Abstract
Quinoa is renowned for its nutritional value and ability to withstand harsh environmental conditions such as salinity. In the present work, we isolated 34 phosphate solubilizing endophytic bacteria associated with the roots of quinoa plants. Based on phosphate solubilization efficiency and biochemical characterization, we selected one isolate named ED1. Ribotyping using partial 16S RNA gene analysis revealed that the selected isolate shares 99.7% identity with Serratia rubidaea. Plant growth promoting (PGP) studies showed that the ED1 strain solubilized complexed forms of phosphate (Ca3(PO4)2). Zinc release from ZnO, Zn3(PO4)2, or ZnCO3 revealed the efficient ZnO solubilization by the ED1 strain. Except for proteases, the strain ED1 produced siderophores, cellulase, ammonia and exhibited oligonitrophilic features. Indole acetic acid (IAA) production was detected with and without the L-tryptophan precursor. Next, we demonstrated that the ED1 strain tolerated 1.5 M NaCl final concentration and exhibited intrinsic resistance to seven antibiotics frequently prescribed for medical use. Moreover, we found that ED1 strain withstood 2 mg/L of Cadmium and 1 mg/L of either Nickel or Copper. Furthermore, we observed that S. rubidaea ED1 stimulated quinoa seeds germination and seedlings growth under salt stress conditions. Lastly, we discuss the advantages versus disadvantages of applying the S. rubidaea ED1 strain as a beneficial agent for salty and/or heavy metals contaminated soils.
1. Introduction
Plant associated endophytic microorganisms are considered as the second functional genome of host plants [1]. In 1995, endophytes were defined as symbiotic microorganisms (bacteria, archaea, fungi, and protists) residing within the plant endosphere without causing any detrimental impact to the host plant [2]. Hardoim et al. (2015) stated that any colonizing microbe of plant tissues could be classified as endophyte regardless of the outcome of its association with the host plant [3]. However, colonization by endophytes is crucial for providing benefits to plants [4]. Indeed, the interaction of endophytes with plants is characterized as a symbiotic relationship because of mutual benefits. The plant provides protective niches to the internal microorganisms which, in turn, act directly by promoting plant growth through nitrogen (N) fixation, phytohormone, and metabolites production, and indirectly by stimulating availability and absorption of macro and micronutrients and water. In addition, endophytic microorganisms induce host tolerance to abiotic stresses such as osmotic stress, exposure to heavy metals and xenobiotic molecules, and assist in the biocontrol of plant pathogens [5,6].
Plant growth promoting microorganisms (PGPM), including endophytes, have an immense potential to boost plants’ performance and to reduce the environmental impacts caused by chemical inputs as increasing agricultural productivity is of the upmost priority around the globe [4,7]. Indeed, bacterial endophytes have been reported to prime plants for faster and more intense defense responses upon pathogen attacks at low physiological cost to the plant [8]. Endophytes also stimulate seedlings’ emergence and enhance plant growth under adverse conditions [9,10]. Inoculation of plant growth promoting endophytic (PGPE) bacteria in plants has low environmental impact, requires low-cost techniques, and could be of immense economic importance.
Plants are complex micro-ecosystems where the different tissue and surface compartments are exploited by a wide variety of microorganisms [11]. PGPE bacteria have been isolated from several plants such as wheat roots [12], sweet potato roots [13], rice roots [14], sugarcane stems [15], bean leaves [16], and quinoa seeds [17]. Plants interact with microbes primarily at the roots. Despite the crucial role of root bacterial communities in plant health and nutrient assimilation, the current understanding of the complex plant–microbe interactions is still in its infancy. At the soil-root interface, roots provide different microhabitats: rhizospheric soil, rhizoplane, and endorhizosphere [18].
Among phytohormones, auxins (e.g., indole acetic acid (IAA)) are the main plant growth regulators having many physiological actions including elongation of primary roots, differentiation of tissues, formation of pigments, stimulation of nitrogen fixation, and resistance to various stresses. Plants associated microorganisms can synthesize IAA via tryptophan-dependent pathways by converting tryptophan to different intermediates. This pathway of IAA biosynthesis has been demonstrated in the majority of soil bacteria [19]. However, some microorganisms are endowed with the ability to produce it via tryptophan-independent routes [20].
Salinity in one of the most devastating stresses in agriculture, especially in arid and semiarid environmental areas [21]. It considerably reduces plant growth and productivity because of nutritional imbalance in plants (increase in Na+ and decrease in K+ uptake), inhibition of protein synthesis, enzyme inactivation, early senescence, decrease in photosynthesis and respiration, and loss of cellular integrity [22,23]. Nowadays, methods to increase tolerance of plants to salinity involve the use of salt tolerant crops, transgenic plant genetic engineering, and traditional breeding [24]. Unfortunately, these strategies are labor-intensive and highly technical [25]. The alternative strategy to sustain plant growth under salty conditions is the use of PGPB (plant growth promoting bacteria). These latter can be found both at the root surface and in endophytic associations with host plant. These bacteria can either directly or indirectly enhance plant growth in normal and stressful conditions [25,26,27]. Several bacterial species have been reported to mitigate salinity stress and stimulate plant growth. They include Klebsiella sp., Burkholderia sp., Enterobacter sp. [23], Microbacterium sp., Alcaligenes sp. [28], Ochrobactrum sp. [28], Bacillus sp. [23,29], Arthrobacter sp. [30], Pseudomonas sp. [31], Bacillus licheniformis and Enterobacter asburiae [32], and finally Serratia sp. [33].
Several studies have tackled the emergence and propagation of antibiotic resistance (ARG) genes in different environmental reservoirs through different anthropogenic activities such as the application of manure, slurry, and soil amendments with regard to their different transfer pathways and threats to public health and ecology [34]. However, ARG-harboring PGPB inoculants are mostly unexplored, while most of these PGPB are still recommended as potential effective biofertilizers. Despite the importance of antibiotic resistance in helping biofertilizers-based bacteria to survive and compete the respective native microflora in open microbial habitats [35], large-scale application of ARG-harboring biofertilizers into the soil can rise new resistances and worsen the spread of ARG in other ecosystems.
Quinoa (Chenopodium quinoa Willd.), an herbaceous plant species belonging to the amaranthaceae family [36], is one of the most promising crops for tomorrow’s food demand and nutritional security. Originally from South America, quinoa was heavily cultivated by the ancient Andean civilization, but it was not until the 1970s that quinoa began to be introduced all over the world. Quinoa is renowned for its ability to withstand harsh environmental conditions such as drought, salinity, wind, and heat [37,38] and its outstanding nutritional value [39] with high content of vitamins, and good quality of proteins and saponins [40]. Quinoa crop has recently gained attention and its cultivation is spreading worldwide. In the developing countries of Africa and Asia, quinoa represents a miracle solution to provide highly nutritious food [41] due to its growth adaptability to various environmental conditions. In Morocco, it has started to be cultivated for its income-generating potential, its adaptability to different soil and climatic conditions, and its potential to improve cropping systems [42]. More importantly, it grows up to an altitude of 4000 m above sea level, resists a wide daily temperature ranges, needs only 300 mm of precipitation per year, and is produced in a short period of time [40]. Given the ability of halophyte quinoa plants to cope with abiotic stresses and extreme environments, we hypothesized that quinoa-associated endophytic bacteria may be involved in the acquisition of such tolerance as well as in promoting plant growth. Herein, we identified a phosphate solubilizing endophytic bacterium inhabiting the roots of quinoa plants cultivated in Morocco and characterize its PGP properties in vitro. We also assessed its ability to withstand extreme conditions such salt and heavy metals stresses.
2. Materials and Methods
2.1. Plant Material and Sampling
Root samples were collected from Chenopodium quinoa plants growing in different locations at the experimental farm (32.219731 E, −7.892268 N) of Mohammed VI Polytechnic University, Ben Guerir, Morocco. Sampling was carried out in June 2018, peak growing season for quinoa. Six plants were randomly selected for root sampling, and roots without visible damage were collected under aseptic conditions and transported to the laboratory for further analysis.
2.2. Isolation of Root-Borne Bacterial Endophytes
Primary and secondary roots were washed under running tap water to remove soil debris, then surface-sterilized with ethanol (70%) for 3 min and sodium hypochlorite (3%) for 1 min, followed by three rinses in sterile distilled water for 3 min each [43,44]. To ensure the success of surface sterilization treatments, the last water wash solutions as well as sterilized roots were respectively deposited on TSA (Trypticase Soy Agar, BIOKAR Diagnostics, BEAUVAIS, France) plates and cultivated overnight [44]. The sterilized root pieces were ground in 0.85% aqueous NaCl using a mortar and pestle. Next, serial dilutions (10−1 to 10−5) were generated, and each dilution was spread on a TSA medium using a glass spreader [16]. Plates were then incubated at 30 °C for 24 h. Emerging colonies were subcultured to obtain pure isolates.
2.3. Plate Assay for Phosphate Solubilizing Activity
The purified bacterial isolates were subjected to phosphate solubilization activity screening. Using nitrocellulose membranes, a total of 34 endophytic bacterial colonies grown on TSA plates were transferred to NBRIP (National Botanical Research Institute’s phosphate) agar medium plate [45] consisting of (g/L) dextrose 10; hydroxyapatite 5 (purum p.a., ≥90% (as Ca3(PO4)2,KT); ammonium sulphate 0.5; potassium chloride 0.2; sodium chloride 0.2; magnesium sulphate 0.1; ferrous sulphate trace; manganese sulphate trace; agar 15; and the pH was adjusted to 6.75 ± 0.25 before autoclaving [46]. Plates were incubated at 30 °C and checked daily for 7 days for the appearance of transparent halos indicating P-solubilizing ability. Based on the growth on selective medium and discrete halo zones appearance, one selected isolate (named ED1) was subcultured several times, purified on the same medium, then stored at −80 °C in cryotubes using 10% dimethyl sulfoxide (DMSO) as a cryoprotective agent.
2.4. Ribotyping Identification of Isolated Strains
The polymerase chain reaction (PCR) was performed on bacterial 16S rRNA gene using primers: forward pA (5′-AGAGTTTGATCCTGGCTCAG-3′) and reverse 926R (5′-CCGYCAATTYMTTTRAGTTT-3′) [47] generating an amplicon of 910 bp. The PCR reaction mixture contained 23 μL DNAase free water, 1 μL of forward and reverse primers at 20 µM final concentration, 25 μL PCR SuperMix (Invitrogen, Carlsbad, CA, USA), and 1 μL of fresh bacterial cultures as a DNA matrix. The amplification process was launched according to the following program: initial denaturation step for 5 min at 94 °C, denaturation for 1 min at 94 °C, hybridization for 1 min at 52 °C, elongation step for 1 min 30 s at 72 °C with 35 cycles, and finally an incubation for 10 min at 72 °C. Then, 5 μL of each PCR sample was checked on agarose gel. The amplicons were sequenced and generated nucleotide sequences were aligned using the ExPASy Bioinformatics Resource Portal (https://www.expasy.org/ (accessed on 20 November 2020)), and compared to available homologous sequences using the BLAST search (Basic Local Alignment Search Tools, NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 20 November 2020))) and the High-Quality Ribosomal RNA database (SILVA (https://www.arb-silva.de/ (accessed on 20 November 2020)) [48]. The phylogenetic dendrogram was generated by the Neighbor-Joining method using Unipro UGENE software, version 1.32.0 [49].
