ACE-ADP: Adversarial Contextual Embeddings Based Named Entity Recognition for Agricultural Diseases and Pests
Abstract
:1. Introduction
1.1. Recent Developments Related to NER Models
1.2. Objectives and Hypotheses
- (1)
- An adversarial contextual embeddings-based model could be applied for agricultural diseases and pests named entity recognition. As far as we know, it was the first time that combined BERT and adversarial training to recognizing the named entities in the field of agricultural diseases and pests;
- (2)
- The BERT, which was fine-tuned on the agricultural corpus, could generate the high-quality text representation so that to enhance the quality of text representation and solve the polysemous problem;
- (3)
- Adversarial training could also be adopted to solve the rare entity recognition problem. Besides, it could also exert its maximum performance when the text representation was of high quality. As far as we know, the previous research had not explicitly raised this point;
- (4)
- ACE-ADP could significantly improve the F1 of CNER-ADP with an improvement of 4.31%, especially for rare entities, in which an F1 was increased by 9.83% on average.
2. Materials and Methods
2.1. Datasets
2.2. Parameter Setting
2.3. Evaluation Metrics
2.4. ACE-ADP Method
2.4.1. Problem Definition
2.4.2. Fine-Tuned BERT
2.4.3. Context Encoder and Decoder
2.4.4. Adversarial Training
- (1)
- Contextual-sensitive. BERT can dynamically generate the context-dependent embeddings according to the contexts, which is beneficial for solving the problem of polysemous words that are often caused by context-independent methods such as word2vec and glove;
- (2)
- Domain-aware. In this paper, domain knowledge can be injected into BERT by fine-tuning, which is essential to handle the NER task in specific domains;
- (3)
- Stronger robustness and generalization. The experimental results in Section 4.4 showed that compared with previous models, our proposed model maintains high robustness and generalization.
3. Results
3.1. Main Results Compared with Other Models
3.2. Ablation Study
3.2.1. Macro-Level Analysis
3.2.2. Effect of BERT
3.2.3. Effect of Adversarial Training
4. Discussion
4.1. Performance for Rare Entities
4.2. Robustness and Generalization
4.3. Convergence
4.4. Visualization of Features
4.5. Parameter Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Algorithm A1 Pseudocode for domain-specific named entity recognition task with adversarial training and contextual embeddings. | |
| Input: Fine-tuned BERT model for a specific field, global learning rate perturbation size , the number of iterations T, a domain-specific sentence S, and their ground-truth labels Y. Output: the predicted labels , the training weights of the model . | |
| 1: | Converting the sentence S into the contextual embeddings by fine-tuned BERT on the texts in the field of agricultural diseases and pests. |
| 2: | For t = 1, …, T do |
| 3: | = BiLSTM(E), according to Equation (8) to Equation (10). |
| 4: | P = , according to Equation (12). |
| 5: | Calculating the by using the CRF algorithm. |
| 6: | |
| 7: | |
| 8: | |
| 9: | |
| 10: | Repeat lines 3–9 |
| 11: | |
| 12: | F1-scores conlleval(Y, ), calculating the overall F1-scores for predicted labels. |
| 13: | If F1-scores then |
| 14: | F1-scores |
| 15: | Save the weights of the model |
| 16: | end for |
| 17: | Output: the best-predicted labels , the best training weights of the model . |
- The sentence is converted into contextual embeddings by using BERT, which is fine-tuned on the texts of agricultural diseases and pests.
- The character-level embeddings are used as input of the BiLSTM to extract the global context features. Note that other contextual encoders such as Gated CNN and RD_CNN can also be used to extract the context features according to the experimental results in Section 3.2.3.
- The possible labels are predicted, and the loss is calculated by the CRF layer.
- Calculating the perturbation according to Equation (17) and adding it to the original character-level embeddings.
- Steps (1) to (4) are repeated until a maximum iteration is reached.
