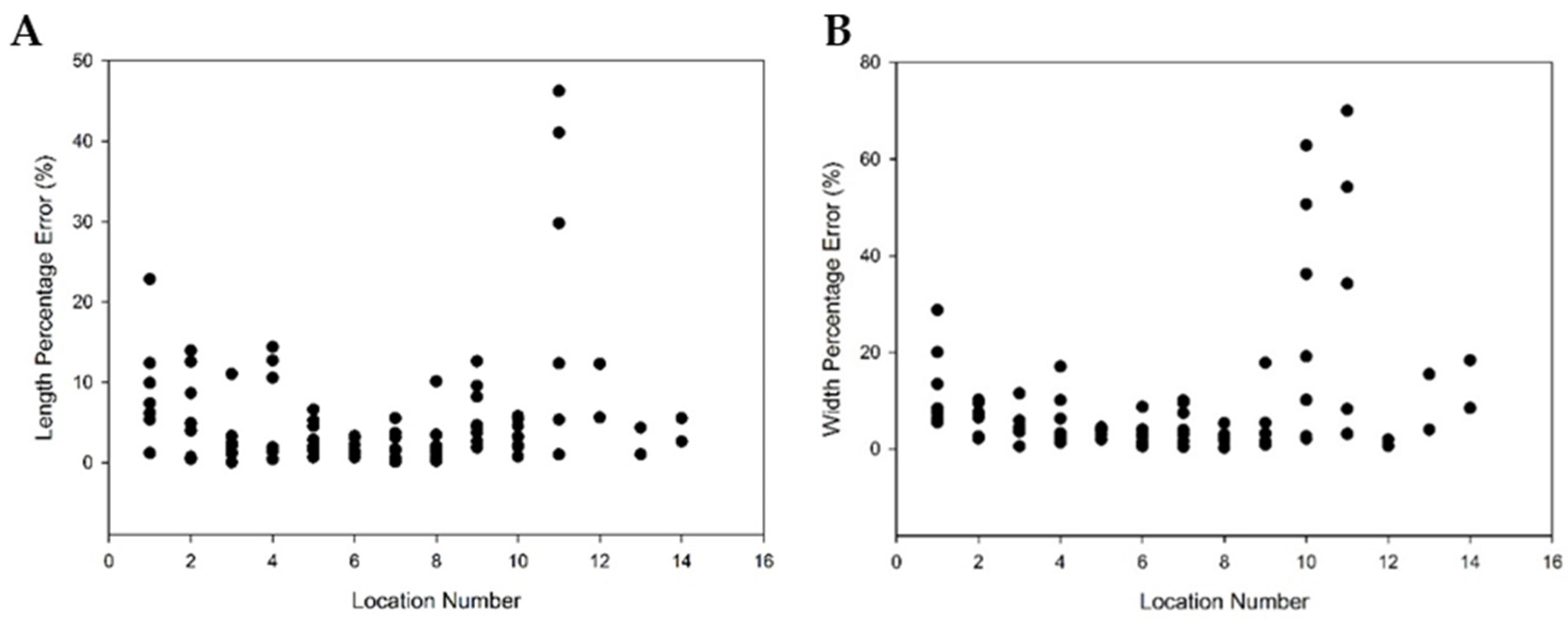
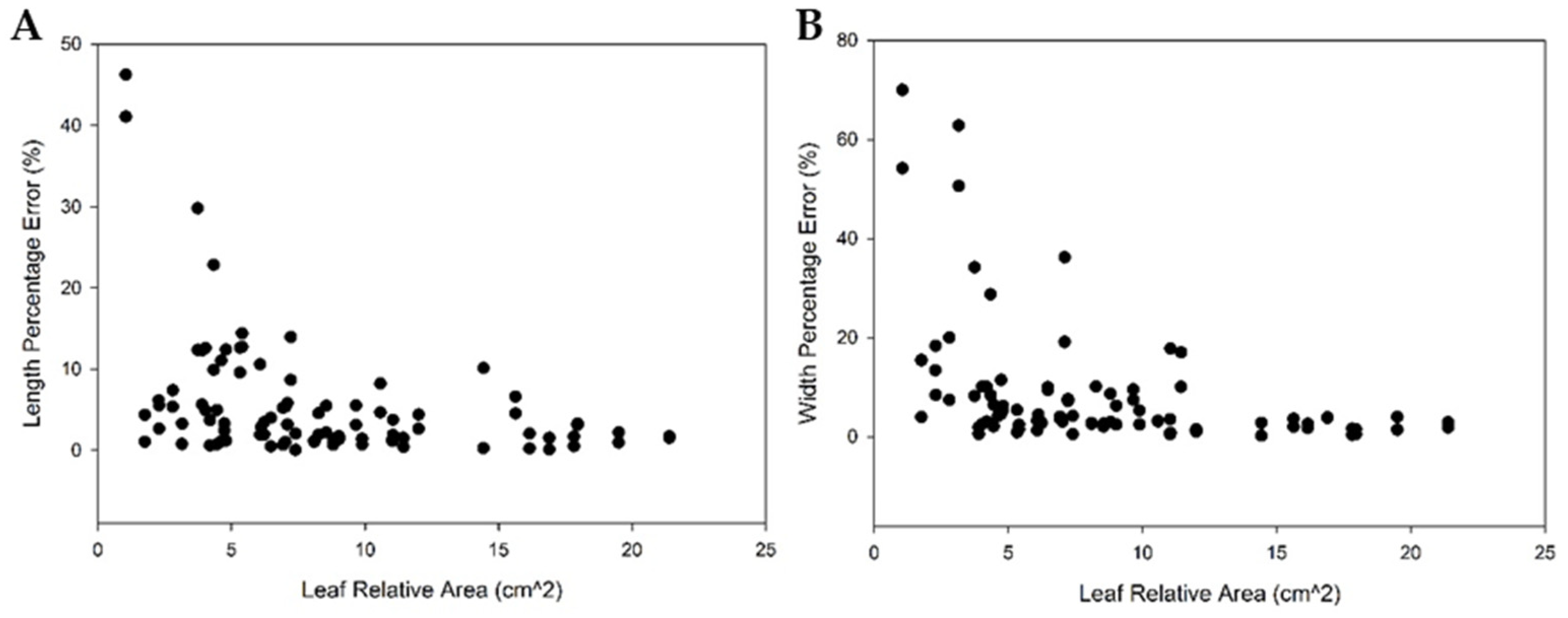
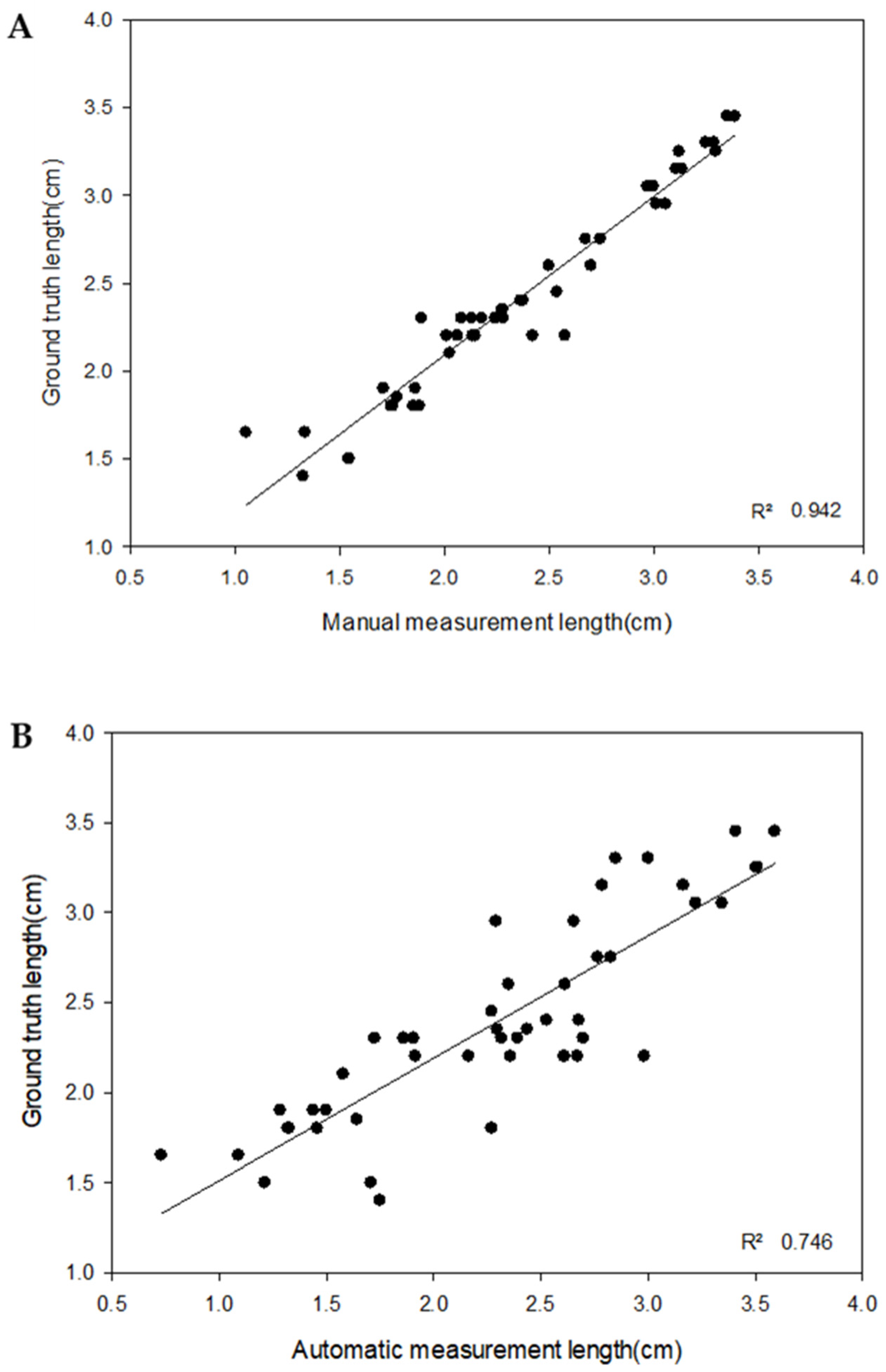
3.2.2. 3D Crop Image Segmentation and Automatic Measurement
4. Discussion
4.1. 3D Reconstruction
4.2. 3D Crop Image Automatic Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Furbank, R.T.; Tester, M. Phenomics–technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef]
- Fahlgren, N.; Gehan, M.A.; Baxter, I. Lights, camera, action: High-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Fátima da Silva, F.; Luz, P.H.C.; Romualdo, L.M.; Marin, M.A.; Zúñiga, A.M.G.; Herling, V.R.; Bruno, O.M. A diagnostic tool for magnesium nutrition in maize based on image analysis of different leaf sections. Crop Sci. 2014, 54, 738–745. [Google Scholar] [CrossRef]
- Li, C.; Adhikari, R.; Yao, Y.; Miller, A.G.; Kalbaugh, K.; Li, D.; Nemali, K. Measuring plant growth characteristics using smartphone based image analysis technique in controlled environment agriculture. Comput. Electron. Agric. 2020, 168, 105123. [Google Scholar] [CrossRef]
- Yahata, S.; Onishi, T.; Yamaguchi, K.; Ozawa, S.; Kitazono, J.; Ohkawa, T.; Yoshida, T.; Murakami, N.; Tsuji, H. A hybrid machine learning approach to automatic plant phenotyping for smart agriculture. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 14–19 May 2017; pp. 1787–1793. [Google Scholar]
- Pound, M.P.; Atkinson, J.A.; Townsend, A.J.; Wilson, M.H.; Griffiths, M.; Jackson, A.S.; Bulat, A.; Tzimiropoulos, G.; Wells, D.M.; Murchie, E.H. Deep machine learning provides state-of-the-art performance in image-based plant phenotyping. Gigascience 2017, 6, gix083. [Google Scholar] [CrossRef]