2.5. In Vitro Assessment of PGP Attributes and Extreme Growth Properties
2.5.1. Heat, Salt, and pH Tolerance
To assess for heat tolerance, bacteria were streaked on TSA plates and further incubated for 24 h at various temperatures: 20, 25, 30, 37, 42, 45, and 50 °C [50]. Salt tolerance was firstly evaluated on a TSA medium by supplementing plates with various concentrations of NaCl (0 to 14%) (w/v) and incubated at 30 °C for 48 h [51]. The salt and heat tolerance of the isolate was confirmed by observing its growth on salt-supplemented and nutrient agar media. B. licheniformis QA1 [32] and Esherichia coli DH5α were used as positive (C+) and negative (C−) controls, respectively. To determine the optimum salinity tolerance of the selected bacterium, we used 48-well microtiter microplates; each well was filled with 500 µL of TSB broth supplemented with various NaCl concentrations (0, 2, 4, 6, 8, 10, 12, 14, and 16%) (w/v). In addition, pH tolerance was assessed by adjusting the pH of the media at different levels (2, 3, 4, 5, 6, 7, 8, 9, 10, 11, and 12). Five microliters of an overnight bacterial culture (OD600nm = 0.8) was inoculated in each well and incubated at 30 °C under shaking at 150 rpm. Growth was measured after 24 h by colorimetry at OD600nm using the VICTOR NivoTM Multimode Plate Reader (Perkin Elmer, Casablanca, Morocco) [52].
2.5.2. Phosphate Solubilization Assay under Salt Stress Conditions
Inorganic phosphate solubilization by bacteria was studied in a liquid medium. Bacterial suspension of OD600nm = 0.8 was inoculated into 50 mL of NBRIP broth containing 0%, 4%, and 8% NaCl concentrations and incubated in an incubator shaker at 30 °C/150 rpm [51]. Solubilization of hydroxyapatite (purum p.a., ≥90% (as Ca3(PO4)2, KT) was measured after 5- and 10-days post incubation (DAI; day after incubation). The bacterial broth was centrifuged at 5000 rpm for 10 min, filtered using 0.22 µm sterile syringe filters, and 5 mL of each supernatant was transferred into 50 mL centrifuge tubes. E. coli DH5α was used as a negative control (C−) [46]. Soluble P concentrations (orthophosphates (PO4)) in each sample were determined using an inductively coupled plasma optical emission spectrometer (ICP-OES) at the Agricultural Innovation and Technology Transfer Center (AITTC) of the Mohammed VI Polytechnic University (UM6P).
2.5.3. Oligonitrophilic Activity
To monitor the oligonitrophilic property of the selected bacterium, we used the nitrogen free Jensen’s medium [53] containing the following composition (in 1 L): Sucrose 20 g, Dipotassium phosphate 1 g, Magnesium sulphate 0.5 g, Sodium chloride 0.5 g, Ferrous sulphate 0.1 g, Sodium molybdate 0.005 g, Calcium carbonate 2 g, and Agar 15 g. Plates were streaked with fresh bacterial culture of isolated bacterium and checked for growth following incubation at 30 °C for 24 h. B. licheniformis QA1 [32] and E. coli DH5α were used as positive (C+) and negative (C−) controls, respectively.
2.5.4. Monitoring Indole Acetic Acid (IAA) Production
To quantify the amounts of IAA produced by the P-solubilizing endophyte, a colorimetric method of Van Urk Salkowski reagent was performed [54]. The isolate was inoculated in LB medium supplemented with 0, 0.2, and 0.5% of L-tryptophan and incubated at 28 ± 2 °C for 11 days under shaking of 150 rpm [55]. The determination of produced IAA was assessed after 3, 7, and 11 days of incubation (DAI). The bacterial cultures were centrifuged at 13,000 rpm for 5 min at 4 °C. Next, two mL of Van Urk Salkowski reagent (1 mL of 0.5 M FeCl3 and 50 mL of 35% HClO4) were mixed with 1 mL of each supernatant. Mixtures were incubated in the dark for 30 min [52]. IAA quantification was determined spectrophotometrically at OD533nm using a standard curve of pure IAA (Sigma Aldrich, Overijse, Belgium) for concentrations in the 0–100 µg/mL range.
2.5.5. Solubilization of Insoluble Zinc Compounds
The ability of the selected endophytes to solubilize insoluble Zn compounds was evaluated on Tris-mineral agar medium [56] containing (in 1 L): D-glucose 10 g, (NH4)2SO4 1 g, KCl 0.2 g, K2HPO4 0.1 g, MgSO4 2 g, pH = 6.75 ± 0.25. Prepared media were separately amended with three sources of insoluble Zn namely zinc oxide (ZnO, 15.23 mM), zinc phosphate (Zn3(PO4)2, 5.0 mM), and zinc carbonate (CO3Zn, 5.2 mM) at 0.1% Zn final concentration [57]. The selected isolate was spot inoculated on each medium and plates were incubated at 30 °C for 10 days. The appearance of a clear zone around colonies indicate positive Zn solubilization. B. licheniformis QA1 [32] and E. coli DH5α were used as positive (C+) and negative (C−) controls, respectively.
2.5.6. Siderophores Production Assay
The production of siderophores by bacterial isolates was monitored according to the method described by Schwyn and Neilands (1987) [58] using Chromium Azurol S medium (CAS) which contains the ternary complex CAS-Fe+3-Hexadecyltrimethyl-Ammonium bromide as an indicator. The isolate was spot inoculated on CAS plates and incubated at 28 ± 2 °C for 5 days. The efficiency to produce siderophores was checked by monitoring the size of the halo zone and the intensity of the color change (blue to yellow or orange). The halo diameter was taken as an indicator of siderophores production intensity. B. licheniformis QA1 [32] and E. coli DH5α were used as positive (C+) and negative (C−) controls, respectively.
2.5.7. Ammonia Production Assay
ED1 isolate was tested for ammonia production by adopting the method described by Cappuccino and Sherman (1992) [59]. Fresh bacterial cultures of selected isolate were inoculated into 10 mL of peptone water and incubated at 28 ± 2 °C with shaking at 150 rpm. Post 48 h incubation, 0.5 mL of Nessler reagent was added to each bacterial culture. A faint yellow color indicated a small amount of ammonia and a deep yellow to brownish color indicated high production of ammonia. The absorbance was measured at OD450nm and the concentrations of ammonia were estimated using a standard curve of ammonium sulphate ((NH₄)₂SO₄) for concentrations in 0–0.3 µmol/mL range [60]. B. licheniformis QA1 [32] and E. coli DH5α were used as positive (C+) and negative (C−) controls, respectively.
2.5.8. Cellulase and Protease Production
To check the production of extracellular cellulase by selected bacterium, we used the mineral–salt agar plates containing 0.4% (NH4)2SO4, 0.6% NaCl, 0.1% K2HPO4, 0.01% MgSO4, 0.01% CaCl2 with 0.5% carboxymethyl cellulose, and 2% agar, pH = 6.75 ± 0.25. The plates were then spot inoculated and incubated at 30 °C. After 2 days of incubation, a Congo red solution (1%) was added to the surface of each plate. 20 min later, the plates were flooded with a 1M NaCl solution and then left to stand for 30 min. The appearance of a clear halo around colonies indicates the degradation of the CMC and reflects the presence of extracellular cellulase [61].
The protease activity was performed according to the method of Kavitha et al. (2013) [62] using the medium having the following composition (g L−1): Pancreatic casein (5); Yeast extract (2.5); Glucose (1); and Agar (15). The pH of medium was adjusted to 6.75 ± 0.25 and autoclaved. After cooling, 100 mL of a 10% skimmed milk solution was aseptically prepared and added to the medium, then seeded by the spot inoculation method. Bacteria with protease activity show a transparent halo around colonies over 48 h of incubation [63]. B. licheniformis QA1 [32] and E. coli DH5α were used as positive (C+) and negative (C−) controls, respectively.
2.6. Antibiotic Sensitivity Assay
Bacterial isolate was tested for its potential resistance to antibiotics on Mueller-Hinton (MH) media (BIOKAR Diagnostics, BEAUVAIS, France) [64] using agar dilution assay, disc diffusion method, and Etest [65]. Freshly prepared MH plates were amended with various concentrations of antibiotics frequently used in the treatment of infectious diseases namely ampicillin (100 µg/mL), kanamycin (50 µg/mL), tetracycline (10 µg/mL), streptomycin (100 µg/mL), chloramphenicol (20 µg/mL), and spectinomycin (60 µg/mL). As for disc diffusion and Etest, 200 µL of 18 h old bacterial culture (OD600nm = 1) was spread over the entire MH agar surfaces using a swab stick and left to dry for five minutes before placing the antibiotic-impregnated discs and/or strips. Antibiotic discs of ceftazidime (CAZ; 30 µg/disc), ciprofloxacin (CIP; 5 µg/disc), meropenem (MRP; 30 µg/disc), ertapenem (ETP; 10 µg/disc), and vancomycin (VA; 30 µg/disc) were placed in triplicate. Etest was adopted using the Liofilchem® Minimum Inhibitory Concentration (MIC) Test Strips with a predefined concentration gradient of imipenem (IMI; 0.002–32 µg/mL), gentamycin (CN; 0.06–1024 µg/mL), and cefotaxime (CTX; 0.002–32 µg/mL). As a negative control, we used the E. coli DH5α strain as it is sensitive to all tested antibiotics [66]. In addition, we used a PGPR (Plant Growth Promoting Rhizobacterium) Bacillus atrophaeus strain S8 (MW295957) from our lab collection as an additional control. Resistance patterns were determined following incubation at 30 °C for 24 h. Positive resistances were considered in antibiotic MH amended plates by observing the growth of the bacterium on each medium. In disk diffusion plates, the zone of inhibition surrounding discs was measured in millimeters (mm) using calipers near the agar surface [67]. However, the minimum inhibitory concentration (MIC) was read directly from the scale in terms of µg/mL, at the intersection point of the edge of the inhibition ellipse and the MIC Test Strip. Susceptibilities and resistances were analyzed using Zone Size Interpretative Chart (ZSIC) [68,69], Liofilchem®—Antibiotic Disc Interpretative and CLSI Standards for Antimicrobial Susceptibility Testing of Clinical and Laboratory Standards Institute (https://clsi.org/ (accessed on 18 October 2020)).
2.7. Monitoring Trace Elements Tolerance
The ED1 isolate was tested for its growth under various concentrations ranging from 0 to 2000 μg/mL of trace metals: CdSO4, CuO4S.5H2O, and N2NiO8. Stock solutions were prepared in distilled water, sterilized through 0.45 μm using sterile syringe filters, and stored at 4 °C. Agar dilution method was followed [70]. Freshly prepared agar plates were amended with increasing soluble heavy metal salts concentrations [71]. The ED1 isolate was streaked in both metal amended and control plates (free metal media). Trace metal tolerance was determined after 48 h of incubation at 30 °C. B. licheniformis QA1 [32] and E. coli DH5α were used as positive (C+) and negative (C−) controls, respectively.
