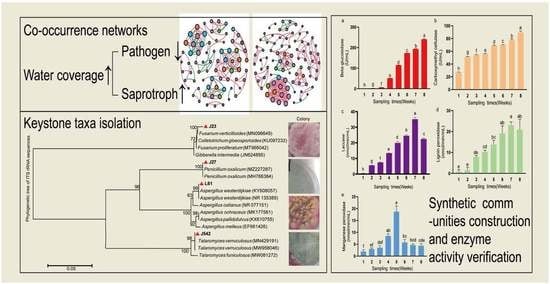
Water-Covered Depth with the Freeze–Thaw Cycle Influences Fungal Communities on Rice Straw Decomposition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Site
2.2. Soil DNA Extraction, Gene Amplification, and Sequencing
2.3. Analysis of Soil and Straw Properties
2.4. Illumina HiSeq Sequence Processing
2.5. Statistical Analysis
2.6. Network Analyses and Keystone Taxa
2.7. Constructing Synthetic Fungal Communities
2.8. Rice Straw Degrading Enzyme Activity
3. Result
3.1. Degradation Degree of Straw
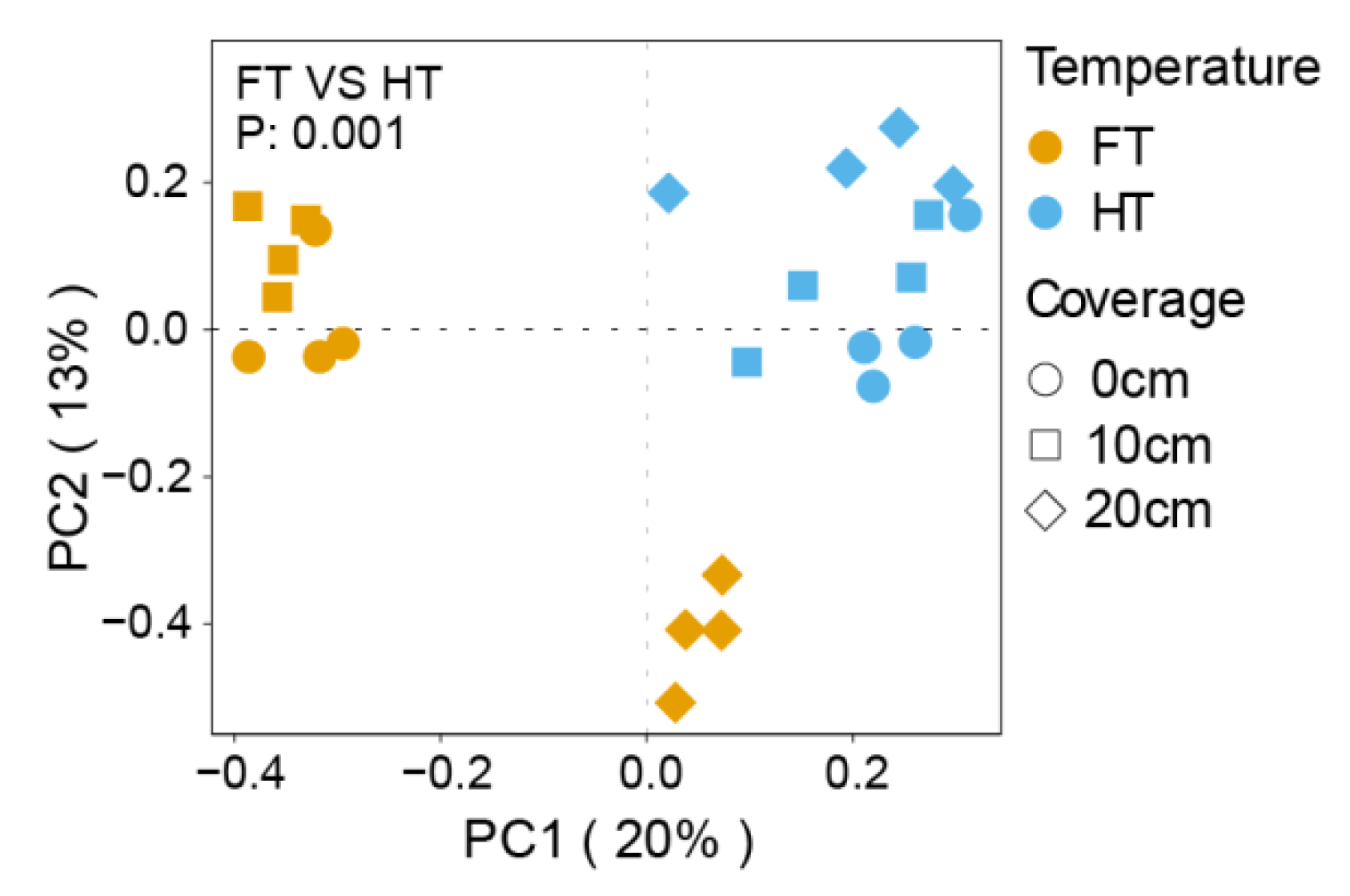
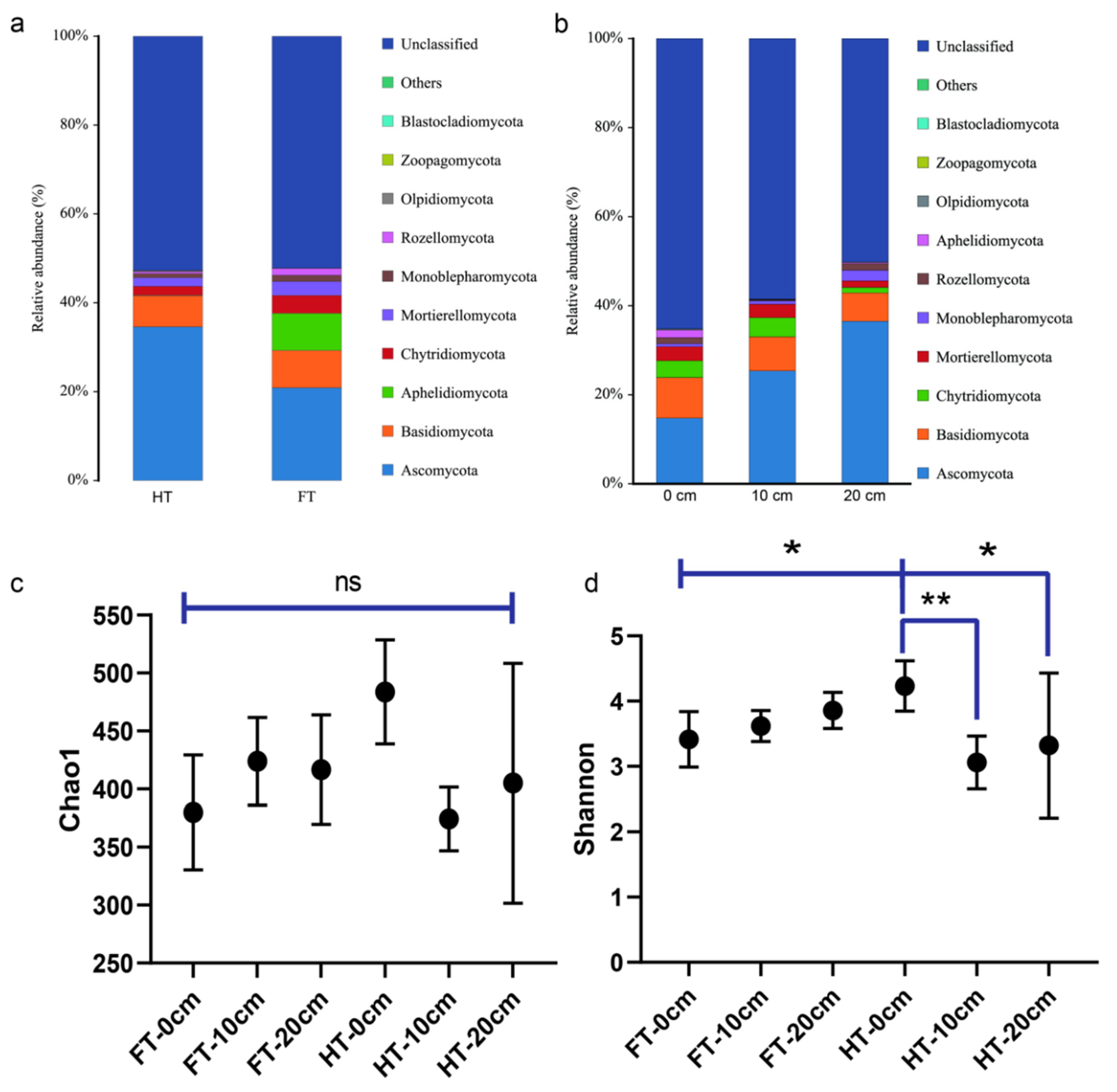
3.2. Study on the Beta Diversity of Microorganisms in the Plants’ Rhizospheres
3.3. RDA Analysis of Fungal Communities and Soil Properties
3.4. Changes in Enzyme Activities of Synthetic Fungal Community
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dashtban, M.; Schraft, H.; Qin, W. Fungal bioconversion of lignocellulosic residues; opportunities & perspectives. Int. J. Biol. Sci. 2009, 5, 578. [Google Scholar] [PubMed]
- Marschner, P.; Kandeler, E.; Marschner, B. Structure and function of the soil microbial community in a long-term fertilizer experiment. Soil Biol. Biochem. 2003, 35, 453–461. [Google Scholar] [CrossRef]
- Arthurson, V. Closing the global energy and nutrient cycles through application of biogas residue to agricultural land–potential benefits and drawback. Energies 2009, 2, 226–242. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Qiu, L.; Yao, Y.; Qin, M. A technology for strongly improving methane production from rice straw: Freeze–thaw pretreatment. RSC Adv. 2018, 8, 22643–22651. [Google Scholar] [CrossRef] [Green Version]
- Zhan, Y.; Liu, W.; Bao, Y.; Zhang, J.; Petropoulos, E.; Li, Z.; Lin, X.; Feng, Y. Fertilization shapes a well-organized community of bacterial decomposers for accelerated paddy straw degradation. Sci. Rep. 2018, 8, 7981. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Li, K.; Zhou, W.; Qiu, S.; Huang, S.; He, P. Changes in soil microbial community, enzyme activities and organic matter fractions under long-term straw return in north-central China. Agric. Ecosyst. Environ. 2016, 216, 82–88. [Google Scholar] [CrossRef]
- McGuire, K.L.; Treseder, K.K. Microbial communities and their relevance for ecosystem models: Decomposition as a case study. Soil Biol. Biochem. 2010, 42, 529–535. [Google Scholar] [CrossRef] [Green Version]