References
- Lu, J.; Tan, L.; Jiang, H. Review on Convolutional Neural Network (CNN) Applied to Plant Leaf Disease Classification. Agriculture 2021, 11, 707. [Google Scholar] [CrossRef]
- Molina-Villegas, A.; Muñiz-Sanchez, V.; Arreola-Trapala, J.; Alcántara, F. Geographic Named Entity Recognition and Disambiguation in Mexican News using Word Embeddings. Expert Syst. Appl. 2021, 176, 114855. [Google Scholar] [CrossRef]
- Yin, M.; Mou, C.; Xiong, K.; Ren, J. Chinese clinical named entity recognition with radical-level feature and self-attention mechanism. J. Biomed. Inform. 2019, 98, 103289. [Google Scholar] [CrossRef]
- Huang, K.; Altosaar, J.; Ranganath, R. ClinicalBert: Modeling clinical notes and predicting hospital readmission. arXiv 2019, arXiv:1904.05342. [Google Scholar]
- Francis, S.; Van Landeghem, J.; Moens, M.F. Transfer learning for named entity recognition in financial and biomedical documents. Information 2019, 10, 248. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2019, 36, 1234–1240. [Google Scholar] [CrossRef]
- Guo, X.; Zhou, H.; Su, J.; Hao, X.; Tang, Z.; Diao, L.; Li, L. Chinese agricultural diseases and pests named entity recognition with multi-scale local context features and self-attention mechanism. Comput. Electron. Agric. 2020, 179, 105830. [Google Scholar] [CrossRef]
- Yasunaga, M.; Kasai, J.; Radev, D. Robust multilingual part-of-speech tagging via adversarial training. arXiv 2017, arXiv:1711.04903. [Google Scholar]
- Du, C.; Sun, H.; Wang, J.; Qi, Q.; Liao, J. Adversarial and domain-aware bert for cross-domain sentiment analysis. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online. 5–10 July 2020; pp. 4019–4028. [Google Scholar]
- Xu, J.; Wen, J.; Sun, X.; Su, Q. A discourse-level named entity recognition and relation extraction dataset for chinese literature text. arXiv 2017, arXiv:1711.07010. [Google Scholar]
- Malarkodi, C.S.; Lex, E.; Devi, S.L. Named Entity Recognition for the Agricultural Domain. Res. Comput. Sci. 2016, 117, 121–132. [Google Scholar]
- Nadeau, D.; Sekine, S. A survey of named entity recognition and classification. Lingvisticae Investig. 2007, 30, 3–26. [Google Scholar] [CrossRef]
- Liu, W.; Yu, B.; Zhang, C.; Wang, H.; Pan, K. Chinese Named Entity Recognition Based on Rules and Conditional Random Field. In Proceedings of the 2018 2nd International Conference on Computer Science and Artificial Intelligence, ShenZhen, China, 8–10 December 2018; pp. 268–272. [Google Scholar]
- WANG Chun-yu, W.F. Study on recognition of chinese agricultural named entity with conditional random fields. J. Hebei Agric. Univ. 2014, 37, 132–135. [Google Scholar] [CrossRef]
- Zhao, P.; Zhao, C.; Wu, H.; Wang, W. Named Entity Recognition of Chinese Agricultural Text Based on Attention Mechanism. Nongye Jixie Xuebao/Trans. Chin. Soc. Agric. Mach. 2021, 52, 185–192. [Google Scholar] [CrossRef]
- Saleem, M.H.; Potgieter, J.; Arif, K.M. Plant Disease Detection and Classification by Deep Learning. Plants 2019, 8, 468. [Google Scholar] [CrossRef] [Green Version]
- Hasan, R.I.; Yusuf, S.M.; Alzubaidi, L. Review of the State of the Art of Deep Learning for Plant Diseases: A Broad Analysis and Discussion. Plants 2020, 9, 1302. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Peng, Y.; Liu, J.; Wu, S. Tomato Leaf Disease Diagnosis Based on Improved Convolution Neural Network by Attention Module. Agriculture 2021, 11, 651. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, K.; Zhao, Y.; Sun, Y.; Ban, W.; Chen, Y.; Zhuang, H.; Zhang, X.; Liu, J.; Yang, T. An Approach for Rice Bacterial Leaf Streak Disease Segmentation and Disease Severity Estimation. Agriculture 2021, 11, 420. [Google Scholar] [CrossRef]
- Hao, X.; Jia, J.; Gao, W.; Guo, X.; Zhang, W.; Zheng, L.; Wang, M. MFC-CNN: An automatic grading scheme for light stress levels of lettuce (Lactuca sativa L.) leaves. Comput. Electron. Agric. 2020, 179, 105847. [Google Scholar] [CrossRef]