2.8. Quinoa Seed Germination Assay
Quinoa seeds of Titicaca variety were used for in vitro seed bacterization to evaluate the early plant growth promoting potential of selected bacterium. Firstly, seeds were sorted and those with damageable aspects were excluded [32]. Seeds were surface sterilized using 3% of sodium hypochlorite for 1 min, soaked in 70% ethanol for 1 min, rinsed 5 times by sterilized distilled water, and air-dried under a laminar flow hood. The bacterial pellets (OD600nm = 0.8) were obtained from an overnight bacterial culture by centrifugation at 5000 rpm for 5 min. The pellets were then resuspended in 10 mL of phosphate-buffered saline (PBS) and used as seed drench [72]. Bacterial suspension was applied as seed drenches for 1 h with gentle shaking [73]. Afterwards, seeds were air-dried and transferred into 9 cm sterile Petri dishes on filter paper with a ratio of 30 seeds per plate. Each treatment was performed in triplicate, and seeds treated with PBS were used as negative control [74]. Each filter paper was aseptically wetted with 3 mL of sterilized distilled saline water solution at 0, 200 (1.16%), or 400 mM (2.32%) NaCl [74]. To prevent evaporation and contamination, the plates were closed with parafilm. The plates were labeled and maintained at 25 °C for 48 h in the dark [73] and germination rates were monitored 24 h and 48 h post-incubation. At the third day, the plates were maintained at room temperature in a day/night cycle (~12/12 h) for an additional 72 h to compute their morphological traits mainly total length, fresh and dry weight. The germination percentage and vigor index were calculated using the following equations [74,75].
n: number of germinated seed and N: total number of seeds.
Germination percentage (%) = (n/N) × 100
Vigor index = Germination percentage (%) × Total seedling length (cm)
2.9. Statistical Analysis
All results obtained were subjected to statistical analysis using IBM SPSS Statistics 20 software. Comparison between means were performed using one-way analysis of variance (ANOVA) followed by the post-hoc analysis with Tukey test. Significant differences were set at p ≤ 0.05. The results were expressed as the mean of 3 replicates ± SD. The experiments performed in this study were done in triplicates using a complete randomized design (CRD).
3. Results
3.1. In Vitro PGP Properties of Selected Quinoa Endophytic Bacteria
3.1.1. P and Zn Solubilization, and Oligonitrophilic Activity
A total of 34 isolated endophytic bacteria were screened for mineral phosphate solubilization on NBRIP agar plates. One isolate named ED1 was further selected based on its remarkable capacity to solubilize inorganic P on plates as evidenced by the halos surrounding colonies (Figure 1). We also checked its ability to solubilize insoluble Zn from three minerals: ZnO, Zn3(PO4)2, and CO3Zn. The ED1 isolate was able to solubilize ZnO at a greater extend compared to the two other forms, while B. licheniformis QA1 and E. coli DH5α only solubilized ZnO (Table 1).
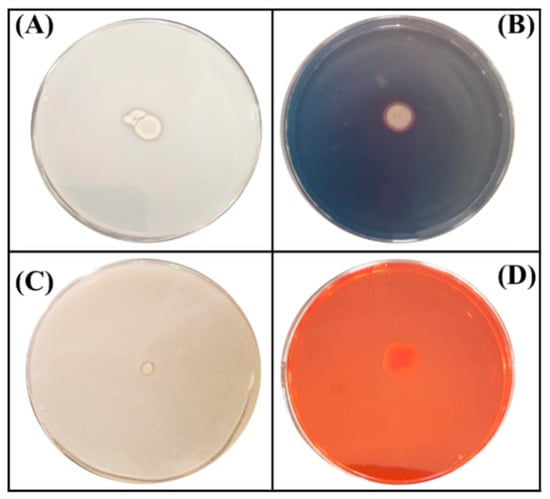
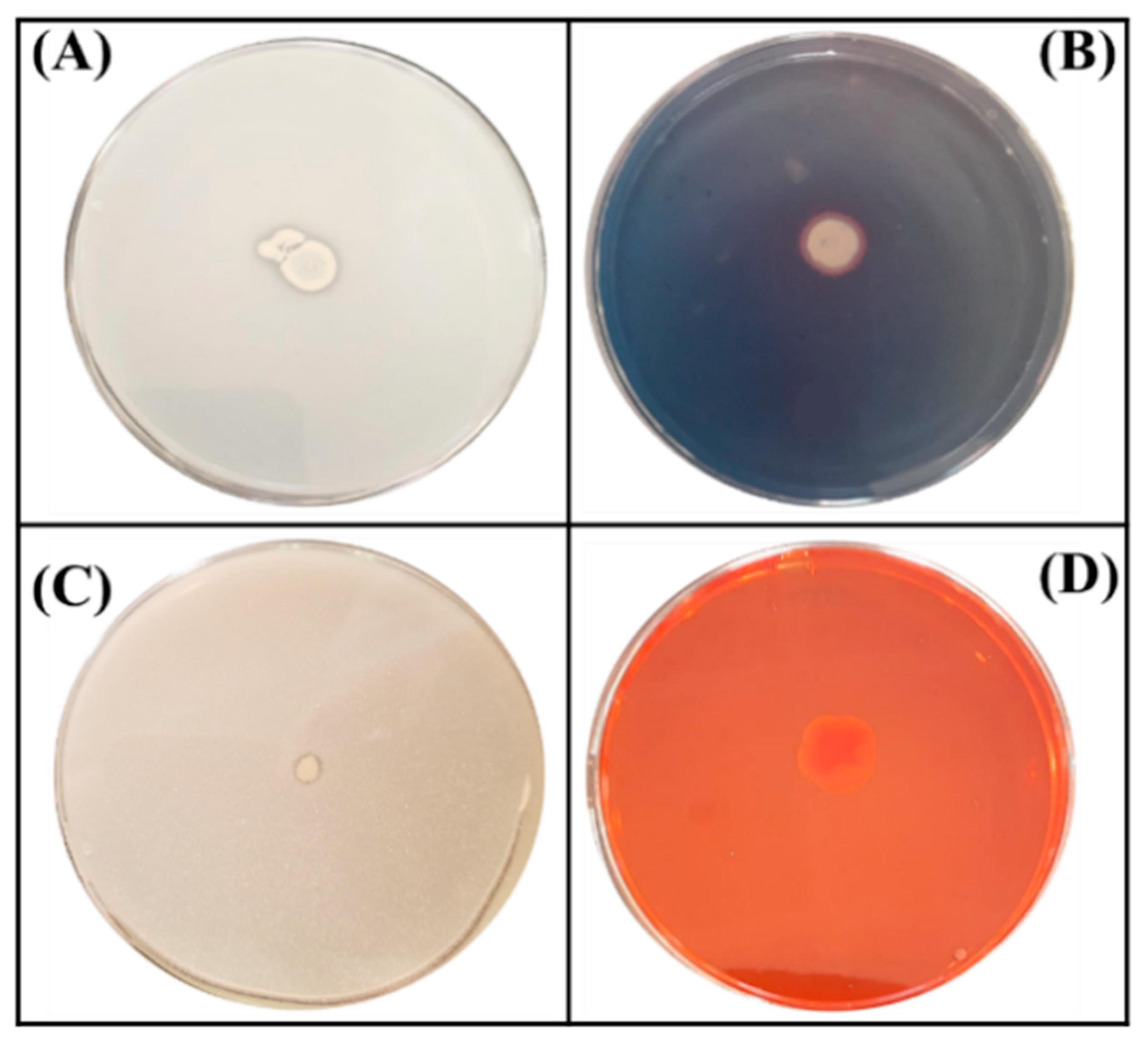
Figure 1.
Schematic illustration of four tests performed on plates for the assessment of plant growth promoting traits of the ED1 isolate. (A) Phosphate solubilization, (B) Siderophores production, (C) Zinc oxide solubilization, (D) Cellulase production.

Table 1.
Summary of relevant plant growth promoting (PGP) traits of the ED1 isolate, the PGPR strain Bacillus licheniformis QA1, and E. coli DH5α (C−).
In contrast to B. licheniformis QA1 and E. coli DH5α used as positive and negative controls, respectively, the ED1 isolate was able to grow on Jensen’s medium which is very poor in nitrogen, indicating its oligonitrophilic activity (Table 1).
3.1.2. Siderophores, Ammonia and Extracellular Enzymes Production by the ED1 Isolate
We next tested the capacity of the ED1 isolate to produce molecules and metabolites of agricultural interest. We showed that it liberated siderophores and produced ammonia and cellulase enzyme (Figure 1). In contrast, no protease activity was detected. In addition, the ED1 isolate was able to grow in peptone water, indicating its capacity to produce ammonia which was 10-fold higher than in E. coli DH5α. Comparatively with the PGPR reference strain B. licheniformis QA1, the selected isolate produced less amounts of siderophores and cellulase but higher concentrations of ammonia (Table 1).
3.2. The ED1 Isolate Is Related to the Genus of Serratia Rubidaea
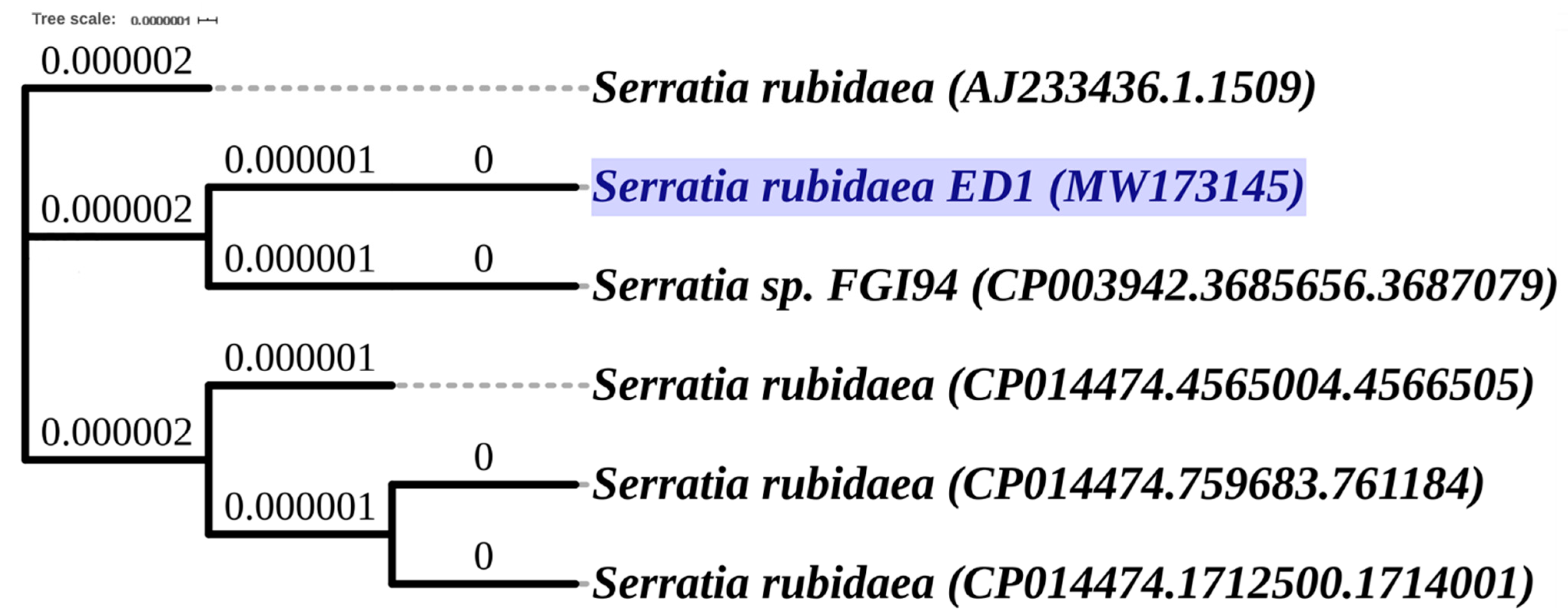
Using the genotyping analysis based on 16S rDNA sequencing and sequences comparison to the NCBI and SILVA databases, we found that the ED1 isolate is 99.7% identical to the genus of Serratia rubidaea. Next, the amplified 16S rDNA sequence was submitted to GenBank and the accession number MW173145 was provided. The phylogenetic tree analysis reflected the same identity with Serratia rubidaea (Figure 2).