- Koh, J.C.; Spangenberg, G.; Kant, S. Automated Machine Learning for High-Throughput Image-Based Plant Phenotyping. Remote Sens. 2021, 13, 858. [Google Scholar] [CrossRef]
- Sugiura, R.; Tsuda, S.; Tamiya, S.; Itoh, A.; Nishiwaki, K.; Murakami, N.; Shibuya, Y.; Hirafuji, M.; Nuske, S. Field phenotyping system for the assessment of potato late blight resistance using RGB imagery from an unmanned aerial vehicle. Biosyst. Eng. 2016, 148, 1–10. [Google Scholar] [CrossRef]
- Zheng, J.; Fu, H.; Li, W.; Wu, W.; Yu, L.; Yuan, S.; Tao, W.Y.W.; Pang, T.K.; Kanniah, K.D. Growing status observation for oil palm trees using Unmanned Aerial Vehicle (UAV) images. ISPRS J. Photogramm. Remote Sens. 2021, 173, 95–121. [Google Scholar] [CrossRef]
- Qiu, R.; Wei, S.; Zhang, M.; Li, H.; Sun, H.; Liu, G.; Li, M. Sensors for measuring plant phenotyping: A review. Int. J. Agric. Biol. Eng. 2018, 11, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Fourcaud, T.; Zhang, X.; Stokes, A.; Lambers, H.; Körner, C. Plant growth modelling and applications: The increasing importance of plant architecture in growth models. Ann. Bot. 2008, 101, 1053–1063. [Google Scholar] [CrossRef] [Green Version]
- Snavely, N.; Seitz, S.M.; Szeliski, R. Photo tourism: Exploring photo collections in 3D. ACM Trans. Graph. 2006, 25, 835–849. [Google Scholar] [CrossRef]
- Li, M.; Zheng, D.; Zhang, R.; Yin, J.; Tian, X. Overview of 3d reconstruction methods based on multi-view. In Proceedings of the 2015 7th International Conference on Intelligent Human-Machine Systems and Cybernetics, Hangzhou, China, 26–27 August 2015; pp. 145–148. [Google Scholar]