- Wegner, C.E.; Liesack, W. Microbial community dynamics during the early stages of plant polymer breakdown in paddy soil. Environ. Microbiol. 2016, 18, 2825–2842. [Google Scholar] [CrossRef]
- Fuchslueger, L.; Bahn, M.; Hasibeder, R.; Kienzl, S.; Fritz, K.; Schmitt, M.; Watzka, M.; Richter, A. Drought history affects grassland plant and microbial carbon turnover during and after a subsequent drought event. J. Ecol. 2016, 104, 1453–1465. [Google Scholar] [CrossRef]
- Hawkes, C.V.; Waring, B.G.; Rocca, J.D.; Kivlin, S.N. Historical climate controls soil respiration responses to current soil moisture. Proc. Natl. Acad. Sci. USA 2017, 114, 6322–6327. [Google Scholar] [CrossRef] [Green Version]
- Fanin, N.; Bertrand, I. Aboveground litter quality is a better predictor than belowground microbial communities when estimating carbon mineralization along a land-use gradient. Soil Biol. Biochem. 2016, 94, 48–60. [Google Scholar] [CrossRef]
- Saad, M.; Oliveira, L.; Cândido, R.; Quintana, G.; Rocha, G.; Gonçalves, A. Preliminary studies on fungal treatment of sugarcane straw for organosolv pulping. Enzym. Microb. Technol. 2008, 43, 220–225. [Google Scholar] [CrossRef]
- Kogo, T.; Yoshida, Y.; Koganei, K.; Matsumoto, H.; Watanabe, T.; Ogihara, J.; Kasumi, T. Production of rice straw hydrolysis enzymes by the fungi Trichoderma reesei and Humicola insolens using rice straw as a carbon source. Bioresour. Technol. 2017, 233, 67–73. [Google Scholar] [CrossRef]
- Lauber, C.L.; Strickland, M.S.; Bradford, M.A.; Fierer, N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 2008, 40, 2407–2415. [Google Scholar] [CrossRef]
- Li, D.; Li, Z.; Zhao, B.; Zhang, J. Relationship between the chemical structure of straw and composition of main microbial groups during the decomposition of wheat and maize straws as affected by soil texture. Biol. Fertil. Soils 2020, 56, 11–24. [Google Scholar] [CrossRef]
- Banerjee, S.; Walder, F.; Büchi, L.; Meyer, M.; Held, A.Y.; Gattinger, A.; Keller, T.; Charles, R.; van der Heijden, M.G. Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 2019, 13, 1722–1736. [Google Scholar] [CrossRef] [Green Version]