- Biswas, P.; Sharan, A. A Noble Approach for Recognition and Classification of Agricultural Named Entities using Word2Vec. Int. J. Adv. Stud. Comput. Sci. Eng. 2021, 9, 1–8. [Google Scholar]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient estimation of word representations in vector space. arXiv 2013, arXiv:1301.3781. [Google Scholar]
- Jawahar, G.; Sagot, B.; Seddah, D. What Does BERT Learn about the Structure of Language? In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, Florence, Italy, 6 July 2019; pp. 3651–3657. [Google Scholar]
- Zhang, S.; Zhao, M. Chinese agricultural diseases named entity recognition based on BERT-CRF. In Proceedings of the 2020 5th International Conference on Mechanical, Control and Computer Engineering (ICMCCE), Harbin, China, 25–27 December 2020; pp. 1148–1151. [Google Scholar]
- Huang, Z.; Xu, W.; Yu, K. Bidirectional LSTM-CRF models for sequence tagging. arXiv 2015, arXiv:1508.01991. [Google Scholar]
- Strubell, E.; Verga, P.; Belanger, D.; McCallum, A. Fast and Accurate Entity Recognition with Iterated Dilated Convolutions. In Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing, Copenhagen, Denmark, 1 September 2017; pp. 2670–2680. [Google Scholar]
- Dauphin, Y.N.; Fan, A.; Auli, M.; Grangier, D. Language modeling with gated convolutional networks. In Proceedings of the International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; pp. 933–941. [Google Scholar]
- Qiu, J.; Wang, Q.; Zhou, Y.; Ruan, T.; Gao, J. Fast and accurate recognition of Chinese clinical named entities with residual dilated convolutions. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 3–6 December 2018; pp. 935–942. [Google Scholar]
- Yan, H.; Deng, B.; Li, X.; Qiu, X. Tener: Adapting transformer encoder for named entity recognition. arXiv 2019, arXiv:1911.04474. [Google Scholar]
- Chen, H.; Lin, Z.; Ding, G.; Lou, J.; Zhang, Y.; Karlsson, B. GRN: Gated relation network to enhance convolutional neural network for named entity recognition. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; pp. 6236–6243. [Google Scholar]
- Zhu, Y.; Wang, G. CAN-NER: Convolutional Attention Network for Chinese Named Entity Recognition. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), Minneapolis, MN, USA, 2–7 June 2019; pp. 3384–3393. [Google Scholar]
- Li, L.; Zhao, J.; Hou, L.; Zhai, Y.; Shi, J.; Cui, F. An attention-based deep learning model for clinical named entity recognition of Chinese electronic medical records. BMC Med. Inform. Decis. Mak. 2019, 19, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Cao, P.; Chen, Y.; Liu, K.; Zhao, J.; Liu, S. Adversarial transfer learning for Chinese named entity recognition with self-attention mechanism. In Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, Brussels, Belgium, 2–4 November 2018; pp. 182–192. [Google Scholar]
- Wang, C.; Chen, W.; Xu, B. Named entity recognition with gated convolutional neural networks. In Chinese Computational Linguistics and Natural Language Processing Based on Naturally Annotated Big Data; Springer: Berlin/Heidelberg, Germany, 2017; pp. 110–121. [Google Scholar]
- Li, X.; Yan, H.; Qiu, X.; Huang, X.-J. FLAT: Chinese NER Using Flat-Lattice Transformer. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Seattle, WA, USA, 5–10 July 2020; pp. 6836–6842. [Google Scholar]
- Cetoli, A.; Bragaglia, S.; O’Harney, A.; Sloan, M. Graph Convolutional Networks for Named Entity Recognition. In Proceedings of the 16th International Workshop on Treebanks and Linguistic Theories, Prague, Czech Republic, 1 July 2017; pp. 37–45. [Google Scholar]
- Gui, T.; Zou, Y.; Zhang, Q.; Peng, M.; Fu, J.; Wei, Z.; Huang, X.-J. A lexicon-based graph neural network for chinese ner. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), Hong Kong, China, 3 November 2019; pp. 1039–1049. [Google Scholar]
- Li, J.; Sun, A.; Han, J.; Li, C. A survey on deep learning for named entity recognition. IEEE Trans. Knowl. Data Eng. 2020. [Google Scholar] [CrossRef]
- Pre-trained models for natural language processing: A survey. arXiv 2020, arXiv:2003.08271.