- Remondino, F.; Spera, M.G.; Nocerino, E.; Menna, F.; Nex, F.; Gonizzi-Barsanti, S. Dense image matching: Comparisons and analyses. In Proceedings of the 2013 Digital Heritage International Congress (DigitalHeritage), Marseille, France, 28 October–1 November 2013; pp. 47–54. [Google Scholar]
- Lou, L.; Liu, Y.; Han, J.; Doonan, J.H. Accurate multi-view stereo 3D reconstruction for cost-effective plant phenotyping. In Proceedings of the International Conference Image Analysis and Recognition, Vilamoura, Portugal, 22–24 October 2014; pp. 349–356. [Google Scholar]
- Lou, L.; Liu, Y.; Sheng, M.; Han, J.; Doonan, J.H. A cost-effective automatic 3D reconstruction pipeline for plants using multi-view images. In Proceedings of the Conference Towards Autonomous Robotic Systems, Birmingham, UK, 1–3 September 2014; pp. 221–230. [Google Scholar]
- Jancosek, M.; Pajdla, T. Multi-view reconstruction preserving weakly-supported surfaces. In Proceedings of the CVPR 2011, Colorado Springs, CO, USA, 20–25 June 2011; pp. 3121–3128. [Google Scholar]
- Ni, Z.; Burks, T.F.; Lee, W.S. 3D reconstruction of small plant from multiple views. In Proceedings of the 2014 Montreal, Quebec, QC, Canada, 13–16 July 2014; p. 1. [Google Scholar]
- Nguyen, T.T.; Slaughter, D.C.; Townsley, B.; Carriedo, L.; Julin, N.; Sinha, N. Comparison of structure-from-motion and stereo vision techniques for full in-field 3d reconstruction and phenotyping of plants: An investigation in sunflower. In Proceedings of the 2016 ASABE Annual International Meeting, Orlando, FL, USA, 17–20 July 2016; p. 1. [Google Scholar]
- Yang, Q.; Tan, K.-H.; Culbertson, B.; Apostolopoulos, J. Fusion of active and passive sensors for fast 3d capture. In Proceedings of the 2010 IEEE International Workshop on Multimedia Signal Processing, Saint Malo, France, 4–6 October 2010; pp. 69–74. [Google Scholar]
