Use of High Resolution Spatiotemporal Gene Expression Data to Uncover Novel Tissue-Specific Promoters in Tomato
Abstract
1. Introduction
2. Materials and Methods
2.1. Promotor Sequence of Tissue-Specific Unigenes
2.2. Promoter Sequence Analysis and Examination of cis-Regulatory Elements
2.3. Plant Materials
2.4. RNA Extraction and Identification of Tissue-Specific Genes
3. Results
3.1. Identification of the Five Most-Abundantly Expressed Tissue-Specific Unigenes from Each of the Five Tomato Pericarp Tissues
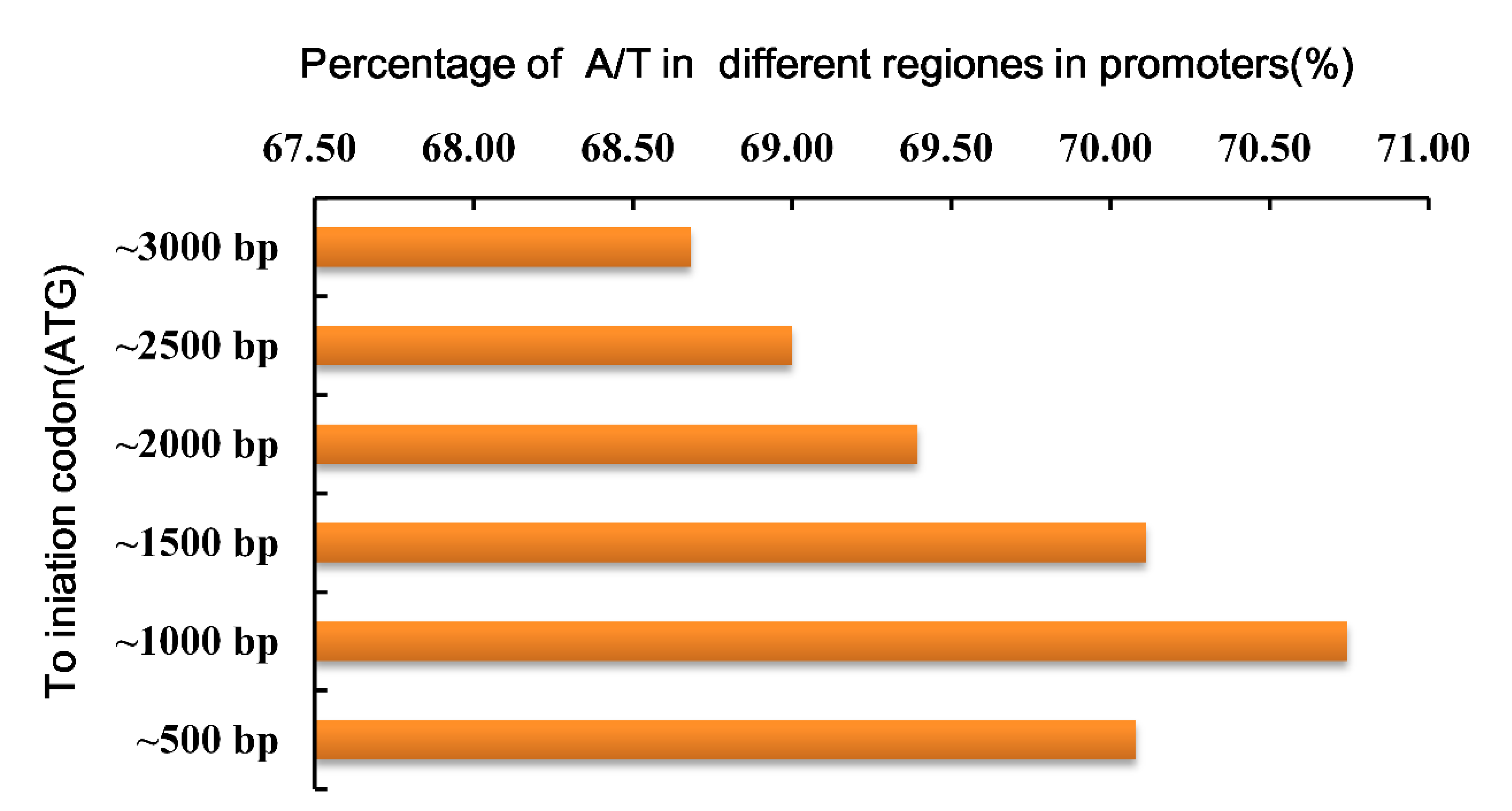
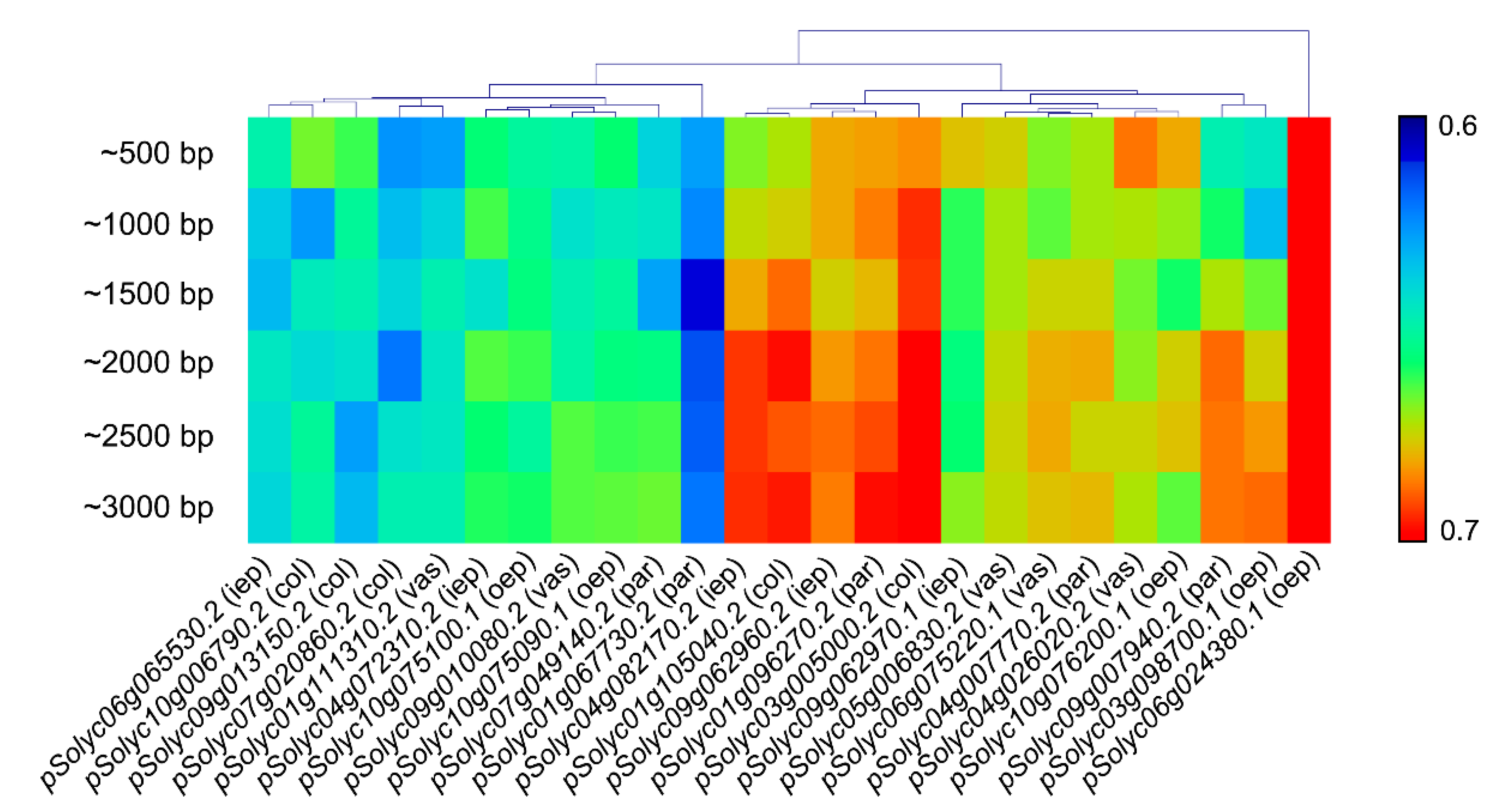
3.2. Promoter A/T Content and Cluster Analysis
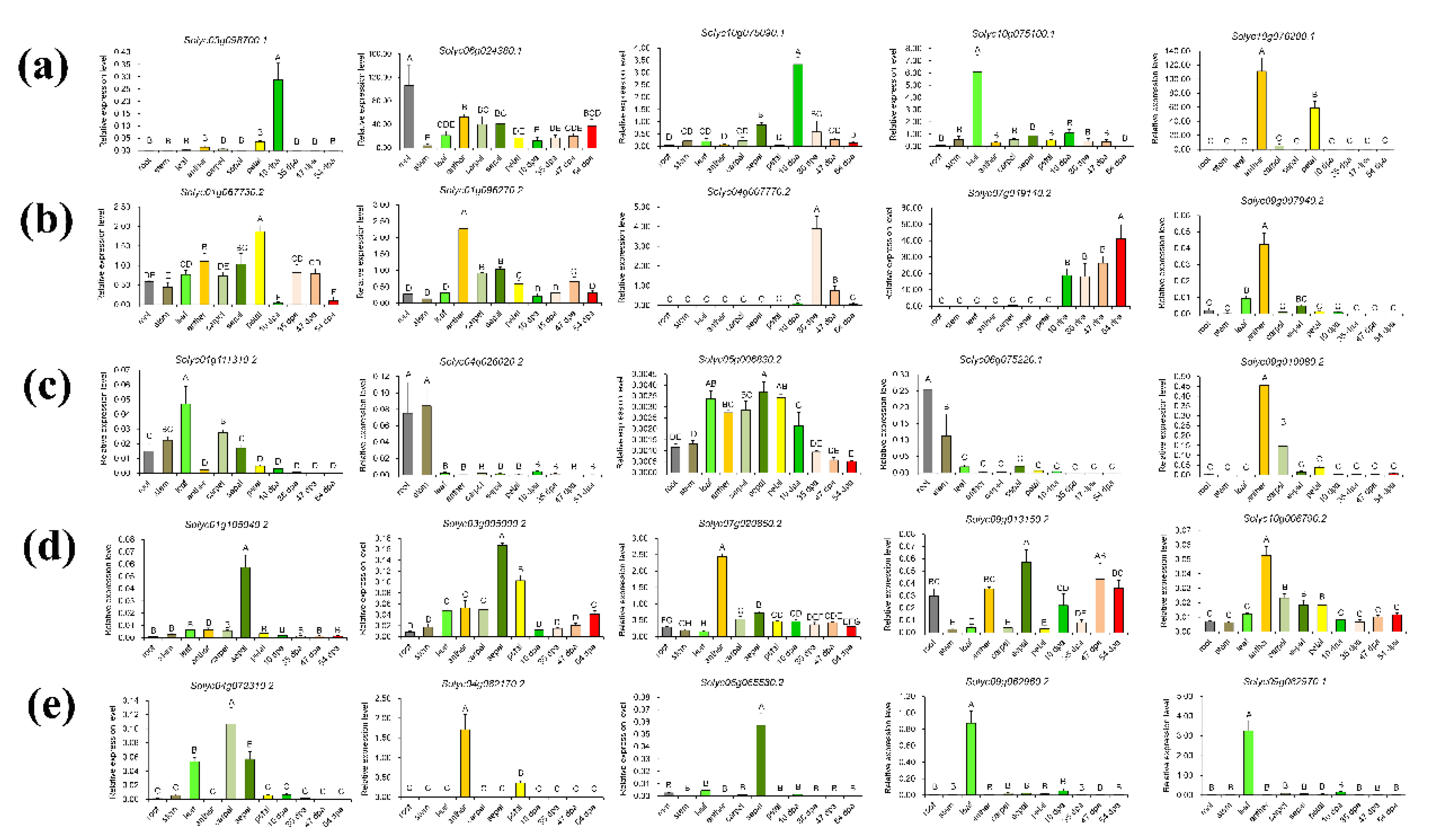
3.3. Expression Profiles of Pericarp Tissue-Specific Unigenes in Other Organs
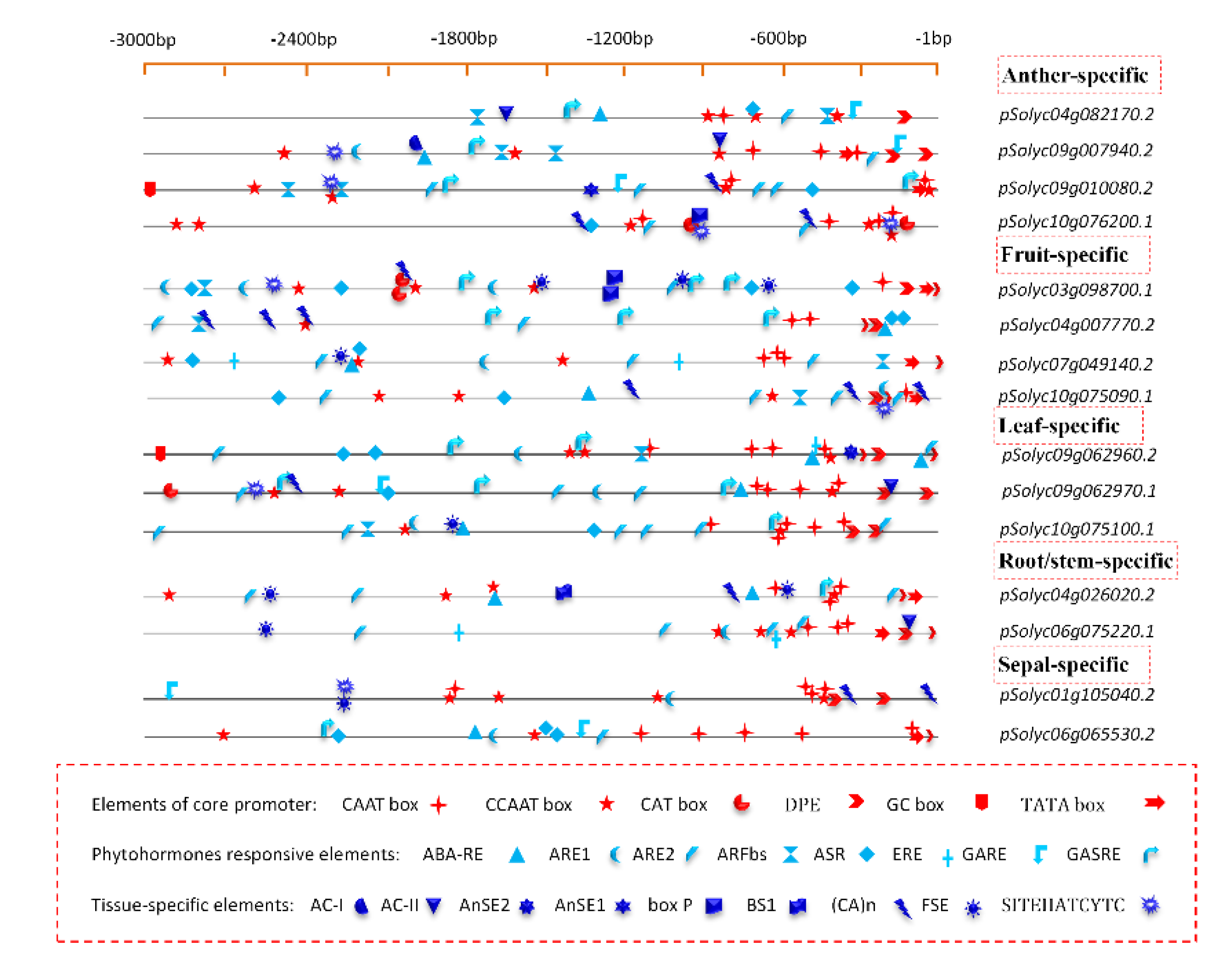
3.4. Distribution of cis-Regulatory Elements in Tissue-Specific Promoters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Chen, L.; Li, W.; Li, Y.; Feng, X.; Du, K.; Wang, G.; Zhao, L. Identified trans-splicing of YELLOW-FRUITED TOMATO 2 encoding the PHYTOENE SYNTHASE 1 protein alters fruit color by map-based cloning, functional complementation and RACE. Plant Mol. Biol. 2019, 100, 647–658. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, A.; Zhao, L.; Shen, G.; Cui, L.; Tang, K. Expression of thymosin α1 concatemer in transgenic tomato (Solanum lycopersicum) fruits. Appl. Biotechnol. Biochem. 2009, 52, 303–312. [Google Scholar] [CrossRef]
- Ma, J.K.; Drake, P.M.; Christou, P. Genetic modification: The production of recombinant pharmaceutical proteins in plants. Nat. Rev. Genet. 2003, 4, 794–805. [Google Scholar] [CrossRef]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.P.; Matros, A.; Peterek, S.; Schijlen, E.G.W.M.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef]
- Deaner, M.; Alper, H.S. Promoter and terminator discovery and engineering. In Synthetic Biology–Metabolic Engineering; Zhao, H., Zeng, A.P., Eds.; Springer: Cham, Switzerland, 2016; pp. 21–44. [Google Scholar]
- Hieno, A.; Naznin, H.A.; Hyakumachi, M.; Sakurai, T.; Tokizawa, M.; Koyama, H.; Sato, N.; Nishiyama, T.; Hasebe, M.; Zimmer, A.D.; et al. ppdb: Plant promoter database version 3.0. Nucleic Acids Res. 2014, 42, D1188–D1192. [Google Scholar] [CrossRef] [PubMed]
- Dreos, R.; Ambrosini, G.; Groux, R.; Périer, R.C.; Bucher, P. The eukaryotic promoter database in its 30th year: Focus on non-vertebrate organisms. Nucleic Acids Res. 2016, 45, D51–D55. [Google Scholar] [CrossRef]
- Hiatt, A.; Caffferkey, R.; Bowdish, K. Production of antibodies in transgenic plants. Nature 1989, 342, 76–78. [Google Scholar] [CrossRef]
- Paine, J.A.; Shipton, C.A.; Chaggar, S.; Howells, R.M.; Kennedy, M.J.; Vernon, G.; Wright, S.Y.; Hinchliffe, E.; Adams, J.L.; Silverstone, A.L.; et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat. Biotechnol. 2005, 23, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Xu, X.; Gong, Q.; Li, Z.; Li, Y.; Wang, S.; Yang, Y.; Ma, W.; Liu, L.; Zhu, B.; et al. Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable tale-binding elements of multiple susceptibility genes in Rice. Mol. Plant 2019, 12, 1434–1446. [Google Scholar] [CrossRef]