- Zhang, R.; Lu, W.; Wang, S.; Peng, X.; Yu, R.; Gao, Y. Chinese clinical named entity recognition based on stacked neural network. Concurr. Comput. Pract. Exp. 2020, e5775. [Google Scholar] [CrossRef]
- Suman, C.; Reddy, S.M.; Saha, S.; Bhattacharyya, P. Why pay more? A simple and efficient named entity recognition system for tweets. Expert Syst. Appl. 2021, 167, 114101. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, H.; Zhang, J.; Ma, J.; Chang, Y. Attention-based multi-level feature fusion for named entity recognition. IJCAI Int. Jt. Conf. Artif. Intell. 2020, 2021, 3594–3600. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, Y.; Wang, Z. Deep neural network-based recognition of entities in Chinese online medical inquiry texts. Futur. Gener. Comput. Syst. 2021, 114, 581–604. [Google Scholar] [CrossRef]
- Chiu, J.P.C.; Nichols, E. Named entity recognition with bidirectional LSTM-CNNs. Trans. Assoc. Comput. Linguist. 2016, 4, 357–370. [Google Scholar] [CrossRef]
- Ma, X.; Hovy, E. End-to-end Sequence Labeling via Bi-directional LSTM-CNNs-CRF. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Berlin, Germany, 3 August 2016; pp. 1064–1074. [Google Scholar]
- Peters, M.E.; Ruder, S.; Smith, N.A. To Tune or Not to Tune? Adapting Pretrained Representations to Diverse Tasks. In Proceedings of the 4th Workshop on Representation Learning for NLP (RepL4NLP-2019), Florence, Italy, 5 August 2019; pp. 7–14. [Google Scholar]
- Song, C.H.; Sehanobish, A. Using Chinese Glyphs for Named Entity Recognition (Student Abstract). In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; Volume 34, pp. 13921–13922. [Google Scholar]
- Xuan, Z.; Bao, R.; Jiang, S. FGN: Fusion glyph network for Chinese named entity recognition. arXiv 2020, arXiv:2001.05272. [Google Scholar]
- Xu, L.; Dong, Q.; Liao, Y.; Yu, C.; Tian, Y.; Liu, W.; Li, L.; Liu, C.; Zhang, X. CLUENER2020: Fine-grained named entity recognition dataset and benchmark for chinese. arXiv 2020, arXiv:2001.04351. [Google Scholar]
- Zhang, Y.; Yang, J. Chinese NER Using Lattice LSTM. In Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Melbourne, Australia, 15–20 July 2018; pp. 1554–1564. [Google Scholar]