- Zhu, J.; Wang, L.; Yang, R.; Davis, J. Fusion of time-of-flight depth and stereo for high accuracy depth maps. In Proceedings of the 2008 IEEE Conference on Computer Vision and Pattern Recognition, Anchorage, AK, USA, 24–26 June 2008; pp. 1–8. [Google Scholar]
- Lee, E.-K.; Ho, Y.-S. Generation of high-quality depth maps using hybrid camera system for 3-D video. J. Vis. Commun. Image Represent. 2011, 22, 73–84. [Google Scholar] [CrossRef]
- Paulus, S.; Behmann, J.; Mahlein, A.-K.; Plümer, L.; Kuhlmann, H. Low-cost 3D systems: Suitable tools for plant phenotyping. Sensors 2014, 14, 3001–3018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasenmüller, O.; Stricker, D. Comparison of kinect v1 and v2 depth images in terms of accuracy and precision. In Proceedings of the Asian Conference on Computer Vision, Taipei, Taiwan, 20–24 November 2016; pp. 34–45. [Google Scholar]
- Yang, L.; Zhang, L.; Dong, H.; Alelaiwi, A.; El Saddik, A. Evaluating and improving the depth accuracy of Kinect for Windows v2. IEEE Sens. J. 2015, 15, 4275–4285. [Google Scholar] [CrossRef]
- Lachat, E.; Macher, H.; Landes, T.; Grussenmeyer, P. Assessment and calibration of a RGB-D camera (Kinect v2 Sensor) towards a potential use for close-range 3D modeling. Remote Sens. 2015, 7, 13070–13097. [Google Scholar] [CrossRef] [Green Version]
- Gai, J.; Tang, L.; Steward, B. Plant recognition through the fusion of 2D and 3D images for robotic weeding. In Proceedings of the 2015 ASABE Annual International Meeting, New Orleans, LA, USA, 26–29 July 2015; p. 1. [Google Scholar]