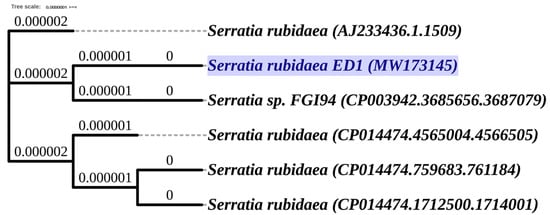
Figure 2.
Phylogenetic tree of selected bacterium Serratia rubidaea ED1 (MW173145) based on the PHYLIP (PHYLogeny Inference Package) Neighbor-Joining method of the 16S rRNA gene sequences using (Unipro UGENE software, version 1.32.0. Branch lengths are displayed as age. The 16S rRNA gene sequences and tree of related species were downloaded from the High-Quality Ribosomal RNA database (SILVA).
3.3. The Strain S. rubidaea ED1 Tolerates Growth under Stressful Conditions
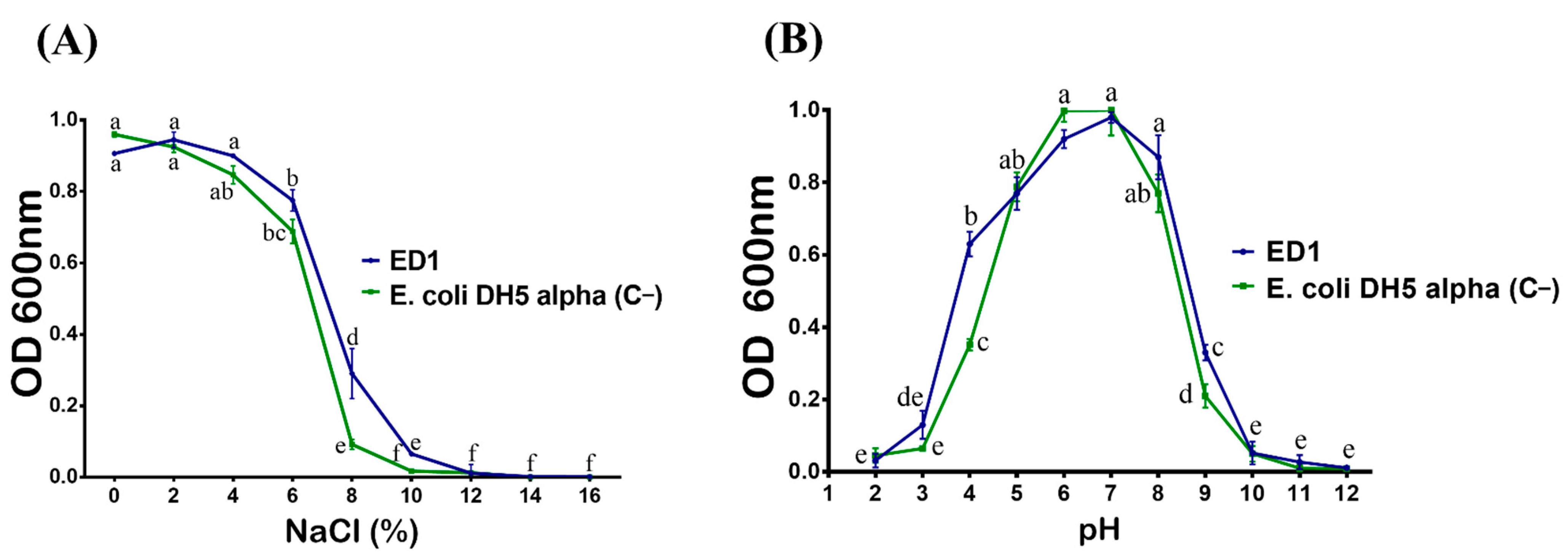
We first checker the tolerance of S. rubidaea ED1 to salt. Bacteria were plated on TSA containing various NaCl concentrations. We found that the ED1 strain grew up to 9% of NaCl, while the PGPR reference strain, B. licheniformis QA1, tolerated up to 11%. However, the growth arrest happened in E. coli DH5α at only 3%. In addition, ED1 is a mesophilic bacterium as its optimal growth ranged from 20 to 42 °C (Table 2). When grown on liquid TSB media with increasing NaCl concentrations, the maximum growth of the S. rubidaea ED1 strain was detected at 4% NaCl. However, although at reduced growth level, it tolerated up to 10% NaCl. Total growth inhibition occurred at 12% NaCl. In contrast to the ED1 strain, the growth of E. coli DH5α proportionally decreased as the concentration of NaCl increased (Figure 3A). Next, we monitored bacterial survival at different pH and found that the optimum growth of both S. rubidaea ED1 strain and E. coli DH5α occurred at pH 7. However, S. rubidaea ED1 could still grow in a range of pH from 4 to 9 (Figure 3B).

Table 2.
Summary of relevant extreme properties of isolated endophytic bacterium Serratia rubidaea ED1, the PGPR reference strain B. licheniformis QA1, and E. coli DH5α (C−).
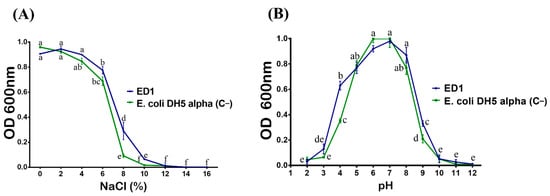
Figure 3.
Effect of NaCl (A) and pH (B) on the growth of the S. rubidaea ED1 strain. Media inoculated with Esherichia coli DH5α were used as a negative control (C−). The values represent means of three replicates (n = 3) ± SD. The different letters in superscript (a–f) indicate the statistically significant difference at 95% between treatments.
The ability of the S. rubidaea ED1 strain to grow on plates containing various trace metals concentrations was also evaluated. Because of their great interest, cadmium, copper, and nickel were chosen. We found that S. rubidaea ED1 supported CdSO4 at 2000 µg/mL, CuO4S.5H2O at 1000 µg/mL, and finally, N2NiO8 at 1000 µg/mL. For both the positive and negative control strains, maximum tolerances were seen at 300 µg/mL, 1000 µg/mL and 1000 µg/mL, of CdSO4, N2NiO8, and CuO4S, respectively (Table 2).
3.4. Effect of NaCl on P solubilization by Strain S. rubidaea ED1
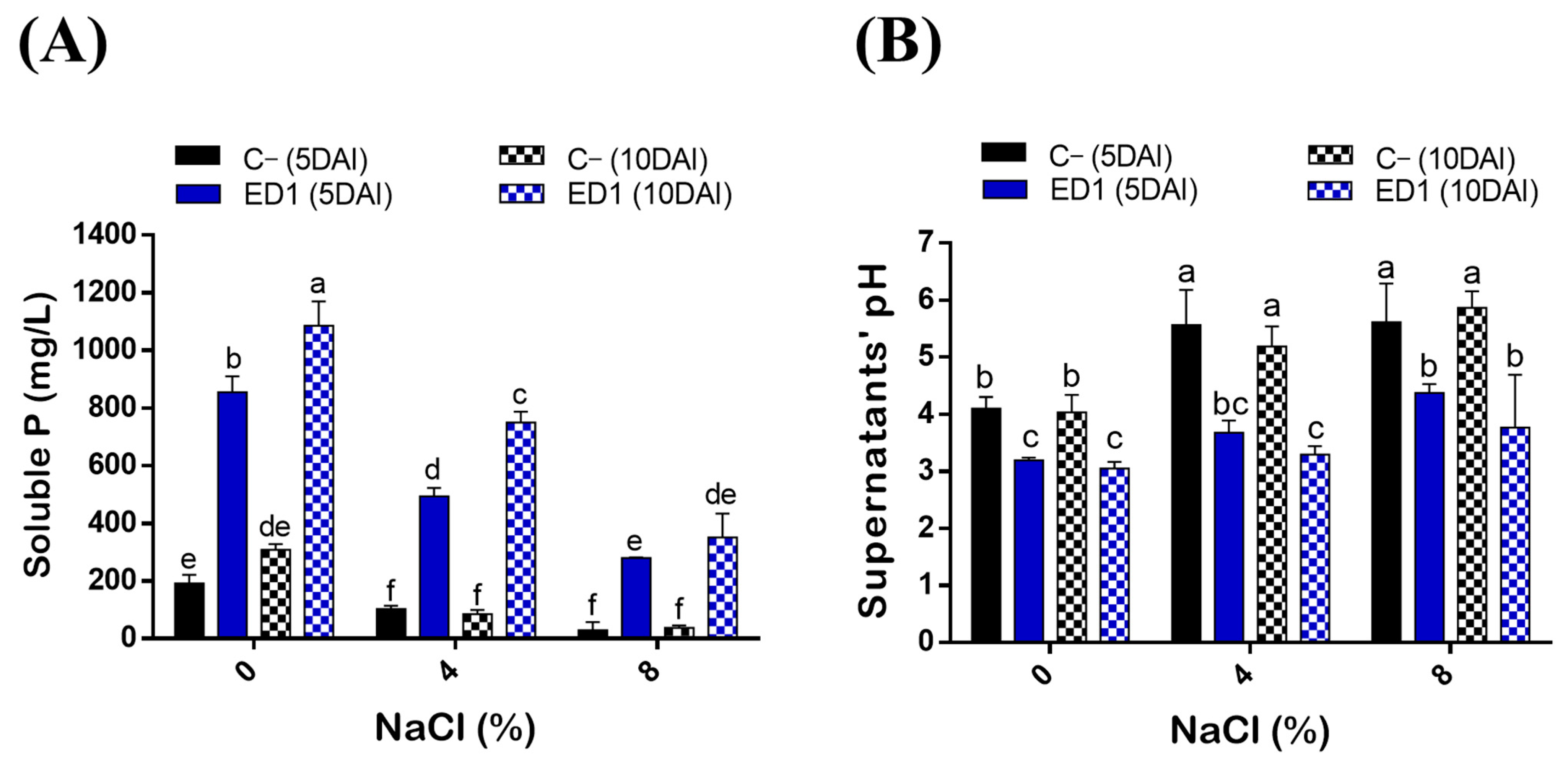
Salinity is considered as one of the main factors influencing P solubilization by bacteria [76]. Here, we assessed the ability of ED1 strain to solubilize hydroxyapatite at increasing NaCl final concentrations: 0%, 4%, and 8%. P solubilization by the S. rubidaea ED1 strain decreased following the increase of NaCl concentration (R2 = 0.87–0.99). In addition, P solubilization by ED1 was significantly increased over the incubation time from 5 to 10 DAI (Figure 4A, Table 3). Soluble PO4 levels significantly deceased following the increase of NaCl concentration (Figure 4A) with correlation coefficients of up to −0.99 (Table 3). The mineral P was solubilized at its maximum at 0% NaCl at 10DAI where it reached 1085.22 ± 84.34 mg/L by ED1 strain and 307.81 ± 20 mg/L by E. coli DH5α (C−). It turned out that compared to the S. rubidaea ED1 strain, 8% NaCl has a greater reducing effect on E. coli DH5α solubilizing capacity at 5DAI. Interestingly, the level of solubilized P under 8% NaCl is the same as the one obtained at 0% NaCl using ED1 strain. More precisely, soluble P reached 350.63 ± 82.40 mg/L in 8% NaCl medium using the S. rubidaea ED1 strain and 307.81 ± 20 mL/L in 0% NaCl medium using E. coli DH5α. In salty media, solubilized P by ED1 increased from 5 to 10DAI, while no statistically significant increase was noticed in E. coli DH5α (C−) (Figure 4A).