- Wagg, C.; Schlaeppi, K.; Banerjee, S.; Kuramae, E.E.; van der Heijden, M.G. Fungal-bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 2019, 10, 4841. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Germain, R.M.; Srivastava, D.S.; Filotas, E.; Dee, L.E.; Gravel, D.; Thompson, P.L.; Isbell, F.; Wang, S.; Kéfi, S. Scaling-up biodiversity-ecosystem functioning research. Ecol. Lett. 2020, 23, 757–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbons, S.M. Keystone taxa indispensable for microbiome recovery. Nat. Microbiol. 2020, 5, 1067–1068. [Google Scholar] [CrossRef]
- Niu, B.; Paulson, J.N.; Zheng, X.; Kolter, R. Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. USA 2017, 114, E2450–E2459. [Google Scholar] [CrossRef] [Green Version]
- Ulrich, A.; Klimke, G.; Wirth, S. Diversity and activity of cellulose-decomposing bacteria, isolated from a sandy and a loamy soil after long-term manure application. Microb. Ecol. 2008, 55, 512–522. [Google Scholar] [CrossRef]
- Feng, Y.; Yu, Y.; Wang, X.; Qu, Y.; Li, D.; He, W.; Kim, B.H. Degradation of raw corn stover powder (RCSP) by an enriched microbial consortium and its community structure. Bioresour. Technol. 2011, 102, 742–747. [Google Scholar] [CrossRef]
- Schimel, J.; Balser, T.C.; Wallenstein, M. Microbial stress-response physiology and its implications for ecosystem function. Ecology 2007, 88, 1386–1394. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.; Zhang, J.; Liu, T.; Song, K.; Xie, J.; Luo, S.; Qu, T.; Zhang, J.; Tian, C.; Zhang, J. Rhizosphere fungal communities of wild and cultivated soybeans grown in three different soil suspensions. Appl. Soil Ecol. 2020, 153, 103586. [Google Scholar] [CrossRef]
- Tian, L.; Zhou, X.; Ma, L.; Xu, S.; Nasir, F.; Tian, C. Root-associated bacterial diversities of Oryza rufipogon and Oryza sativa and their influencing environmental factors. Arch. Microbiol. 2017, 199, 563. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [Green Version]
- Ostertagova, E.; Ostertag, O.; Kováč, J. Methodology and application of the Kruskal-Wallis test. Proc. Appl. Mech. Mater. 2014, 611, 115–120. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. Fast R functions for robust correlations and hierarchical clustering. J. Stat. Softw. 2012, 46, 11. [Google Scholar] [CrossRef] [Green Version]
- Grandjean, M. Gephi: Introduction to Network Analysis and Visualisation. 2015. Available online: https://https://gephi.org/ (accessed on 21 June 2021).