- Liu, X.; Li, S.; Yang, W.; Mu, B.; Jiao, Y.; Zhou, X.; Zhang, C.; Fan, Y.; Chen, R. Synthesis of seed-specific bidirectional promoters for metabolic engineering of anthocyanin-rich maize. Plant Cell Physiol. 2018, 59, 1942–1955. [Google Scholar] [CrossRef]
- Takaiwa, F.; Ogo, Y.; Wakasa, Y. Specific region affects the difference in accumulation levels between apple food allergen Mal d 1 and birch pollen allergen Bet v 1 which are expressed in vegetative tissues of transgenic rice. Plant Mol. Biol. 2018, 98, 439–454. [Google Scholar] [CrossRef]
- Conway, T.; Sewell, G.W.; Osman, Y.A.; Ingram, L.O. Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J. Bacteriol. 1987, 169, 2591–2597. [Google Scholar] [CrossRef]
- Ow, D.W.; Jacobs, J.D.; Howell, S.H. Functional regions of the cauliflower mosaic virus 35S RNA promoter determined by use of the firefly luciferase gene as a reporter of promoter activity. Proc. Natl. Acad. Sci. USA 1987, 84, 4870–4874. [Google Scholar] [CrossRef]
- Muschietti, J.; Dircks, L.; Vancanneyt, G.; McCormick, S. LAT52 protein is essential for tomato pollen development: Pollen expressing antisense LAT52 RNA hydrates and germinates abnormally and cannot achieve fertilization. Plant J. 1994, 6, 321–338. [Google Scholar] [CrossRef]
- Hirai, T.; Kim, Y.-W.; Kato, K.; Hiwasa-Tanase, K.; Ezura, H. Uniform accumulation of recombinant miraculin protein in transgenic tomato fruit using a fruit-ripening-specific E8 promoter. Transgenic Res. 2011, 20, 1285–1292. [Google Scholar] [CrossRef]
- Taylor, W.E.; Straus, D.B.; Grossman, A.D.; Burton, Z.F.; Gross, C.A.; Burgess, R.R. Transcription from a heat-inducible promoter causes heat shock regulation of the sigma subunit of E. coli RNA polymerase. Cell 1984, 38, 371–381. [Google Scholar] [CrossRef]
- Cordingley, M.G.; Riegel, A.T.; Hager, G.L. Steroid-dependent interaction of transcription factors with the inducible promoter of mouse mammary tumor virus in vivo. Cell 1987, 48, 261–270. [Google Scholar] [CrossRef]
- Johns, A.M.B.; Love, J.; Aves, S.J. Four inducible promoters for controlled gene expression in the oleaginous yeast Rhodotorula toruloides. Front. Microbiol. 2016, 7, 1666. [Google Scholar] [CrossRef] [PubMed]
- Odell, J.T.; Nagy, F.; Chua, N.H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 1985, 313, 810–812. [Google Scholar] [CrossRef]
- Benfey, P.N.; Chua, N.H. The cauliflower mosaic virus 35S promoter: Combinatorial regulation of transcription in plants. Science 1990, 250, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, H.; Takenami, S.; Kubo, Y.; Ueda, K.; Ueda, A.; Yamaguchi, M.; Hirata, K.; Demura, T.; Kanaya, S.; Kato, K. A computational and experimental approach reveals that the 5′-proximal region of the 5′-UTR has a cis-regulatory signature responsible for heat stress-regulated mRNA translation in Arabidopsis. Plant Cell Physiol. 2013, 54, 474–483. [Google Scholar] [CrossRef]
- Christensen, A.H.; Quail, P.H. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 1996, 5, 213–218. [Google Scholar] [CrossRef]
- Rooke, L.; Byrne, D.; Salgueiro, S. Marker gene expression driven by the maize ubiquitin promoter in transgenic wheat. Ann. Appl. Biol. 2000, 136, 167–172. [Google Scholar] [CrossRef]