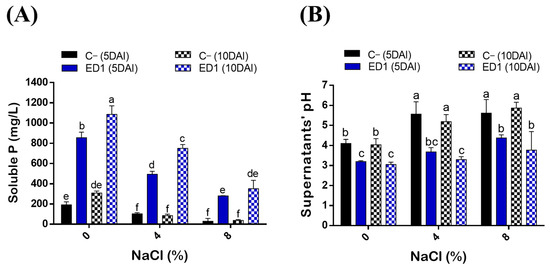
Figure 4.
Quantitative determination of solubilized P (PO4) in the National Botanical Research Institute’s phosphate (NBRIP) broth under salt stress conditions. (A) Solubilized P concentrations by the S. rubidaea ED1 strain and E. coli DH5α (C−) at 0%, 4%, and 8% NaCl final concentrations. (B) pH variation of final culture supernatants. DAI means day after incubation. The values represent means of replicates (n = 3) ± SD. The different letters in superscript (a–f) indicate the statistically significant difference at 95% between treatments.

Table 3.
Analysis of the variation of soluble P concentrations released by S. rubidaea ED1 and E. coli DH5α strain as a function of NaCl levels and pH values.
The solubilization of P by the endophyte strain was accompanied by a significant drop in pH of the culture supernatants in both saline and non-saline NBRIP media (Figure 4B). Highly negative correlations were noticed between soluble p values and final pH of the supernatants (r = 0.9–0.99, R2 = 0.82–0.98) (Table 3). The maximum pH decline, 3.04 ± 0.11, was observed using ED1 in the non-saline medium at 10DAI and it proportionally increased with NaCl concentrations over the incubation period. For instance, at 8% NaCl, the pH at 10DAI was 5.86 ± 0.29 in E. coli DH5α, and 3.76 ± 0.92 in S. rubidaea ED1 strain (Figure 4B).
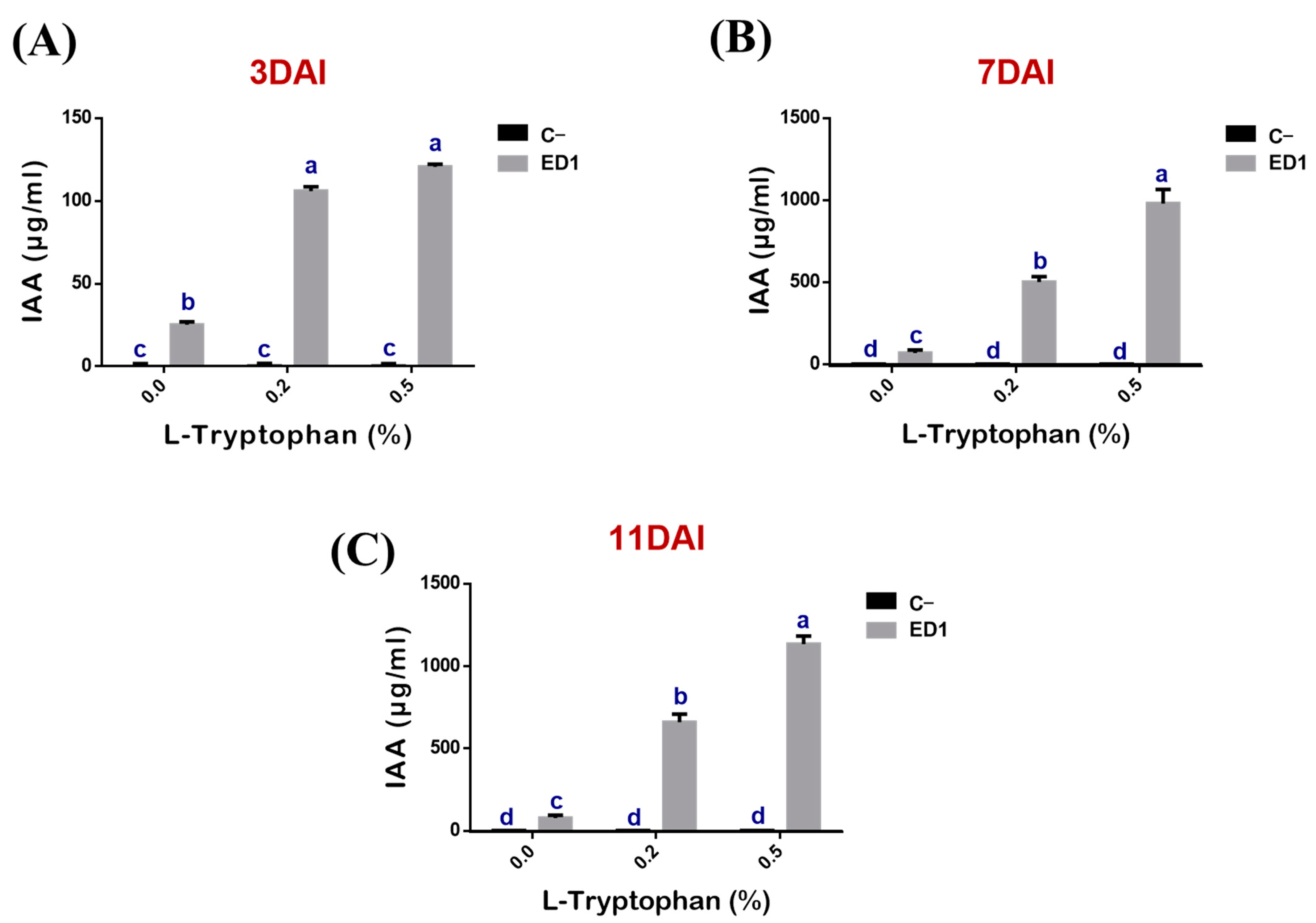
3.5. Indole Acetic Acid Production by the Serratia Rubidaea ED1 Strain Is Partly L-Tryptophan Dependent
The production of IAA by the S. Rubidaea ED1 strain correlated proportionally to added L-tryptophan concentrations (r = 0.88, 0.99, 0.98, R2 = 0.77, 0.99 and 0.96) and to period of incubation (r = 0.95, 0.99, 0.99, R2 = 0.91, 0.98 and 0.99) (Figure 5). IAA production was significantly higher using 0.5% L-tryptophan at day 11 (Figure 5C) compared to 0.2% L-tryptophan at day 7. In the negative control media, IAA did not exceed 0.17 ± 0.09 μg/mL over the three tested incubation periods (Figure 5A–C). Unexpectedly, IAA was produced even in the absence of L-tryptophan as the measured quantities were: 25.23 ± 1.81 μg/mL at day 3 (Figure 5A), 66.79 ± 18.88 μg/mL at day 7 (Figure 5B), and 76.47 ± 17.7 μg/mL at day 11 (Figure 5C).
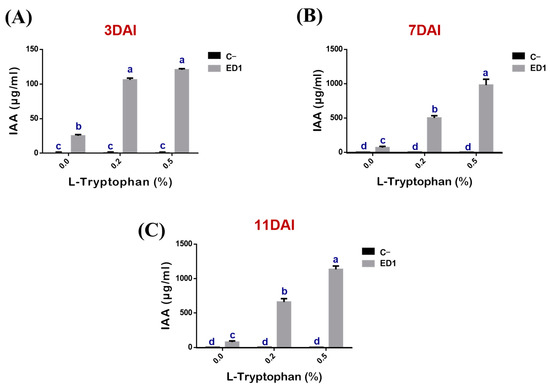
Figure 5.
Kinetic of indole acetic acid (IAA) production by S. rubidaea ED1 with or without L-tryptophan addition. IAA determination was performed after 3 (A), 7 (B), and 11 (C) days of incubation. Uninoculated media were used as a negative control (C−). The values represent means of three replicates (n = 3) ± SD. The different letters in superscript (a–d) indicate the statistically significant difference at 95% between treatments.
3.6. Strain Serratia rubidaea ED1 Exhibited Intrinsic Antibiotic Resistance
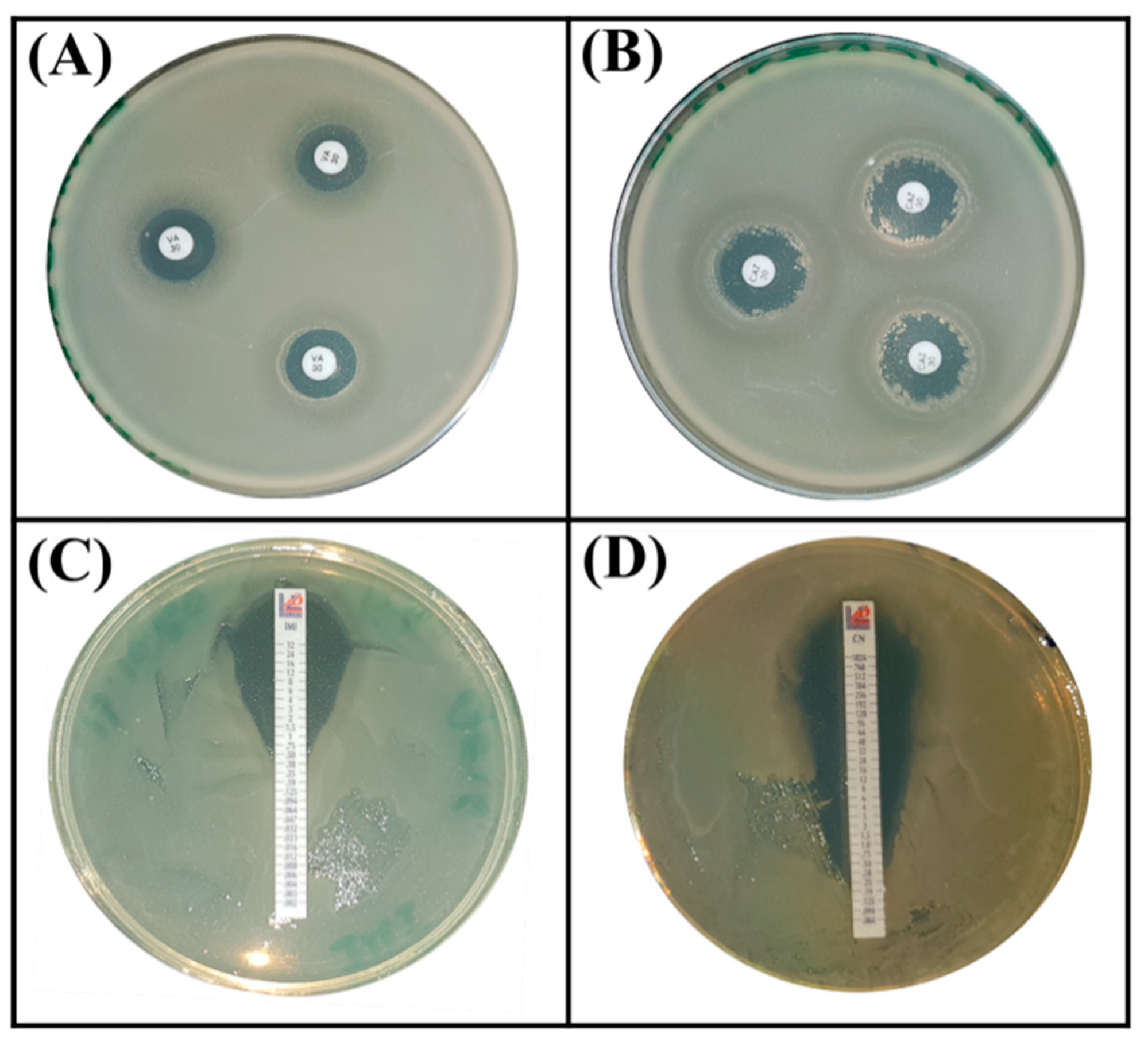
Next, we evaluated the intrinsic resistance phenotype to antibiotics by choosing those that are frequently used in humans to tackle infectious diseases. Based on the Zone Size Interpretative Chart (ZSIC) and MIC strips (Figure 6), it turned out that the S. rubidaea ED1 strain is resistant to several antibiotics such as ciprofloxacin, ertapenem, meropenem, ampicillin, chloramphenicol, tetracycline, and spectinomycin, while Bacillus atrophaeus S8 strain resists only ceftazidim and ertapenem (Table 4, Figure 6).