- Banerjee, S.; Schlaeppi, K.; van der Heijden, M.G. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 2018, 16, 567–576. [Google Scholar] [CrossRef]
- Parmar, H.; Hassan, M.M.; Bodar, N.; Umrania, V.; Patel, S.; Lakhani, H. In vitro antagonism between phytopathogenic fungi Sclerotium rolfsii and Trichoderma strains. Int. J. Appl. Sci. Biotechnol. 2015, 3, 16–19. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Zhang, H.; Wang, Z.; Yang, D. Screening of decolorization fungus to Rose-Bengal and optimization of decolorization conditions. Environ. Sci. Technol. 2017, 40, 103–108. [Google Scholar]
- Lu, B.; Zang, S.; Sun, L. The effects of freezingthawing process on soil active organic carbon and nitrogen mineralization in Daxing’anling Mountain forests. Acta Sci. Circumstantiae 2019, 39, 1664–1672. [Google Scholar]
- Van Kuijk, S.J.; Sonnenberg, A.S.; Baars, J.J.; Hendriks, W.H.; José, C.; Rencoret, J.; Gutiérrez, A.; de Ruijter, N.C.; Cone, J.W. Chemical changes and increased degradability of wheat straw and oak wood chips treated with the white rot fungi Ceriporiopsis subvermispora and Lentinula edodes. Biomass Bioenergy 2017, 105, 381–391. [Google Scholar] [CrossRef]
- Chen, C.-H.; Yao, J.-Y.; Yang, B.; Lee, H.-L.; Yuan, S.-F.; Hsieh, H.-Y.; Liang, P.-H. Engineer multi-functional cellulase/xylanase/β-glucosidase with improved efficacy to degrade rice straw. Bioresour. Technol. Rep. 2019, 5, 170–177. [Google Scholar] [CrossRef]
- Lin, Z.; Guodong, C.; Yongjian, D. Studies on frozen ground of China. J. Geogr. Sci. 2004, 14, 411–416. [Google Scholar] [CrossRef]
- Koponen, H.T.; Bååth, E. Soil bacterial growth after a freezing/thawing event. Soil Biol. Biochem. 2016, 100, 229–232. [Google Scholar] [CrossRef]
- Barnard, R.L.; Osborne, C.A.; Firestone, M.K. Changing precipitation pattern alters soil microbial community response to wet-up under a Mediterranean-type climate. ISME J. 2015, 9, 946–957. [Google Scholar] [CrossRef] [Green Version]
- Yergeau, E.; Kowalchuk, G.A. Responses of Antarctic soil microbial communities and associated functions to temperature and freeze–thaw cycle frequency. Environ. Microbiol. 2008, 10, 2223–2235. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.L.; Grogan, P.; Walker, V.K. Prospecting for ice association: Characterization of freeze–thaw selected enrichment cultures from latitudinally distant soils. Can. J. Microbiol. 2012, 58, 402–412. [Google Scholar] [CrossRef] [Green Version]