- Li, S.; Zhao, Z.; Hu, R.; Li, W.; Liu, T.; Du, X. Analogical Reasoning on Chinese Morphological and Semantic Relations. In Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers), Melbourne, Australia, 15–20 July 2018; pp. 138–143. [Google Scholar]
- Prechelt, L. Early stopping-but when? In Neural Networks: Tricks of the Trade; Springer: Berlin/Heidelberg, Germany, 1998; pp. 55–69. [Google Scholar]
- Bekoulis, G.; Deleu, J.; Demeester, T.; Develder, C. Joint entity recognition and relation extraction as a multi-head selection problem. Expert Syst. Appl. 2018, 114, 34–45. [Google Scholar] [CrossRef] [Green Version]
- Miyato, T.; Dai, A.M.; Goodfellow, I. Adversarial training methods for semi-supervised text classification. arXiv 2016, arXiv:1605.07725. [Google Scholar]
- der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Zhao, S.; Cai, Z.; Chen, H.; Wang, Y.; Liu, F.; Liu, A. Adversarial training based lattice LSTM for Chinese clinical named entity recognition. J. Biomed. Inform. 2019, 99, 103290. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cheng, H.; He, P.; Chen, W.; Wang, Y.; Poon, H.; Gao, J. Adversarial training for large neural language models. arXiv 2020, arXiv:2004.08994. [Google Scholar]
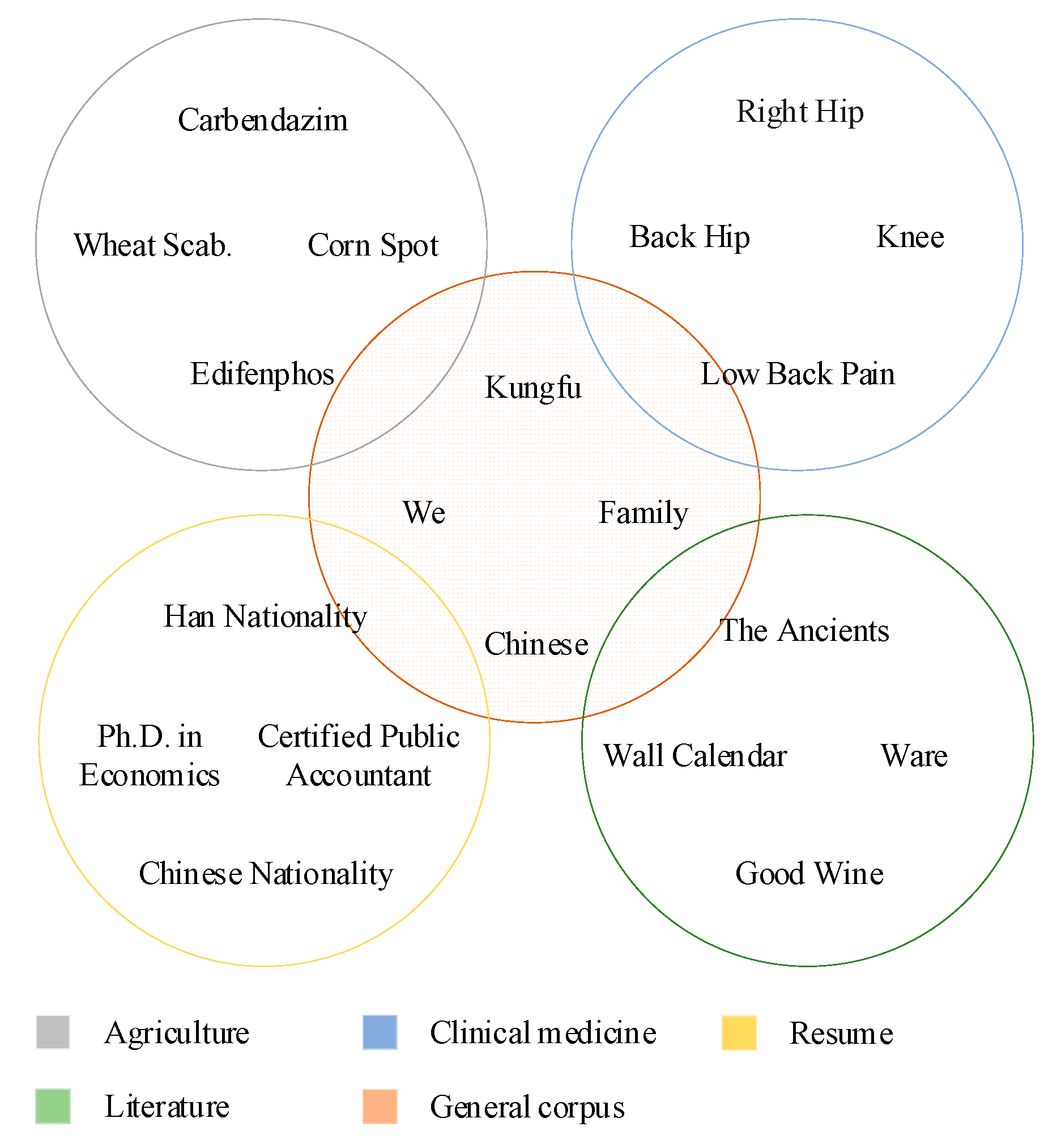








| Dataset | Domain | Samples | Entities | Class | Categories |
|---|---|---|---|---|---|
| AgCNER | Agriculture | 24,696 | 248,171 | 11 | Crop, Disease, Drug, Fertilizer, Part/Organs, Period, Pest, Pathogeny, Crop Cultivar, Weed, Other |
| CLUENER | News | 12,091 | 26,320 | 10 | Person, Organization, Position, Company, Address, Game, Government, Scene, Book, Movie |
| CCKS2017 | Clinic | 2231 | 63,063 | 5 | Body, Symptoms, Check, Disease, Treatment |
| Resume | Resume | 4740 | 16,565 | 8 | Country, Educational institution, Location, Personal name, Organization, Profession, Ethnicity, Background and Job, Title |
| Hyper-Parameter | Value | |
|---|---|---|
| Character embedding | 768 | |
| Hidden units | 256 | |
| Dropout | 0.25 | |
| Optimizer | Adam | |
| Batch_size | fine-tuning | 8 |
| model training | 32 | |
| Max_epoch | Word2vec | 100 |
| BERT | 50 | |
| Algorithms | CLUENER | AgCNER | CCKS2017 | Resume | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | |
| BERT-IDCNN-CRF | 78.37 | 77.60 | 77.98 ± 0.11 | 94.39 | 95.08 | 94.74 ± 0.07 | 90.55 | 93.52 | 92.01 ± 0.13 | 95.47 | 96.59 | 96.03 ± 0.13 |
| BERT-Gated CNN-CRF | 75.85 | 77.98 | 76.90 ± 0.34 | 94.32 | 95.20 | 94.76 ± 0.08 | 89.43 | 92.93 | 91.15 ± 0.14 | 95.75 | 96.81 | 96.27 ± 0.16 |
| AR-CCNER | 78.34 | 77.74 | 78.04 ± 0.28 | 94.60 | 94.73 | 94.67 ± 0.06 | 90.23 | 93.36 | 91.77 ± 0.28 | 95.89 | 97.22 | 96.55 ± 0.27 |
| FGN [48] | 79.50 | 79.71 | 79.60 ± 0.15 | 94.33 | 94.56 | 94.45 ± 0.03 | 90.44 | 93.09 | 91.75 ± 0.16 | 96.67 | 97.09 | 96.88 ± 0.10 |
| TENER | 72.94 | 74.21 | 73.57 ± 0.17 | 93.01 | 95.22 | 94.10 ± 0.09 | 91.24 | 93.08 | 92.15 ± 0.13 | 94.91 | 95.03 | 94.97 ± 0.21 |
| Flat-Lattice [35] | 79.25 | 80.68 | 79.96 ± 0.13 | 93.52 | 94.31 | 93.91 ± 0.08 | 91.55 | 93.40 | 92.46 ± 0.16 | 95.22 | 95.72 | 95.47 ± 0.18 |
| ACE-ADP | 93.03 | 94.36 | 93.68 ± 0.18 | 98.30 | 98.32 | 98.31 ± 0.02 | 95.17 | 96.27 | 95.72 ± 0.13 | 96.22 | 97.44 | 96.83 ± 0.17 |
| # | Algorithms | CLUENER | AgCNER | CCKS2017 | Resume | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | ||
| 1 | ACE-ADP | 93.03 | 94.36 | 93.68 ± 0.18 | 98.30 | 98.32 | 98.31 ± 0.02 | 95.17 | 96.27 | 95.72 ± 0.13 | 96.22 | 97.44 | 96.83 ± 0.17 |
| 2 | -BERT | 68.43 | 67.15 | 67.78 ± 0.29 | 94.01 | 93.89 | 93.95 ± 0.06 | 90.27 | 91.86 | 91.05 ± 0.23 | 91.25 | 93.15 | 92.19 ± 0.15 |
| 3 | -fine-tuning | 92.02 | 93.16 | 92.58 ± 0.13 | 95.99 | 96.23 | 96.11 ± 0.17 | 95.01 | 97.15 | 96.07 ± 0.16 | 95.78 | 96.85 | 96.38 ± 0.09 |
| 4 | -AT | 78.83 | 77.39 | 78.11 ± 0.02 | 94.59 | 95.16 | 94.88 ± 0.04 | 90.30 | 92.84 | 91.56 ± 0.14 | 95.12 | 96.60 | 95.86 ± 0.28 |
| 5 | -BERT-AT | 68.48 | 66.95 | 67.70 ± 0.41 | 94.18 | 93.99 | 94.08 ± 0.06 | 89.16 | 91.42 | 90.27 ± 0.13 | 92.09 | 93.56 | 92.82 ± 0.12 |
| Algorithms | CLUENER | AgCNER | CCKS2017 | Resume | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| W | O | F | W | O | F | W | O | F | W | O | F | |
| BiLSTM | 67.70 ±0.41 | 76.77 ±0.35 | 78.11 ±0.18 | 94.08 ±0.06 | 94.19 ±0.07 | 94.88 ±0.02 | 90.27 ±0.13 | 91.62 ±0.16 | 91.56 ±0.13 | 92.82 ±0.12 | 94.88 ±0.11 | 95.86 ±0.17 |