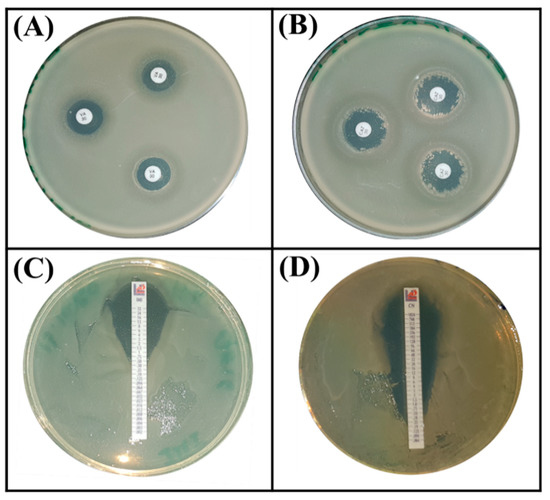
Figure 6.
Antibiotic resistance assays using Serratia rubidaea ED1 strain on MHA (Mueller Hinton Agar, BIOKAR Diagnostics, BEAUVAIS, France) plates supplemented by antibiotic disc method (A,B) and Etest (C,D). (A) vancomycin, (B) ceftazidime, (C) imipenem, (D) gentamycin.

Table 4.
Antibiotic resistance profile of the S. rubidaea ED1 strain using disc diffusion, Etest, and antibiotic amended agar methods.
3.7. Strain Serratia rubidaea ED1 Enhanced Germination Rate and Seedlings Growth under Salty Amendment
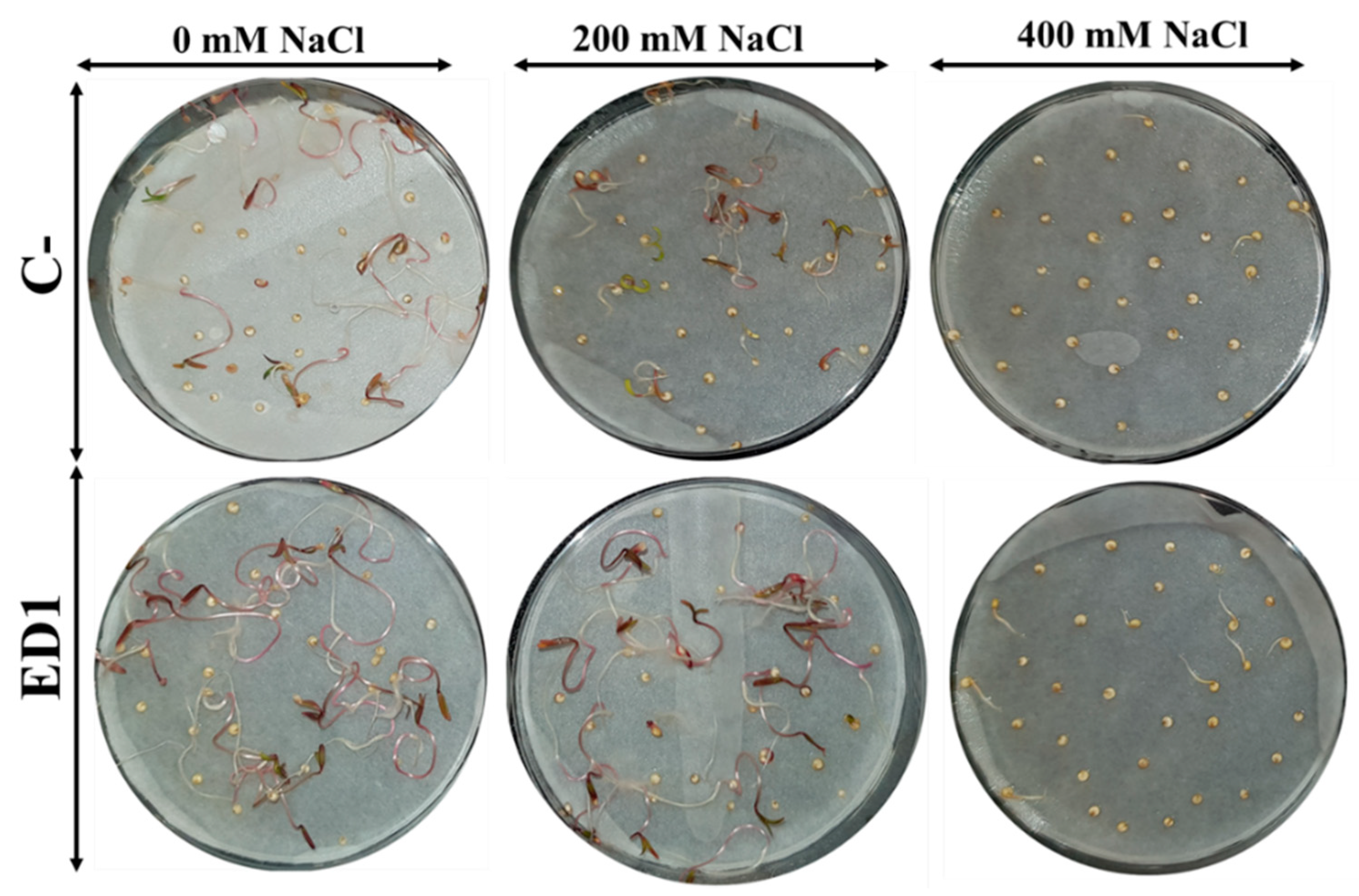
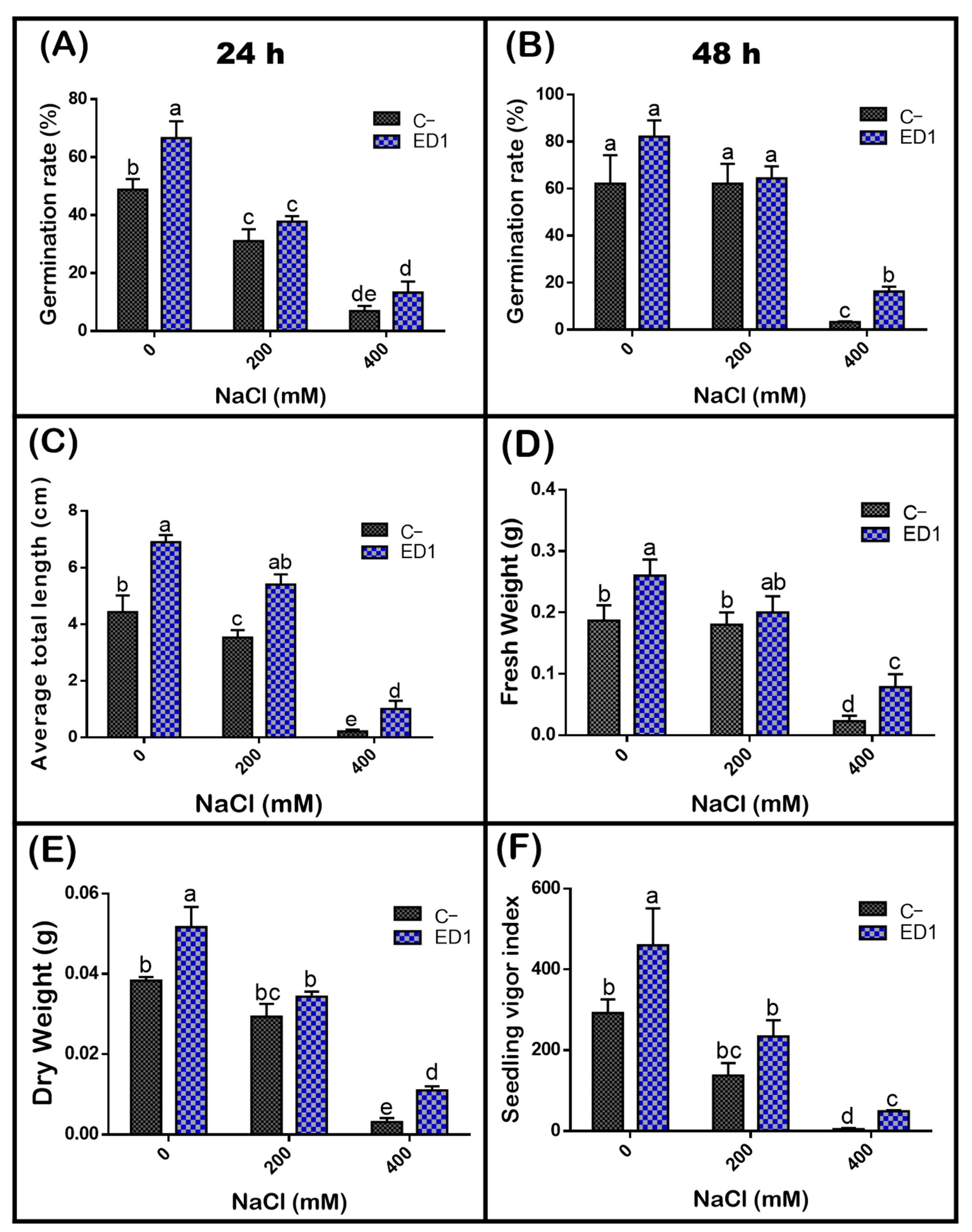
The quinoa seed germination rate of both inoculated and control seeds was gradually reduced along with an increasing of NaCl concentration, especially 24 h post incubation. The S. rubidaea ED1 strain significantly enhanced seed germination rate under non-saline (0 mM NaCl) and saline (400 mM or 2.32% NaCl) conditions by 36.35% (after 24 h incubation) and 390.39% (after 48 h incubation), respectively (Figure 7). Compared to non-saline treatment, 400 mM NaCl decreased seed germination by 58.87% in controls seeds and by 45.8% using S. rubidaea ED1 strain (Figure 8A,B, Table 5). At a moderate NaCl concentration (200 mM or 1.16% NaCl), the percentage of germination increased in a comparable manner to the control (Figure 8A,B). Inoculation with Serratia rubidaea ED1 increased the total length of seedlings under both non-saline and saline treatments. ED1 strain enhanced seedling length by 57.76, 52.97, and 376.19% under 0, 200, and 400 mM NaCl, respectively (Figure 8C, Table 5). Likewise, quinoa seedlings showed 34.78 and 253.39% increase in dry weights under 0 and 400 mM NaCl treatments, respectively. Finally, the vigor index was significantly affected using the ED1 strain, especially under 0 and 400 mM NaCl treatment (p < 0.05) (Figure 8F).
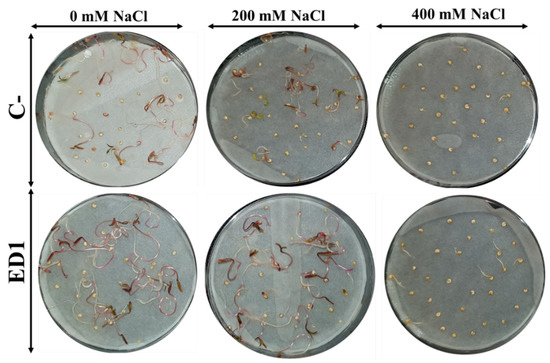
Figure 7.