- Gurr, S.J.; Rushton, P.J. Engineering plants with increased disease resistance: How are we going to express it? Trends Biotechnol. 2005, 23, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Lin, S.; Zhang, Q.; Zhang, Z.; Zhang, Z.; Lei, Y.; Hou, L. Isolation and analysis of a TIR-specific promoter from poplar. For. Stud. China 2007, 9, 95–106. [Google Scholar] [CrossRef]
- Cornejo, M.J.; Luth, D.; Blankenship, K.M.; Anderson, O.D.; Blechl, A.E. Activity of a maize ubiquitin promoter in transgenic rice. Plant Mol. Biol. 1993, 23, 567–581. [Google Scholar] [CrossRef] [PubMed]
- Pooggin, M.M.; Fütterer, J.; Skryabin, K.G.; Hohn, T. Ribosome shunt is essential for infectivity of cauliflower mosaic virus. Proc. Natl. Acad. Sci. USA 2001, 98, 886–891. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lu, L.; Zhang, L.; Wang, A.; Wang, N.; Liang, Z.; Lu, X.; Tang, K. Molecular evolution of the E8 promoter in tomato and some of its relative wild species. J. Biosci. 2009, 34, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Yi, N.; Kim, Y.S.; Jeong, M.H.; Oh, S.J.; Jeong, J.S.; Park, S.H.; Jung, H.; Choi, Y.D.; Kim, J.K. Functional analysis of six drought-inducible promoters in transgenic rice plants throughout all stages of plant growth. Planta 2010, 232, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.K.C.; Drossard, J.; Lewis, D.; Altmann, F.; Boyle, J.; Christou, P.; Cole, T.; Dale, P.; van Dolleweerd, C.J.; Isitt, V.; et al. Regulatory approval and a first-in-human phase I clinical trial of a monoclonal antibody produced in transgenic tobacco plants. Plant Biotechnol. J. 2015, 13, 1106–1120. [Google Scholar] [CrossRef] [PubMed]
- Deikman, J.; Fischer, R.L. Interaction of a DNA binding factor with the 5′-flanking region of an ethylene-responsive fruit ripening gene from tomato. EMBO J. 1988, 7, 3315–3320. [Google Scholar] [CrossRef] [PubMed]
- Deikman, J.; Kline, R.; Fischer, R.L. Organization of ripening and ethylene regulatory regions in a fruit-specific promoter from tomato (Lycopersicon esculentum). Plant Physiol. 1992, 100, 2013–2017. [Google Scholar] [CrossRef]
- Deikman, J.; Xu, R.; Kneissl, M.L.; Ciardi, J.A.; Kim, K.N.; Pelah, D. Separation of cis elements responsive to elements responsive to ethylene, ethylene, fruit development, and ripening in the 5′-flanking region of the ripening-related E8 gene. Plant Mol. Biol. 1998, 37, 1001–1011. [Google Scholar] [CrossRef] [PubMed]
- Van Haaren, M.J.; Houck, C.M. A functional map of the fruit-specific promoter of the tomato 2A11 gene. Plant Mol. Biol. 1993, 21, 625–640. [Google Scholar] [CrossRef]
- Xu, R.; Goldman, S.; Coupe, S.; Deikman, J. Ethylene control of E4 transcription during tomato fruit ripening involves two cooperative cis elements. Plant Mol. Biol. 1996, 31, 1117–1127. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.M.; Cooper, N.G.; Robinson, D.L.; Korban, S.S. Sequence and in silico characterization of the tomato polygalacturonase (PG) promoter and terminator regions. Plant Mol. Biol. Rep. 2009, 27, 250–256. [Google Scholar] [CrossRef]