- Looby, C.I.; Treseder, K.K. Shifts in soil fungi and extracellular enzyme activity with simulated climate change in a tropical montane cloud forest. Soil Biol. Biochem. 2018, 117, 87–96. [Google Scholar] [CrossRef]
- Jin, X.; Harman, G.; Taylor, A. Conidial biomass and desiccation tolerance of Trichoderma harzianum produced at different medium water potentials. Biol Control. 1991, 1, 237–243. [Google Scholar] [CrossRef]
- Drotz, S.H.; Sparrman, T.; Nilsson, M.B.; Schleucher, J.; Öquist, M.G. Both catabolic and anabolic heterotrophic microbial activity proceed in frozen soils. Proc. Natl. Acad. Sci. USA 2010, 107, 21046–21051. [Google Scholar] [CrossRef] [Green Version]
- Lennon, J.T.; Jones, S.E. Microbial seed banks: The ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 2011, 9, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Larsen, K.S.; Jonasson, S.; Michelsen, A. Repeated freeze–thaw cycles and their effects on biological processes in two arctic ecosystem types. Appl. Soil Ecol. 2002, 21, 187–195. [Google Scholar] [CrossRef]
- Williams, M.A.; Xia, K. Characterization of the water soluble soil organic pool following the rewetting of dry soil in a drought-prone tallgrass prairie. Soil Biol. Biochem. 2009, 41, 21–28. [Google Scholar] [CrossRef]
- Fukami, T. Historical contingency in community assembly: Integrating niches, species pools, and priority effects. Annu. Rev. Ecol. Evol. Syst. 2015, 46, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Meisner, A.; Jacquiod, S.; Snoek, B.L.; Ten Hooven, F.C.; Van Der Putten, W.H. Drought legacy effects on the composition of soil fungal and prokaryote communities. Front. Microbiol. 2018, 9, 294. [Google Scholar] [CrossRef] [Green Version]
- Gao, C.; Montoya, L.; Xu, L.; Madera, M.; Hollingsworth, J.; Purdom, E.; Singan, V.; Vogel, J.; Hutmacher, R.B.; Dahlberg, J.A. Fungal community assembly in drought-stressed sorghum shows stochasticity, selection, and universal ecological dynamics. Nat. Commun. 2020, 11, 34. [Google Scholar] [CrossRef] [Green Version]
- De Vries, F.T.; Griffiths, R.I.; Bailey, M.; Craig, H.; Girlanda, M.; Gweon, H.S.; Hallin, S.; Kaisermann, A.; Keith, A.M.; Kretzschmar, M. Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 2018, 9, 3033. [Google Scholar] [CrossRef] [Green Version]