Effect of the Serratia rubidaea ED1 strain on quinoa seed germination and early growth seedlings under saline treatments. Seeds inoculated with phosphate-buffered saline (PBS) were used as a negative control (C−).
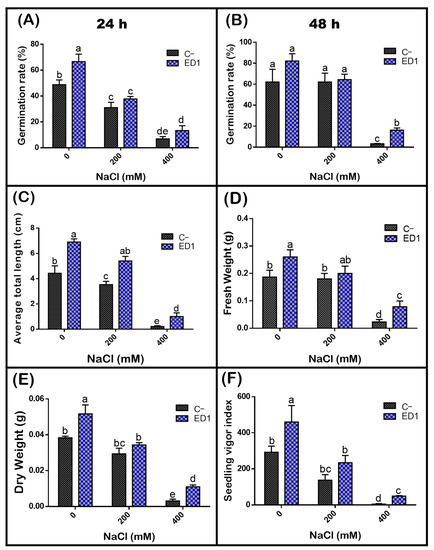
Figure 8.
Effect of the S. rubidaea ED1 strain on quinoa seed germination parameters under different salinity treatments (0, 200, and 400 mM NaCl) after 48 h of incubation at 25 °C. C− (negative control; Seeds inoculated with phosphate-buffered saline (PBS)), ED1 (Seed treated with the ED1 strain): (A) Germination rate after 24 h of incubation, (B) germination rate after 48 h of incubation, (C) average of total length of seedlings, (D) average of fresh weight of seedlings, (E) average of dry weight of seedlings, and (F) seedling vigor index. The values represent means of three replicates (n = 3) ± SD. The different letters in superscript (a–e) indicate the statistically significant difference at 95% between treatments.

Table 5.
Inoculation of quinoa seeds with the Serratia rubidaea ED1 strain enhanced the rate of seedling growth parameters.
4. Discussion
Alongside nitrogen (N) and potassium (K), phosphorus (P) is one of the most limiting nutrients for crop yields, growth, and development. P deficiency in plants is usually translated by inhibited stem and root development, poor flowering, lack of seed and fruit formation etc. In last decades, the use of PGPB-based bio-inoculants as an alternative to chemical fertilizers has emerged [77]. In the present study, we identified an endophytic S. rubidaea ED1 bacterium from quinoa roots endosphere, a plant endowed with many phyto-beneficial attributes facing extreme growth stresses. Some PGP results of Serratia rubidaea ED1 were expected as they were reported in Serratia rubidaea alongside S. plymuthica, S. liquefaciens, S. proteamaculans, and S. nematodiphila, isolated from various environments niches [78]. PGP attributes of Serratia rubidaea (NR114716) associated with Origanum plants was reported for IAA and siderophores production, P solubilization, ACC-deaminase activity, and N fixation [79]. In addition, this strain improved growth performance parameters and biochemicals, and was considered as a potential bio-factor to engineer the essential oil constituents from aromatic plants [79]. The first and foremost activity we tested here was mineral P and Zn solubilization. Although reports on P and Zn solubilization by bacterial endophytes are scarce, we showed that S. rubidaea ED1 strain solubilized both mineral compounds of P and Zn. Maximum Zn solubilization was recorded in ZnO compared to Zn3(PO4)2, and ZnCO3 (Table 1). The mechanisms underlying this specificity remain unknown and very recently, endophytic Serratia sp. ZoB14 has been screened for dual P and ZnO solubilization [80]. Next, we showed that S. rubidaea ED1 grew on a N-free solid medium supporting its oligonitrophilic feature (Table 1) as previously reported in Serratia liquefaciens and Serratia marcescens [81,82]. Indeed, oligonitrophilic bacteria have broad substrate specificities that allow them to maintain their structure in low N environments [83]. However, other environmental factors, such as soil pH, could greatly influence the oligonitrophilic function [84]. This confirms that Serratia rubidaea ED1 strain is more prone to oligonitrophilic conditions, e.g., plants’ endosphere.
Around the world, 20% of cultivated and 33% of irrigated arable lands are affected by high salinity [85]. Induced salt tolerance in plants has been shown to be associated with various PGPR strains [32,86,87,88]. We demonstrated here that Serratia rubidaea ED1 grew up to 10% NaCl (Figure 3). Taking advantage to this result, we next studied P solubilization under salty conditions. The quantity of orthophosphates (PO4) released from hydroxyapatite by S. rubidaea ED1 decreased with the increase of NaCl concentration. However, substantial amount of soluble P (up to 750 mg/L) was detected at 10DAI using 4% NaCl final concentration. Maximum soluble P level, 1085.22 ± 84.34 mg/L, was seen in 0% NaCl media at 10DAI (Figure 4). In salty media, the increased P release between days 5 and 10 suggests the adaptation of Serratia rubidaea ED1 to the salty medium, as previously reported for Bacillus megaterium [51]. Our result is in-line with the reported data on the endophytic Pseudomonas fluorescens that solubilized up to 1312 mg/L in non-saline NBRIP medium [46]. As expected, P solubilization was accompanied by a pH decrease which remained almost unchanged under salt treatment. This finding is likely attributed to organic acids production [89].
Bacteria from any environment are exposed to abiotic stress, exemplified by salinity, pH, temperature, trace elements, nutrient deficiency, and biotic stress such as toxic metabolites, pathogens, competing microorganisms [90]. To cope with these stresses, bacteria use several adaptative strategies to modify their signalization pathways [51]. Salt level is one of the main parameters affecting the growth rate and metabolism of microorganisms and several reports indicated the positive impact of halotolerant microorganisms on plant growth. At present, PGPB are suggested to enhance the productivity of plants facing salinity and can be exploited to sustain saline agroecosystems [20]. In this study, salt tolerance of S. rubidaea ED1 to NaCl was about 10%, while total growth arrest happened at 12% under hyper osmotic conditions (Figure 3). Salt-tolerant bacteria activate osmoadaptation pathways mainly by synthesizing compatible solutes and/or accumulation of K+ to overcome the Na+ ions associated toxicity [91]. The excess of sodium ions affects the bacterial enzymes activity [92]. In addition, bacterial exopolysaccharides (EPS), antioxidant enzymes, and ACC deaminase activity are among the strongest salt stress-adapting mechanisms [21].
The capacity of selected endophytic bacterium to solubilize unavailable nutrients such as P and Zn and grow in low nitrogen medium is supportive to further investigate its plant growth potential by boosting plant nutrition. To further confirm the halotolerance property of S. rubidaea ED1 strain, we performed a seed germination assay and revealed that inoculation of quinoa seeds enhanced seeds germination and seedlings growth under 400 mM (2.32%) NaCl. Noticeably, germination rate in both inoculated and non-inoculated seeds were inversely proportional to the increase of NaCl (Figure 7 and Figure 8), as previously reported [93]. Indeed, inoculation of quinoa seeds showed a prominent effect on vigor index (904.50% increase) (Table 5), pointing out the mitigation of salt stress in both assays. To our knowledge, this finding represents the first report stating that Serratia rubidaea promoted seeds germination and seedlings growth under salty treatments. Comparatively, a study on Ligustrum sinense seeds was conducted by increasing NaCl concentrations (0–500 mM) using strains: Isoptericola dokdonensis, Arthrobacter soli, Streptomyces pactum, and Bacillus flexus. The latter induced seeds germination by reaching 100% under 300–500 mM NaCl treatment [93]. Sorghum seeds inoculated with endorhizospheric Serratia sp. promoted seedlings growth [94]. The increase in seeds germination is likely attributed to the production of phytohormones such IAA [95]. Other studies reported the promoting role of Bacillus and Pseudomonas in seed germination, root development, and plant growth [96] of various crops including wheat, rice, and sugarcane [97]. Indeed, stimulation of seeds germination in the presence of IAA producing bacteria has been described [75]. Here, IAA production by Serratia rubidaea ED1 was seen even in the absence of L-Tryptophan supplementation (Figure 5). However, in L-tryptophan-supplemented broth, production of IAA increased significantly (Figure 5), corroborating previous finding on Serratia species including S. rubidaea highlighting that IAA signaling pathways are either auxin-dependent, auxin-independent, or tryptophan-independent [78,98]. Besides, other halotolerant bacteria species isolated from halophytic plant in coastal soils in Korea, enhanced plant growth under saline stress. In this case, plant growth was linked to the activity of the ACC deaminase that reduced ethylene production [99]. Additionally, it was reported that the halophyte Salicornia brachiate induced plant growth by withstanding a high level of salt [100]. More mechanisms by which microorganisms improve physiological response of plants under salt stress were described [101,102]. These include osmolytes accumulation which regulate water homeostasis, hormonal root–shoot signaling inducing salt tolerance in plants, modulation of the source-sink relationships for plant energetics, nutrients and toxic ions uptake by root by modifying host plant physiology, physical barriers around the roots, or by reducing foliar accumulation of toxic ions [101,102].
In another part of this study, we checked the ability of S. rubidaea ED1 to resist pH and to withstand trace elements. It was found to grow from pH 4 to 9 but at a significantly low rate compared to optimal pH 7 (Figure 3). Compared to other PGP strains such as Bacillus megaterium, Staphylococcus haemolyticus, and Bacillus licheniformis [52], ED1 strain tolerates slightly higher acidity, suggesting its ability to grow in acidic soils [103]. Accumulation of trace elements in the environment represent a major concern to public health and agriculture [104]. We found that ED1 strain survives at 2 mg/L of CdSO4, and 1 mg/L of either CuO4S.5H2O or N2NiO8 (Table 2). In the same range, Planomicrobium chinense and Bacillus cereus tolerate Cd (1.5 mg/L) and Ni (1 mg/L) by accumulating heavy metals upon chelating agents release that affect their mobility and availability which consequently enhance phytoremediation and nutrient transformation [105]. For example, Serratia marcescens was shown to assist phytoremediation of metals via the reduction of the toxic chromium (VI) to the less soluble and less toxic chromium (III) [106].
To be an effective biofertilizer, PGPR must survive, resist, and colonize root surface and/or interior to establish itself at population densities required to induce beneficial effects on the host plant. We assessed the intrinsic antibiotic resistance of selected strain on MH agar plates. Surprisingly, this topic is very often neglected in agriculture. Here, we report that S. rubidaea ED1 is resistant to seven antibiotics, frequently used to treat infection diseases in humans such as ciprofloxacin, ertapenem, meropenem, ampicillin, chloramphenicol, tetracycline, and spectinomycin (Table 4). Our finding supports previous findings for another Serratia rubidaea strain conferring resistance to ampicillin, tetracycline, gentamycin, penicillin, vancomycin, and streptomycin [107,108,109]. The novelty here is that the list of antibiotics can be extended to five additional antibiotics, namely, ciprofloxacin, ertapenem, meropenem, chloramphenicol, and spectinomycin. It is well established that soil, food, and water are the habitats of the Serratia rubidaea strain. Interestingly, it was also found among clinical specimens as a nonpathogenic bacterium [110]. However, infection by Serratia rubidaea was reported in immunocompromised patients [111,112]. In 2016, the first complete sequence of the genome of Serratia rubidaea (CP014474) was isolated from a patient in China [113]. Sequence analysis revealed the presence of several antibiotic resistance genes (ARG) encoding metallo-beta-lactamase, chloramphenicol, and aminoglycoside phosphotransferase. Other Serratia strains such as S. marcescens FGI94, S. fonticola DSM 4576, and S. ficaria showed also multi-resistances to antibiotics [114,115,116]. Thus, the emerging question is how to consider the two-blade weapons of PGP bacterial strains exhibiting intrinsic resistance to several antibiotics. On the one hand, they can be used either as markers to assess bacteria survival either in vitro or in vivo [117,118], and to help bacteria surviving and competing in native and open microbial environmental niches [35]. On the other hand, their use may represent potential risks by disseminating ARG to neighboring bacteria, plant, animals, and later, to humans [119]. Having said that, the alterable functional and adaptive behavior of resistant strains originating from different environments can differ in genome expression profiles, accessory genome sequences, antibiotic resistance pattern, and virulence activities. In addition, polyphasic approaches, mainly multi-locus sequence typing, discriminatory genotyping methods such as recN sequencing, and genome hybridization, showed that a same strain (e.g., Pseudomonas aeruginosa) could be a singleton among a large group of closely related strains, clustering distantly from the typical clinical isolates [90,120,121]. The occurrence of S. rubidaea in human specimens is rare, and there are no data suggesting that it is of a clinical significance [122]. Recently, rhamnolipid biosurfactant production by Serratia rubidaea SNAU02 was shown to act as an anti-Fusarium wilt of eggplant [123]. Production of prodigiosin and pyrrolnitrin by Serratia rubidaea C27 represents the main antifungal activity mechanisms [124]. In addition, S. rubidaea species were isolated from infected pepper fruits as a causal agent of splotches. This phyto-pathogenicity was attributed to the liberation of extracellular enzymes playing a role during the host infection including protease, lipase, polygalacturonase, and alkaline phosphatase that degrade plant cell wall and membrane constituents. However, their production is inducible and usually dependent on several environmental factors [125].