- Twell, D.; Wing, R.; Yamaguchi, J.; McCormick, S. Isolation and expression of an anther-specific gene from tomato. Mol. Gen. Genet. 1989, 217, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Twell, D.; Yamaguchi, J.; Wing, R.A.; Ushiba, J.; McCormick, S. Promoter analysis of genes that are coordinately expressed during pollen development reveals pollen-specific enhancer secluences and shared regulatory elements. Genes Dev. 1991, 5, 496–507. [Google Scholar] [CrossRef]
- Eyal, Y.; Curie, C.; McCormick, S. Pollen specificity elements reside in 30 bp of the proximal promoters of two pollen-expressed genes. Plant Cell 1995, 7, 373–384. [Google Scholar]
- Luo, H.; Lee, J.Y.; Hu, Q.; Nelson-Vasilchik, K.; Eitas, T.K.; Lickwar, C.; Kausch, A.P.; Chandlee, J.M.; Hodges, T.K. RTS, a rice anther-specific gene is required for male fertility and its promoter sequence directs tissue-specific gene expression in different plant species. Plant Mol. Biol. 2006, 62, 397–408. [Google Scholar] [CrossRef] [PubMed]
- van Tunen, A.J.; Mur, L.A.; Brouns, G.S.; Rienstra, J.D.; Koes, R.E.; Mol, J.N. Pollen- and anther-specific chi promoters from petunia: Tandem promoter regulation of the chiA gene. Plant Cell 1990, 2, 393–401. [Google Scholar]
- Bäumlein, H.; Nagy, I.; Villarroel, R.; Inzé, D.; Wobus, U. Cis-analysis of a seed protein gene promoter: The conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. Plant J. 1992, 2, 233–239. [Google Scholar]
- Damaj, M.B.; Kumpatla, S.P.; Emani, C.; Beremand, P.D.; Reddy, A.S.; Rathore, K.S.; Buenrostro-Nava, M.T.; Curtis, I.S.; Thomas, T.L.; Mirkov, T.E. Sugarcane DIRIGENT and O-METHYLTRANSFERASE promoters confer stem-regulated gene expression in diverse monocots. Planta 2010, 231, 1439–1458. [Google Scholar] [CrossRef] [PubMed]
- Moyle, R.L.; Birch, R.G. Sugarcane Loading Stem Gene promoters drive transgene expression preferentially in the stem. Plant Mol. Biol. 2013, 82, 51–58. [Google Scholar] [CrossRef]
- Prashant, S.; Sunita, M.S.L.; Sirisha, V.L.; Bhaskar, V.V.; Rao, A.M.; Narasu, M.L.; Kishor, P.B.K. Isolation of cinnamoyl CoA reductase and cinnamyl alcohol dehydrogenase gene promoters from Leucaena leucocephala, a leguminous tree species, and characterization of tissue-specific activity in transgenic tobacco. Plant Cell Tiss. Org. Cult. 2012, 108, 421–436. [Google Scholar] [CrossRef]
- Cortés, A.J.; Restrepo-Montoya, M.; Bedoya-Canas, L.E. Modern strategies to assess and breed forest tree adaptation to changing climate. Front. Plant Sci. 2020, 11, 583323. [Google Scholar] [CrossRef] [PubMed]
- Cortés, A.J.; López-Hernández, F. Harnessing Crop Wild Diversity for Climate Change Adaptation. Genes 2021, 12, 783. [Google Scholar] [CrossRef] [PubMed]
- Manning, K.; Tör, M.; Poole, M.; Hong, Y.; Thompson, J.A.; King, J.G.; Giovannoni, J.J.; Seymour, B.G. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006, 38, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Matas, A.J.; Yeats, T.H.; Buda, G.J.; Zheng, Y.; Chatterjee, S.; Tohge, T.; Ponnala, L.; Adato, A.; Aharoni, A.; Stark, R.; et al. Tissue-and cell-type specific transcriptome profiling of expanding tomato fruit provides insights into metabolic and regulatory specialization and cuticle formation. Plant Cell 2011, 23, 3893–3910. [Google Scholar] [CrossRef]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef]