| IDCNN | 66.58 ±0.38 | 76.33 ±0.23 | 77.98 ±0.11 | 93.99 ±0.06 | 93.91 ±0.13 | 94.74 ±0.07 | 91.46 ±0.29 | 91.20 ±0.31 | 92.01 ±0.13 | 92.71 ±0.42 | 94.44 ±0.33 | 96.03 ±0.13 |
| Gated CNN | 66.26 ±0.25 | 75.23 +0.22 | 76.90 ±0.34 | 93.56 ±0.11 | 93.72 ±0.02 | 94.76 ±0.08 | 91.02 ±0.28 | 89.86 ±0.12 | 91.15 ±0.14 | 89.25 ±0.35 | 93.12 ±0.23 | 96.27 ±0.16 |
| RD_CNN | 66.16 ±0.15 | 75.73 ±0.21 | 77.95 ±0.18 | 93.20 ±0.08 | 93.89 ±0.04 | 94.82 ±0.05 | 89.18 ±0.23 | 90.03 ±0.19 | 91.52 ±0.17 | 89.56 ±0.23 | 93.39 ±0.17 | 95.87 ±0.19 |
| AR-CCNER | 68.67 ±0.35 | 77.08 ±0.26 | 78.04 ±0.28 | 94.46 ±0.08 | 94.12 ±0.06 | 94.67 ±0.06 | 91.45 ±0.30 | 91.10 ±0.15 | 91.77 ±0.28 | 93.09 ±0.25 | 95.01 ±0.19 | 96.55 ±0.27 |
| CNN-BiLSTM-CRF | 68.45 ±0.37 | 76.88 ±0.22 | 78.18 ±0.12 | 94.07 ±0.12 | 94.53 ±0.02 | 94.78 ±0.05 | 92.03 ±0.16 | 91.49 ±0.24 | 91.28 ±0.25 | 93.84 ±0.18 | 95.18 ±0.16 | 95.26 ±0.24 |
| Algorithms | CLUENER | AgCNER | CCKS2017 | Resume | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| W | O | F | W | O | F | W | O | F | W | O | F | |
| BiLSTM | 67.78 ±0.29 | 92.58 ±0.13 | 93.68 ±0.18 | 93.95 ±0.06 | 96.11 ±0.17 | 98.31 ±0.02 | 91.05 ±0.23 | 96.07 ±0.16 | 95.72 ±0.13 | 92.19 ±0.15 | 96.38 ±0.09 | 96.83 ±0.17 |
| IDCNN | 66.12 ±0.25 | 94.72 ±0.21 | 94.45 ±0.17 | 93.71 ±0.08 | 96.98 ±0.14 | 98.23 ±0.05 | 91.27 ±0.19 | 96.12 ±0.13 | 95.25 ±0.17 | 93.13 ±0.12 | 96.91 ±0.11 | 96.16 ±0.12 |
| Gated CNN | 66.07 ±0.14 | 95.03 ±0.16 | 96.33 ±0.13 | 93.48 ±0.03 | 97.48 ±0.15 | 98.42 ±0.08 | 90.88 ±0.12 | 96.27 ±0.11 | 96.19 ±0.14 | 91.57 ±0.16 | 96.73 ±0.11 | 97.57 ±0.15 |
| RD_CNN | 65.88 ±0.16 | 94.51 ±0.18 | 96.68 ±0.13 | 92.86 ±0.07 | 97.35 ±0.14 | 98.95 ±0.05 | 90.18 ±0.16 | 95.56 ±0.10 | 95.61 ±0.15 | 91.20 ±0.17 | 96.01 ±0.13 | 97.34 ±0.14 |
| AR-CCNER | 62.30 ±0.36 | 91.64 ±0.24 | 89.66 ±0.25 | 92.80 ±0.11 | 97.50 ±0.12 | 97.70 ±0.06 | 90.96 ±0.20 | 96.08 ±0.16 | 95.97 ±0.12 | 90.36 ±0.15 | 96.74 ±0.12 | 97.26 ±0.14 |
| CNN-BiLSTM-CRF | 67.23 ±0.26 | 90.81 ±0.19 | 89.82 ±0.22 | 93.67 ±0.11 | 96.57 ±0.12 | 97.66 ±0.12 | 91.63 ±0.16 | 95.33 ±0.19 | 95.14 ±0.17 | 93.07 ±0.16 | 96.77 ±0.13 | 96.91 ±0.16 |
| Algorithms | AgCNER | Resume | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dev | Test | Dev | Test | |||||||||
| P | R | F1 | P | R | F1 | P | R | F1 | P | R | F1 | |