- Gai, J.; Tang, L.; Steward, B. Plant localization and discrimination using 2D + 3D computer vision for robotic intra-row weed control. In Proceedings of the 2016 ASABE Annual International Meeting, Orlando, FL, USA, 17–20 July 2016; p. 1. [Google Scholar]
- Shah, D.; Tang, L.; Gai, J.; Putta-Venkata, R. Development of a mobile robotic phenotyping system for growth chamber-based studies of genotype x environment interactions. IFAC-PapersOnLine 2016, 49, 248–253. [Google Scholar] [CrossRef]
- Sun, G.; Wang, X. Three-dimensional point cloud reconstruction and morphology measurement method for greenhouse plants based on the kinect sensor self-calibration. Agronomy 2019, 9, 596. [Google Scholar] [CrossRef] [Green Version]
- Vázquez-Arellano, M.; Reiser, D.; Paraforos, D.S.; Garrido-Izard, M.; Burce, M.E.C.; Griepentrog, H.W. 3-D reconstruction of maize plants using a time-of-flight camera. Comput. Electron. Agric. 2018, 145, 235–247. [Google Scholar] [CrossRef]
- Microsoft. Available online: https://developer.microsoft.com/en-us/windows/kinect/hardware (accessed on 14 September 2017).
- Microsoft. Available online: https://msdn.microsoft.com/en-us/library/dn782025.aspx (accessed on 20 October 2017).
- Cignoni, P.; Callieri, M.; Corsini, M.; Dellepiane, M.; Ganovelli, F.; Ranzuglia, G. Meshlab: An open-source mesh processing tool. In Proceedings of the Eurographics Italian Chapter Conference, Salerno, Italy, 2–4 July 2008; pp. 129–136. [Google Scholar]
- PhotoScan. Available online: http://www.agisoft.com/ (accessed on 14 September 2017).
- Wu, C. VisualSFM: A Visual Structure from Motion System. Available online: http://ccwu.me/vsfm/doc.html (accessed on 14 September 2017).