- Visser, M.; Zubakov, D.; Ballantyne, K.N.; Kayser, M. mRNA-based skin identification for forensic applications. Int. J. Legal. Med. 2011, 125, 253–263. [Google Scholar] [CrossRef]
- Taylor, B.J.M.; Wu, Y.L.; Rada, C. Active RNAP pre-initiation sites are highly mutated by cytidine deaminases in yeast, with AID targeting small RNA genes. eLife 2014, 3, e03553. [Google Scholar] [CrossRef]
- Shirsat, A.; Wilford, N.; Croy, R.; Boulter, D. Sequences responsible for the tissue specific promoter activity of a pea legumin gene in tobacco. Mol. Gen. Genet. 1989, 215, 326–331. [Google Scholar] [CrossRef]
- Porto, M.S.; Pinheiro, M.P.N.; Batista, V.G.L.; dos Santos, R.C.; de Albuquerque, M.; Filho, A.; de Lima, L.M. Plant promoters: An approach of structure and function. Mol. Biotechnol. 2014, 56, 38–49. [Google Scholar] [CrossRef]
- Burke, T.W.; Kadonaga, J.T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996, 10, 711–724. [Google Scholar] [CrossRef]
- Zhu, Q.; Dabi, T.; Lamb, C. TATA box and initiator functions in the accurate transcription of a plant minimal promoter in vitro. Plant Cell 1995, 7, 1681–1689. [Google Scholar]
- Munir, S.; Liu, H.; Xing, Y.; Hussain, S.; Ouyang, B.; Zhang, Y.; Li, H.; Ye, Z. Overexpression of calmodulin-like (ShCML44) stress-responsive gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses. Sci. Rep. 2016, 6, 31772. [Google Scholar] [CrossRef]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. Dimerization and DNA binding of auxin response factors. Plant J. 1999, 19, 309–319. [Google Scholar] [CrossRef]
- Itzhaki, H.; Maxson, J.M.; Woodson, W.R. An ethylene-responsive enhancer element is involved in the senescence-related expression of the carnation glutathione-S-transferase (GST1) gene. Proc. Natl. Acad. Sci. USA 1994, 91, 8925–8929. [Google Scholar] [CrossRef]
- Gubler, F.; Jacobsen, J.V. Gibberellin-responsive elements in the promoter of a barley high-pI alpha-amylase gene. Plant Cell 1992, 4, 1435–1441. [Google Scholar] [CrossRef]
- Patzlaff, A.; Newman, L.J.; Dubos, C.; Whetten, R.W.; Smith, C.; McInnis, S.; Bevan, M.W.; Sederoff, R.R.; Campbell, M.M. Characterisation of PtMYB1, an R2R3-MYB from pine xylem. Plant Mol. Biol. 2003, 53, 597–608. [Google Scholar] [CrossRef]
- Da Costa e Silva, O.; Klein, L.; Schmelzer, E.; Trezzini, G.F.; Hahlbrock, K. BPF-1, a pathogen-induced DNA-binding protein involved in the plant defense response. Plant J. 1993, 4, 125–135. [Google Scholar] [CrossRef]
- Feuillet, C.; Lauvergeat, V.; Deswarte, C.; Pilate, G.; Boudet, A.; Grima-Pettenati, J. Tissue-and cell-specific expression of a cinnamyl alcohol dehydrogenase promoter in transgenic poplar plants. Plant Mol. Biol. 1995, 27, 651–667. [Google Scholar] [CrossRef]
- Ito, H.; Hiraga, S.; Tsugawa, H.; Matsui, H.; Honma, M.; Otsuki, Y.; Murakami, T.; Ohashi, Y. Xylem-specific expression of wound-inducible rice peroxidase genes in transgenic plants. Plant Sci. 2000, 155, 85–100. [Google Scholar] [CrossRef]
- Lacombe, E.; Van Doorsselaere, J.; Boerjan, W.; Boudet, A.M.; Grima-Pettenati, J. Characterization of cis-elements required for vascular expression of the Cinnamoyl CoA Reductase gene and for protein-DNA complex formation. Plant J. 2000, 23, 663–676. [Google Scholar] [CrossRef]