- Guhr, A.; Borken, W.; Spohn, M.; Matzner, E. Redistribution of soil water by a saprotrophic fungus enhances carbon mineralization. Proc. Natl. Acad. Sci. USA 2015, 112, 14647–14651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, X.; Nielsen, L.L.; Simpson, M.J. Responses of soil organic matter and microorganisms to freeze–thaw cycles. Soil Biol. Biochem. 2007, 39, 2027–2037. [Google Scholar] [CrossRef]
- Bhattarai, S.P.; Midmore, D.J.; Su, N. Sustainable Irrigation to Balance Supply of Soil Water, Oxygen, Nutrients and Agro-Chemicals. In Biodiversity, Biofuels, Agroforestry and Conservation Agriculture; Springer: Berlin, Germany, 2010; pp. 253–286. [Google Scholar]
- Stark, J.M.; Firestone, M.K. Mechanisms for soil moisture effects on activity of nitrifying bacteria. Appl. Environ. Microbiol. 1995, 61, 218–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devêvre, O.C.; Horwáth, W.R. Decomposition of rice straw and microbial carbon use efficiency under different soil temperatures and moistures. Soil Biol. Biochem. 2000, 32, 1773–1785. [Google Scholar] [CrossRef]
- Shahbaz, M.; Kumar, A.; Kuzyakov, Y.; Börjesson, G.; Blagodatskaya, E. Priming effects induced by glucose and decaying plant residues on SOM decomposition: A three-source 13C/14C partitioning study. Soil Biol. Biochem. 2018, 121, 138–146. [Google Scholar] [CrossRef]
- Otero-Jiménez, V.; del Pilar Carreño-Carreño, J.; Barreto-Hernandez, E.; van Elsas, J.D.; Uribe-Vélez, D. Impact of rice straw management strategies on rice rhizosphere microbiomes. Appl. Soil Ecol. 2021, 167, 104036. [Google Scholar] [CrossRef]
- Chang, J.; Sun, Y.; Tian, L.; Ji, L.; Luo, S.; Nasir, F.; Kuramae, E.E.; Tian, C. The structure of rhizosphere fungal communities of wild and domesticated rice: Changes in diversity and co-occurrence patterns. Front. Microbiol. 2021, 12, 45. [Google Scholar] [CrossRef]
- Sharma, B.; Agrawal, R.; Singhania, R.R.; Satlewal, A.; Mathur, A.; Tuli, D.; Adsul, M. Untreated wheat straw: Potential source for diverse cellulolytic enzyme secretion by Penicillium janthinellum EMS-UV-8 mutant. Bioresour. Technol. 2015, 196, 518–524. [Google Scholar] [CrossRef]
- Singhania, R.R.; Saini, J.K.; Saini, R.; Adsul, M.; Mathur, A.; Gupta, R.; Tuli, D.K. Bioethanol production from wheat straw via enzymatic route employing Penicillium janthinellum cellulases. Bioresour. Technol. 2014, 169, 490–495. [Google Scholar] [CrossRef]