- Ferstl, D.; Reinbacher, C.; Ranftl, R.; Rüther, M.; Bischof, H. Image guided depth upsampling using anisotropic total generalized variation. In Proceedings of the IEEE International Conference on Computer Vision, Sydney, Australia, 1–8 December 2013; pp. 993–1000. [Google Scholar]
- Tsai, R. A versatile camera calibration technique for high-accuracy 3D machine vision metrology using off-the-shelf TV cameras and lenses. IEEE Robot. Autom. 1987, 3, 323–344. [Google Scholar] [CrossRef] [Green Version]
- Hamzah, R.A.; Salim, S.I.M. Software calibration for stereo camera on stereo vision mobile robot using Tsai’s method. Int. J. Comput. Theory Eng. 2010, 2, 390. [Google Scholar] [CrossRef] [Green Version]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A k-means clustering algorithm. J. R. Stat. Soc. Ser. C Appl. Stat. 1979, 28, 100–108. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. Syst. 1979, 9, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Woebbecke, D.; Meyer, G.; Von Bargen, K.; Mortensen, D. Shape features for identifying young weeds using image analysis. Trans. ASABE 1995, 38, 271–281. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Pio, R. Euler angle transformations. IEEE Trans. Automat. Contr. 1966, 11, 707–715. [Google Scholar] [CrossRef]
- Yemez, Y.; Schmitt, F. 3D reconstruction of real objects with high resolution shape and texture. Image Vis. Comput. 2004, 22, 1137–1153. [Google Scholar] [CrossRef]
- Golbach, F.; Kootstra, G.; Damjanovic, S.; Otten, G.; van de Zedde, R. Validation of plant part measurements using a 3D reconstruction method suitable for high-throughput seedling phenotyping. Mach. Vis. Appl. 2016, 27, 663–680. [Google Scholar] [CrossRef] [Green Version]
- Krioukov, D. Clustering implies geometry in networks. Phys. Rev. Lett. 2016, 116, 208302. [Google Scholar] [CrossRef] [PubMed]
- McCormick, R.F.; Truong, S.K.; Mullet, J.E. 3D sorghum reconstructions from depth images identify QTL regulating shoot architecture. Plant Physiol. 2016, 172, 823–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paproki, A.; Sirault, X.; Berry, S.; Furbank, R.; Fripp, J. A novel mesh processing based technique for 3D plant analysis. BMC Plant Biol. 2012, 12, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, M.; Cui, J.; Jeong, E.S.; Cho, S.I. Development of High-resolution 3D Phenotyping System Using Depth Camera and RGB Camera. In Proceedings of the 2017 ASABE Annual International Meeting, Spokane, WA, USA, 16–19 July 2017; p. 1. [Google Scholar]






| Color | Resolution | 1920 × 1080 pixels |
| Field of view | 84 × 54° | |
| Depth/Infrared | Resolution | 512 × 424 pixels |
| Field of view | 70 × 60° | |
| Frame rate | 30 Hz | |
| Depth measurement | Time of flight | |
| Range of depth | 0.5~4.5 m | |
| Dimension (length × width × height) | 24.9 × 6.6 × 6.7 cm | |
| Weight | Approximately 1.4 kg | |
| Crop | ||||
|---|---|---|---|---|
| A | B | C | D | |
| No. of total leaves | 30 | 24 | 24 | 26 |
| No. of leaves that failed | 2 | 3 | 4 | 4 |
| Successful rate (%) | 93.3 | 87.5 | 83.3 | 84.6 |
| Crop | Error (cm) | Percentage Error (%) | Standard Deviation | Total Error (cm) | Total Percentage Error (%) | Total Standard Deviation | |
|---|---|---|---|---|---|---|---|
| Leaf length | A | 0.139 | 4.24 | 0.111 | 0.183 | 5.49 | 0.200 |
| B | 0.233 | 8.89 | 0.265 | ||||
| C | 0.189 | 4.63 | 0.197 | ||||
| D | 0.187 | 4.63 | 0.221 | ||||
| Leaf width | A | 0.101 | 6.30 | 0.0677 | 0.124 | 8.63 | 0.127 |
| B | 0.149 | 15.4 | 0.173 | ||||
| C | 0.173 | 8.90 | 0.162 | ||||
| D | 0.084 | 4.92 | 0.0805 | ||||
| Crop height | A | 0.068 | 0.313 | - | 0.344 | 1.53 | 0.259 |
| B | 0.314 | 0.670 | |||||
| C | 0.693 | 2.74 | |||||
| D | 0.300 | 1.21 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Cho, S.-I. High-Resolution 3D Crop Reconstruction and Automatic Analysis of Phenotyping Index Using Machine Learning. Agriculture 2021, 11, 1010. https://doi.org/10.3390/agriculture11101010
Yang M, Cho S-I. High-Resolution 3D Crop Reconstruction and Automatic Analysis of Phenotyping Index Using Machine Learning. Agriculture. 2021; 11(10):1010. https://doi.org/10.3390/agriculture11101010
Chicago/Turabian StyleYang, Myongkyoon, and Seong-In Cho. 2021. "High-Resolution 3D Crop Reconstruction and Automatic Analysis of Phenotyping Index Using Machine Learning" Agriculture 11, no. 10: 1010. https://doi.org/10.3390/agriculture11101010
APA StyleYang, M., & Cho, S.-I. (2021). High-Resolution 3D Crop Reconstruction and Automatic Analysis of Phenotyping Index Using Machine Learning. Agriculture, 11(10), 1010. https://doi.org/10.3390/agriculture11101010