- Ellerström, M.; Stålberg, K.; Ezcurra, I.; Rask, L. Functional dissection of a napin gene promoter: Identification of promoter elements required for embryo and endosperm-specific transcription. Plant Mol. Biol. 1996, 32, 1019–1027. [Google Scholar] [CrossRef]
- Yamagata, H.; Yonesu, K.; Hirata, A.; Aizono, Y. TGTCACA motif is a novel cis-regulatory enhancer element involved in fruit-specific expression of the cucumisin gene. J. Biol. Chem. 2002, 277, 11582–11590. [Google Scholar] [CrossRef]
- Welchen, E.; Gonzalez, D.H. Differential expression of the Arabidopsis cytochrome c genes Cytc-1 and Cytc-2. Evidence for the involvement of TCP-domain protein-binding elements in anther-and meristem-specific expression of the Cytc-1 gene. Plant Physiol. 2005, 139, 88–100. [Google Scholar] [CrossRef][Green Version]
- Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar] [CrossRef]
- Wellmer, F.; Riechmann, J.L.; Alves-Ferreira, M.; Meyerowitz, E.M. Genome-wide analysis of spatial gene expression in Arabidopsis flowers. Plant Cell 2004, 16, 1314–1326. [Google Scholar] [CrossRef]
- Moyle, R.L.; Birch, R.G. Diversity of sequences and expression patterns among alleles of a sugarcane loading stem gene. Theor. Appl. Genet. 2013, 126, 1775–1782. [Google Scholar] [CrossRef]
- Koltunow, A.M.; Truettner, J.; Cox, K.H.; Wallroth, M.; Goldberg, R.B. Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 1990, 2, 1201–1224. [Google Scholar] [CrossRef]
- Bird, C.R.; Smith, C.J.S.; Ray, J.A.; Moureau, P.; Bevan, M.W.; Bird, A.S.; Hughes, S.; Morris, P.C.; Grierson, D.; Schuch, W. The tomato polygalacturonase gene and ripening-specific expression in transgenic plants. Plant Mol. Biol. 1988, 11, 651–662. [Google Scholar] [CrossRef]
- Bate, N.; Twell, D. Functional architecture of a late pollen promoter: Pollen specific transcription is developmentally regulated by multiple stage specific and codependent activator elements. Plant Mol. Biol. 1998, 37, 859–869. [Google Scholar] [CrossRef]
- Zouine, M.; Maza, E.; Djari, A.; Lauvernier, M.; Frasse, P.; Smouni, A.; Pirrello, J.; Bouzayen, M. TomExpress, a unified tomato RNA-Seq platform for visualization of expression data, clustering and correlation networks. Plant J. 2017, 92, 727–735. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, M.B.; Powell, K.S.; Van Damme, E.; Hilder, V.A.; Gatehouse, A.M.R.; Boulter, D.; Gatehouse, J.A. Use of the rice sucrose synthase-1 promoter to direct phloem-specific expression of β-glucuronidase and snowdrop lectin genes in transgenic tobacco plants. J. Exp. Bot. 1994, 45, 623–631. [Google Scholar] [CrossRef]
- Adrian, B. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar]
- Jones, P.A. The role of dna methylation in mammalian epigenetics. Science 2001, 293, 1068–1070. [Google Scholar] [CrossRef]
- Zhou, L.; Tian, S.; Qin, G. RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019, 20, 156. [Google Scholar] [CrossRef]
- Arenas, S.; Cortés, A.J.; Mastretta-Yanes, A.; Jaramillo-Correa, P.J. Evaluating the accuracy of genomic prediction for the management and conservation of relictual natural tree populations. Tree Genet. Genomes 2021, 17, 1–19. [Google Scholar] [CrossRef]