- Fujii, T.; Inoue, H.; Yano, S.; Sawayama, S. Strain Improvement for Industrial Production of Lignocellulolytic Enzyme by Talaromyces cellulolyticus. In Fungal Cellulolytic Enzymes: Microbial Production and Application; Fang, X., Qu, Y., Eds.; Springer Singapore: Singapore, 2018; pp. 135–154. [Google Scholar]
- Gu, S.; Liu, C.; Zhang, W.; Qu, M.; Li, Y.; Zang, Y.; Xiong, X.; Pan, K.; Zhao, X. Characteristics of a recombinant Fusarium verticillioides cutinase and its effects on enzymatic hydrolysis of rice straw. Int. J. Biol. Macromol. 2021, 171, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-J.; Jang, Y.-S.; Lee, Y.-M.; Lee, J.-J.; Lee, H.-B.; Kim, G.-H.; Kim, J.-J. Rice straw-decomposing fungi and their cellulolytic and xylanolytic enzymes. J. Microbiol. Biotechnol. 2011, 21, 1322–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, X.; Song, J.; Liu, G.-Q. Bioethanol production from rice straw through an enzymatic route mediated by enzymes developed in-house from Aspergillus fumigatus. Energy 2020, 190, 116395. [Google Scholar] [CrossRef]
- Jahromi, M.; Liang, J.; Rosfarizan, M.; Goh, Y.; Shokryazdan, P.; Ho, Y. Efficiency of rice straw lignocelluloses degradability by Aspergillus terreus ATCC 74135 in solid state fermentation. Afr. J. Biotechnol. 2011, 10, 4428–4435. [Google Scholar]
- Tang, Y.; Zhang, M.; Chen, A.; Zhang, W.; Wei, W.; Sheng, R. Impact of fertilization regimes on diazotroph community compositions and N2-fixation activity in paddy soil. Agric. Ecosyst. Environ. 2017, 247, 1–8. [Google Scholar] [CrossRef]
- Montoya, J.M.; Pimm, S.L.; Solé, R.V. Ecological networks and their fragility. Nature 2006, 442, 259–264. [Google Scholar] [CrossRef]
- Kitano, H. Biological robustness. Nat. Rev. Genet. 2004, 5, 826–837. [Google Scholar] [CrossRef]
- Kaisermann, A.; Maron, P.; Beaumelle, L.; Lata, J. Fungal communities are more sensitive indicators to non-extreme soil moisture variations than bacterial communities. Appl. Soil Ecol. 2015, 86, 158–164. [Google Scholar] [CrossRef]
- Freilich, M.A.; Wieters, E.; Broitman, B.R.; Marquet, P.A.; Navarrete, S.A. Species co-occurrence networks: Can they reveal trophic and non-trophic interactions in ecological communities? Ecology 2018, 99, 690–699. [Google Scholar] [CrossRef]
- Mamet, S.D.; Redlick, E.; Brabant, M.; Lamb, E.G.; Helgason, B.L.; Stanley, K.; Siciliano, S.D. Structural equation modeling of a winnowed soil microbiome identifies how invasive plants re-structure microbial networks. ISME J. 2019, 13, 1988–1996. [Google Scholar] [CrossRef]