| ACE-ADP | 98.30 | 98.32 | 98.31 | 98.50 | 98.47 | 98.49 | 96.43 | 97.79 | 97.11 | 96.63 | 97.66 | 97.14 |
| IDCNN | 98.15 | 98.30 | 98.23 | 98.18 | 98.25 | 98.21 | 95.52 | 96.69 | 96.10 | 94.55 | 96.13 | 95.33 |
| Gated CNN | 98.07 | 98.76 | 98.42 | 98.17 | 98.75 | 98.46 | 96.57 | 98.47 | 97.51 | 96.86 | 99.00 | 97.92 |
| RD_CNN | 98.71 | 99.20 | 98.95 | 98.69 | 99.19 | 98.94 | 96.87 | 98.65 | 97.75 | 97.11 | 98.93 | 98.01 |
| AR-CCNER | 97.38 | 98.03 | 97.70 | 97.80 | 97.88 | 97.84 | 95.91 | 97.79 | 96.84 | 97.10 | 98.33 | 97.71 |
| CNN-BiLSTM-CRF | 97.51 | 97.81 | 97.66 | 97.63 | 97.58 | 97.61 | 95.91 | 96.38 | 96.14 | 95.64 | 96.79 | 96.22 |
| FGN | 94.33 | 94.56 | 94.45 | 94.26 | 94.62 | 94.44 | 93.13 | 95.82 | 94.46 | 92.12 | 94.73 | 93.41 |
| Flat-Lattice | 93.52 | 94.31 | 93.91 | 93.71 | 94.11 | 93.91 | 94.74 | 96.26 | 95.49 | 94.90 | 95.83 | 95.36 |
| TENER | 92.88 | 95.09 | 93.97 | 93.03 | 95.09 | 94.05 | 94.45 | 95.09 | 94.77 | 93.71 | 94.52 | 94.11 |
| Rotation Angles | ||||
|---|---|---|---|---|
| Word2vec |  |  |  |  |
| Original BERT |  |  |  |  |
| Fine-tuned BERT |  |  |  |  |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, X.; Hao, X.; Tang, Z.; Diao, L.; Bai, Z.; Lu, S.; Li, L. ACE-ADP: Adversarial Contextual Embeddings Based Named Entity Recognition for Agricultural Diseases and Pests. Agriculture 2021, 11, 912. https://doi.org/10.3390/agriculture11100912
Guo X, Hao X, Tang Z, Diao L, Bai Z, Lu S, Li L. ACE-ADP: Adversarial Contextual Embeddings Based Named Entity Recognition for Agricultural Diseases and Pests. Agriculture. 2021; 11(10):912. https://doi.org/10.3390/agriculture11100912
Chicago/Turabian StyleGuo, Xuchao, Xia Hao, Zhan Tang, Lei Diao, Zhao Bai, Shuhan Lu, and Lin Li. 2021. "ACE-ADP: Adversarial Contextual Embeddings Based Named Entity Recognition for Agricultural Diseases and Pests" Agriculture 11, no. 10: 912. https://doi.org/10.3390/agriculture11100912
APA StyleGuo, X., Hao, X., Tang, Z., Diao, L., Bai, Z., Lu, S., & Li, L. (2021). ACE-ADP: Adversarial Contextual Embeddings Based Named Entity Recognition for Agricultural Diseases and Pests. Agriculture, 11(10), 912. https://doi.org/10.3390/agriculture11100912