- Jeong, J.S.; Kim, Y.S.; Redillas, M.C.F.R.; Jang, G.; Jung, H.; Bang, S.W.; Choi, Y.D.; Ha, S.; Reuzeau, C.; Kim, J. OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 2013, 11, 101–114. [Google Scholar] [CrossRef]
- Yang, C.; Wang, D.; Zhang, C.; Ye, C.; Kong, N.; Ma, H.; Chen, Q. Comprehensive Analysis and Expression Profiling of PIN, AUX/LAX, and ABCB Auxin Transporter Gene Families in Solanum tuberosum under Phytohormone Stimuli and Abiotic Stresses. Biology 2021, 10, 127. [Google Scholar] [CrossRef]
- Wang, R.; Zhu, M.; Ye, R.; Liu, Z.; Zhou, F.; Chen, H.; Lin, Y. Novel green tissue-specific synthetic promoters and cis-regulatory elements in rice. Sci. Rep. 2015, 5, 1–11. [Google Scholar] [CrossRef]

| Gene ID | Annotation | Locus | Promoter Region (−3000 to −1) |
|---|---|---|---|
| Outer epidermis | |||
| Solyc03g098700.1 | putative Kunitz-type tuber invertase inhibitor precursor (Solanum tuberosum) | SL2.50ch03:60981830..60981171 | SL2.50ch03:60984830..60981831 |
| Solyc06g024380.1 | PREDICTED: uncharacterized protein LOC101232191 (Cucumis sativus) | SL3.0ch06:11249034..11249552 | SL3.0ch06:11246034..11249033 |
| Solyc10g075090.1 | PREDICTED: non-specific lipid-transfer protein 1-like (Solanum lycopersicum) | SL2.50ch10:58801024..58800507 | SL2.50ch10:58804024..58801025 |
| Solyc10g075100.1 | non-specific lipid transfer protein precursor (S. lycopersicum) | SL2.50ch10:58811212..58810582 | SL2.50ch10:8814212..58811213 |
| Solyc10g076200.1 | non-specific lipid-transfer protein 2-like (S. lycopersicum) | SL2.50ch10:59051770..59052216 | SL2.50ch10:59048770..59051769 |
| Collenchyma | |||
| Solyc01g105040.2 | PREDICTED: uncharacterized protein LOC101251628 (S. lycopersicum) | SL2.50ch01:93331608..93329788 | SL2.50ch01:93334592..93331593 |
| Solyc03g005000.2 | PREDICTED: protease HtpX homolog 2-like isoform 1 (S. lycopersicum) | SL2.50ch03:16712..19799 | SL2.50ch03:13817..16816 |
| Solyc07g020860.2 | thioredoxin peroxidase 1 (S. lycopersicum) | SL2.50ch07:14295272..14300912 | SL2.50ch07:14292470..14295469 |
| Solyc09g013150.2 | PREDICTED: probable anion transporter 3, chloroplastic-like (S. lycopersicum) | SL2.50ch09:5558428..5552677 | SL2.50ch09:5561282..5558283 |
| Solyc10g006790.2 | PREDICTED: probable serine/threonine-protein kinase abkC-like (S. lycopersicum) | SL2.50ch10:1231572..1237111 | SL2.50ch10:1228572..1231571 |
| Parenchyma | |||
| Solyc01g067730.2 | PREDICTED: acyl carrier protein 1, chloroplastic-like (S. lycopersicum) | SL2.50ch01:76674532..76676885 | SL2.50ch01:76671781..76674780 |
| Solyc01g096270.2 | PREDICTED: cytochrome b5-like (S. lycopersicum) | SL2.50ch01:87349969..87350651 | SL2.50ch01:87347174..87350173 |
| Solyc04g007770.2 | PREDICTED: major latex-like protein (S. lycopersicum) | SL2.50ch04:1458908..1456588 | SL2.50ch04: 1461693..1458694 |
| Solyc07g049140.2 | PREDICTED: metallocarboxypeptidase inhibitor, fruit-specific protein (S. lycopersicum) | SL2.50ch07:59369096..59367495 | SL2.50ch07:59371828..59368829 |
| Solyc09g007940.2 | PREDICTED: adenosine kinase 2-like (S. lycopersicum) | SL2.50ch09:1440058..1444221 | SL2.50ch09:1437058..1440057 |
| Vascular | |||
| Solyc01g111310.2 | LAX2 protein (S. lycopersicum) | SL2.50ch01:97595955..97592414 | SL2.50ch01:97598905..97595906 |