- Sharma, S.; Szele, Z.; Schilling, R.; Munch, J.C.; Schloter, M. Influence of freeze-thaw stress on the structure and function of microbial communities and denitrifying populations in soil. Appl. Environ. Microbiol. 2006, 72, 2148–2154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meisner, A.; Snoek, B.L.; Nesme, J.; Dent, E.; Jacquiod, S.; Classen, A.T.; Priemé, A. Soil microbial legacies differ following drying-rewetting and freezing-thawing cycles. ISME J. 2021, 15, 1207–1221. [Google Scholar] [CrossRef] [PubMed]
- Tilston, E.; Sparrman, T.; Öquist, M. Unfrozen water content moderates temperature dependence of sub-zero microbial respiration. Soil Biol. Biochem. 2010, 42, 1396–1407. [Google Scholar] [CrossRef]

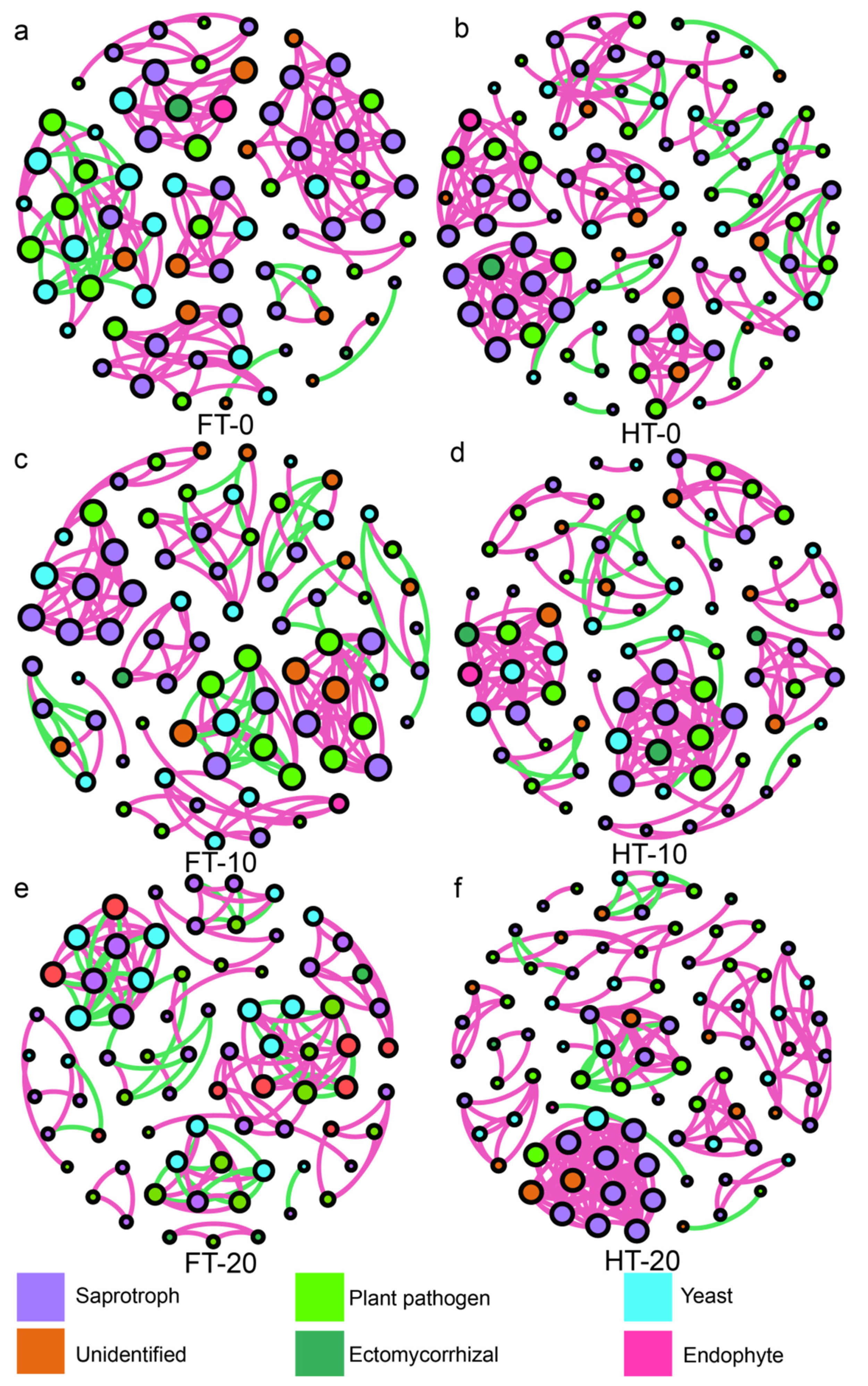
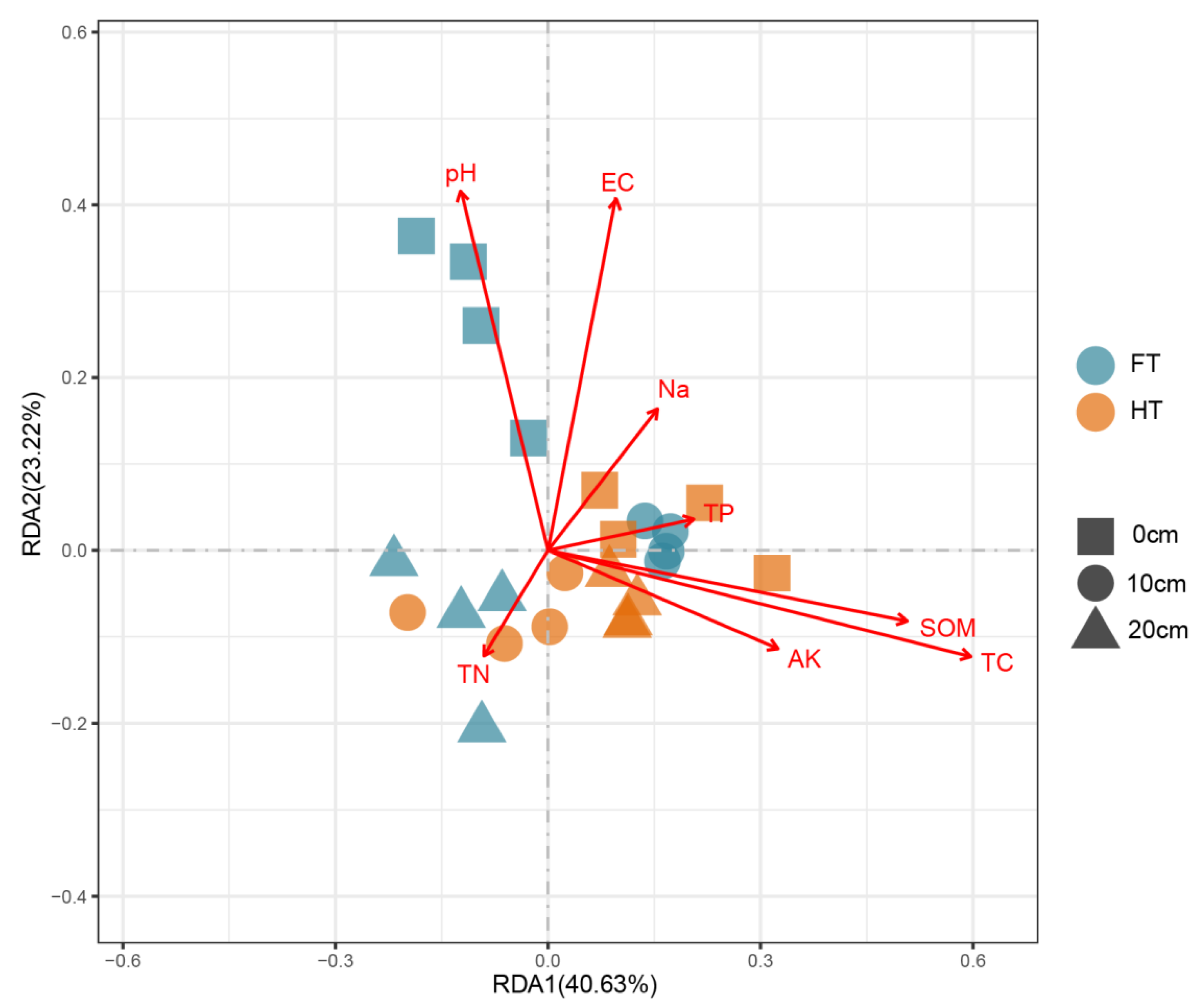
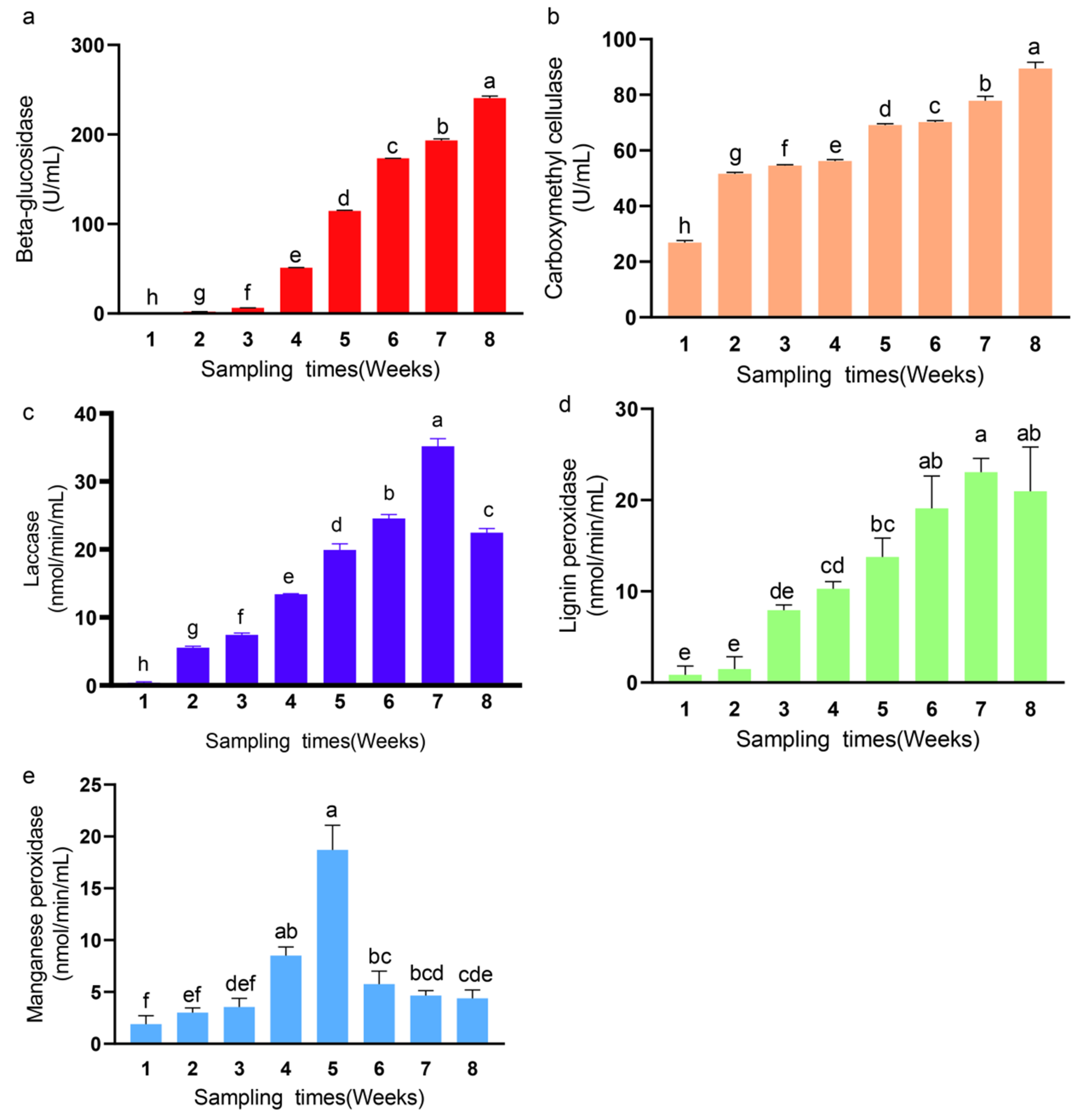
| Treatments | Water Coverage | Nodes/Edges | Average Degree | Average Weighted Degree | Modularity | Keystone Taxa |
|---|---|---|---|---|---|---|
| FT | 0 cm | 74/151 | 4.081 | 2.892 | 1.071 | OTU_433(Penicillium oxalicum J27); OTU_127(Fusarium sp.J23) |
| 10 cm | 76/165 | 4.342 | 2.237 | 1.2 | ||
| 20 cm | 74/154 | 4.162 | 1.514 | 1.619 | ||
| HT | 0 cm | 95/189 | 3.979 | 2.8 | 1.002 | OTU_318(Aspergillus sp.L61); OTU_409(Talaromyces sp.J542) |
| 10 cm | 78/161 | 4.128 | 3.256 | 0.894 | ||
| 20 cm | 86/198 | 4.605 | 3.814 | 0.842 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, X.; Yao, Z.; Wang, X.; Xu, S.; Tian, C.; Tian, L. Water-Covered Depth with the Freeze–Thaw Cycle Influences Fungal Communities on Rice Straw Decomposition. Agriculture 2021, 11, 1113. https://doi.org/10.3390/agriculture11111113
Lin X, Yao Z, Wang X, Xu S, Tian C, Tian L. Water-Covered Depth with the Freeze–Thaw Cycle Influences Fungal Communities on Rice Straw Decomposition. Agriculture. 2021; 11(11):1113. https://doi.org/10.3390/agriculture11111113
Chicago/Turabian StyleLin, Xiaolong, Zongmu Yao, Xinguang Wang, Shangqi Xu, Chunjie Tian, and Lei Tian. 2021. "Water-Covered Depth with the Freeze–Thaw Cycle Influences Fungal Communities on Rice Straw Decomposition" Agriculture 11, no. 11: 1113. https://doi.org/10.3390/agriculture11111113
APA StyleLin, X., Yao, Z., Wang, X., Xu, S., Tian, C., & Tian, L. (2021). Water-Covered Depth with the Freeze–Thaw Cycle Influences Fungal Communities on Rice Straw Decomposition. Agriculture, 11(11), 1113. https://doi.org/10.3390/agriculture11111113