| Solyc04g026020.2 | PREDICTED: sieve element-occluding protein 3 (S. lycopersicum) | SL2.50ch04:19817933..19814414 | SL2.50ch04:19820933..19817934 |
| Solyc05g006830.2 | PREDICTED: thioredoxin H2-like (S. lycopersicum) | SL2.50ch05:1454232..1453126 | SL2.50ch05:1457094..1454095 |
| Solyc06g075220.1 | PREDICTED: fasciclin-like arabinogalactan protein 11-like (S. lycopersicum) | SL2.50ch06:46677877..46678626 | SL2.50ch06:46674877..46677876 |
| Solyc09g010080.2 | invertase 5, an extracellular invertase (S. lycopersicum) | SL2.50ch09:3475480..3479343 | SL2.50ch09:3472522..3475521 |
| Inner epidermis | |||
| Solyc04g072310.2 | PREDICTED: uncharacterized protein LOC101262106 (S. lycopersicum) | SL2.50ch04:59344118..59341881 | SL2.50ch04:59346918..59343919 |
| Solyc04g082170.2 | PREDICTED: alcohol dehydrogenase-like 7-like (S. lycopersicum) | SL2.50ch04:65946520..65949299 | SL2.50ch04:65943520..65946519 |
| Solyc06g065530.2 | PREDICTED: GDSL esterase/lipase At1g29670-like (S. lycopersicum) | SL2.50ch06:40913948..40910209 | SL2.50ch06:40916912..40913913 |
| Solyc09g062960.2 | PREDICTED: uncharacterized protein LOC101264365 (S. lycopersicum) | SL2.50ch09:60864690..60864224 | SL2.50ch09:60867472..60864473 |
| Solyc09g062970.1 | PREDICTED: uncharacterized protein LOC101264365 (S. lycopersicum) | SL2.50ch09:60877001..60877273 | SL2.50ch09:60874001-60877000 |
| TSP ID * | Ratio (TSP/CP **) | |||
|---|---|---|---|---|
| pE8 | pPG | pLAT52 | 2×35S | |
| Anther-specific | ||||
| pSolyc04g082170.2 | 0.23 | 0.54 | ||
| pSolyc09g007940.2 | 0.01 | 0.01 | ||
| pSolyc09g010080.2 | 0.06 | 0.14 | ||
| pSolyc10g076200.1 | 15.32 | 35.07 | ||
| Fruit-specific | ||||
| pSolyc03g098700.1 | 0.03 | 0.20 | 0.02 | |
| pSolyc04g007770.2 | 0.46 | 3.35 | 0.38 | |
| pSolyc07g049140.2 | 10.05 | 72.98 | 8.22 | |
| pSolyc10g075090.1 | 0.42 | 3.03 | 0.34 | |
| Leaf-specific | ||||
| pSolyc09g062960.2 | 0.28 | |||
| pSolyc09g062970.1 | 1.02 | |||
| pSolyc10g075100.1 | 1.91 | |||
| Root-/stem-specific | ||||
| pSolyc04g026020.2 | 0.02/0.03 | |||
| pSolyc06g075220.1 | 0.08/0.04 | |||
| Sepal-specific | ||||
| pSolyc01g105040.2 | 0.05 | |||
| pSolyc06g065530.2 | 0.02 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Li, Y.; Wang, Y.; Li, W.; Feng, X.; Zhao, L. Use of High Resolution Spatiotemporal Gene Expression Data to Uncover Novel Tissue-Specific Promoters in Tomato. Agriculture 2021, 11, 1195. https://doi.org/10.3390/agriculture11121195
Chen L, Li Y, Wang Y, Li W, Feng X, Zhao L. Use of High Resolution Spatiotemporal Gene Expression Data to Uncover Novel Tissue-Specific Promoters in Tomato. Agriculture. 2021; 11(12):1195. https://doi.org/10.3390/agriculture11121195
Chicago/Turabian StyleChen, Lulu, Yuhang Li, Yuting Wang, Wenzhen Li, Xuechao Feng, and Lingxia Zhao. 2021. "Use of High Resolution Spatiotemporal Gene Expression Data to Uncover Novel Tissue-Specific Promoters in Tomato" Agriculture 11, no. 12: 1195. https://doi.org/10.3390/agriculture11121195
APA StyleChen, L., Li, Y., Wang, Y., Li, W., Feng, X., & Zhao, L. (2021). Use of High Resolution Spatiotemporal Gene Expression Data to Uncover Novel Tissue-Specific Promoters in Tomato. Agriculture, 11(12), 1195. https://doi.org/10.3390/agriculture11121195