5. Conclusions
The present study corroborates previous findings and bring new insights on the role of endophytic bacteria in mitigating stresses in agriculture. Our results shed light on the role of the newly isolated S. rubidaea ED1 strain to mitigate salt stress. Further investigations on plant growth promotion under large-scale experiments would be paramount to uncover various mechanisms and potential applications of Serratia rubidaea ED1 in agriculture. Besides, there is a growing need to remediate the heavy metals from contaminated soils using various microbial accumulators capable of growing in heavy metals contaminated soils [126]. Hence, to follow-up the present work, more studies should address the potential application of S. rubidaea ED1 in sustainable rehabilitation of heavy metals contaminated arable lands and to spotlight the underpinning mechanisms. Lastly, our work raises questions on a neglected issue related to the risk in using PGPR bacteria conferring resistance to many antibiotics for crop improvement.
Author Contributions
Conceptualization, L.B. & A.A.; Funding acquisition, L.B.; Investigation, I.M. and L.B.; Methodology, I.M. & L.B., Project administration, L.B.; Supervision, L.B. & M.H.; Validation L.B. & A.A.; Writing—original draft, I.M.; Writing—review & editing, A.A. & L.B. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by welcome grants from the Mohammed VI Polytechnic University (UM6P) of Ben Guerir, Morocco.
Acknowledgments
We thank Fatiha Laalouhmi for technical assistance and Mohamed Hijri for helpful discussion and facilities.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rosenblueth, M.; Martínez-Romero, E. Bacterial endophytes and their interactions with hosts. Mol. Plant-Microbe Interact. 2006, 19, 827–837. [Google Scholar] [CrossRef]
- Wilson, D. Endophyte: The evolution of a term, and clarification of its use and definition. Oikos 1995, 274–276. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial endophyte colonization and distribution within plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; DeBolt, S.; Dreyer, J.; Scott, D.; Williams, M.A. Characterization of culturable bacterial endophytes and their capacity to promote plant growth from plants grown using organic or conventional practices. Front. Plant Sci. 2015, 6, 490. [Google Scholar] [CrossRef]
- Ma, Y.; Prasad, M.N.V.; Rajkumar, M.; Freitas, H. Plant growth promoting rhizobacteria and endophytes accelerate phytoremediation of metalliferous soils. Biotechnol. Adv. 2011, 29, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.M.; Schenk, P.M. Inner plant values: Diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 2017, 8, 2552. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Medina, A.; Flors, V.; Heil, M.; Mauch-Mani, B.; Pieterse, C.M.J.; Pozo, M.J.; Ton, J.; van Dam, N.M.; Conrath, U. Recognizing Plant Defense Priming. Trends Plant Sci. 2016, 21, 818–822. [Google Scholar] [CrossRef]
- Chanway, C. Inoculation of tree roots with plant growth promoting soil bacteria: An emerging technology for reforestation. For. Sci. 1997, 43, 99–112. [Google Scholar] [CrossRef]
- Bent, E.; Chanway, C.P. The growth-promoting effects of a bacterial endophyte on lodgepole pine are partially inhibited by the presence of other rhizobacteria. Can. J. Microbiol. 1998, 44, 980–988. [Google Scholar] [CrossRef]
- McInroy, J.; Kloepper, J. Novel Bacterial Taxa Inhabiting Internal Tissues of Sweet Corn and Cotton; CSIRO: Melbourne, Australia, 1994; pp. 190–238. [Google Scholar]
- Liu, H.; Carvalhais, L.C.; Schenk, P.M.; Dennis, P.G. Effects of jasmonic acid signalling on the wheat microbiome differ between body sites. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Marques, J.M.; da Silva, T.F.; Vollú, R.E.; de Lacerda, J.R.M.; Blank, A.F.; Smalla, K.; Seldin, L. Bacterial endophytes of sweet potato tuberous roots affected by the plant genotype and growth stage. Appl. Soil Ecol. 2015, 96, 273–281. [Google Scholar] [CrossRef]
- Ferrando, L.; Fernández Scavino, A. Strong shift in the diazotrophic endophytic bacterial community inhabiting rice (Oryza sativa) plants after flooding. FEMS Microbiol. Ecol. 2015, 91. [Google Scholar] [CrossRef] [PubMed]
- Magnani, G.; Cruz, L.; Weber, H.; Bespalhok, J.; Daros, E.; Baura, V.; Yates, M.; Monteiro, R.; Faoro, H.; Pedrosa, F. Culture-independent analysis of endophytic bacterial communities associated with Brazilian sugarcane. Genet. Mol. Res 2013, 12, 4549–4558. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira Costa, L.E.; de Queiroz, M.V.; Borges, A.C.; de Moraes, C.A.; de Araújo, E.F. Isolation and characterization of endophytic bacteria isolated from the leaves of the common bean (Phaseolus vulgaris). Braz. J. Microbiol. 2012, 43, 1562–1575. [Google Scholar] [CrossRef]
- Ortuño, N.; Claros, M.; Gutiérrez, C.; Angulo, M.; Castillo, J. Bacteria associated with the cultivation of quinoa in the Bolivian Altiplano and their biotechnological potential. Rev. Agric. 2014, 53, 53–61. [Google Scholar]
- Reinhold-Hurek, B.; Bünger, W.; Burbano, C.S.; Sabale, M.; Hurek, T. Roots shaping their microbiome: Global hotspots for microbial activity. Annu. Rev. Phytopathol. 2015, 53, 403–424. [Google Scholar] [CrossRef]
- Ramos-Solano, B.; García, J.A.L.; Garcia-Villaraco, A.; Algar, E.; Garcia-Cristobal, J.; Mañero, F.J.G. Siderophore and chitinase producing isolates from the rhizosphere of Nicotiana glauca Graham enhance growth and induce systemic resistance in Solanum lycopersicum L. Plant Soil 2010, 334, 189–197. [Google Scholar] [CrossRef]
- Bunsangiam, S.; Sakpuntoon, V.; Srisuk, N.; Ohashi, T.; Fujiyama, K.; Limtong, S. Biosynthetic pathway of indole-3-acetic acid in basidiomycetous yeast Rhodosporidiobolus fluvialis. Mycobiology 2019, 47, 292–300. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.; Bellingrath-Kimura, S.D.; Mishra, J.; Arora, N.K. Salt-Tolerant Plant Growth Promoting Rhizobacteria for Enhancing Crop Productivity of Saline Soils. Front. Microbiol. 2019, 10, 2791. [Google Scholar] [CrossRef]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ. 2002, 25, 239–250. [Google Scholar] [CrossRef]
- Upadhyay, S.; Singh, J.; Singh, D. Exopolysaccharide-producing plant growth-promoting rhizobacteria under salinity condition. Pedosphere 2011, 21, 214–222. [Google Scholar] [CrossRef]
- Ashraf, M.; Athar, H.R.; Harris, P.J.C.; Kwon, T.R. Some Prospective Strategies for Improving Crop Salt Tolerance. In Advances in Agronomy; Academic Press: Cambridge, MA, USA, 2008; Volume 97, pp. 45–110. [Google Scholar]
- Niu, X.; Song, L.; Xiao, Y.; Ge, W. Drought-Tolerant Plant Growth-Promoting Rhizobacteria Associated with Foxtail Millet in a Semi-arid Agroecosystem and Their Potential in Alleviating Drought Stress. Front. Microbiol. 2018, 8. [Google Scholar] [CrossRef]
- Bashan, Y. Inoculants of plant growth-promoting bacteria for use in agriculture. Biotechnol. Adv. 1998, 16, 729–770. [Google Scholar] [CrossRef]
- Cassán, F.; Maiale, S.; Masciarelli, O.; Vidal, A.; Luna, V.; Ruiz, O. Cadaverine production by Azospirillum brasilense and its possible role in plant growth promotion and osmotic stress mitigation. Eur. J. Soil Biol. 2009, 45, 12–19. [Google Scholar] [CrossRef]
- Bal, H.B.; Nayak, L.; Das, S.; Adhya, T.K. Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant Soil 2013, 366, 93–105. [Google Scholar] [CrossRef]
- Goswami, D.; Dhandhukia, P.; Patel, P.; Thakker, J.N. Screening of PGPR from saline desert of Kutch: Growth promotion in Arachis hypogea by Bacillus licheniformis A2. Microbiol. Res. 2014, 169, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, S.; Singh, D. Effect of salt-tolerant plant growth-promoting rhizobacteria on wheat plants and soil health in a saline environment. Plant Biol. 2015, 17, 288–293. [Google Scholar] [CrossRef]
- Bal, H.B.; Das, S.; Dangar, T.K.; Adhya, T.K. ACC deaminase and IAA producing growth promoting bacteria from the rhizosphere soil of tropical rice plants. J. Basic Microbiol. 2013, 53, 972–984. [Google Scholar] [CrossRef] [PubMed]
- Mahdi, I.; Fahsi, N.; Hafidi, M.; Allaoui, A.; Biskri, L. Plant Growth Enhancement using Rhizospheric Halotolerant Phosphate Solubilizing Bacterium Bacillus licheniformis QA1 and Enterobacter asburiae QF11 Isolated from Chenopodium quinoa Willd. Microorganisms 2020, 8, 948. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.P.; Jha, P.N. Alleviation of salinity-induced damage on wheat plant by an ACC deaminase-producing halophilic bacterium Serratia sp. SL-12 isolated from a salt lake. Symbiosis 2016, 69, 101–111. [Google Scholar] [CrossRef]
- Wellington, E.M.H.; Boxall, A.B.A.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W.; et al. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef]
- Cray, J.A.; Bell, A.N.; Bhaganna, P.; Mswaka, A.Y.; Timson, D.J.; Hallsworth, J.E. The biology of habitat dominance; can microbes behave as weeds? Microb. Biotechnol. 2013, 6, 453–492. [Google Scholar] [CrossRef] [PubMed]
- Vega-Galvez, A.; Miranda, M.; Vergara, J.; Uribe, E.; Puente, L.; Martinez, E.A. Nutrition facts and functional potential of quinoa (Chenopodium quinoa willd.), an ancient Andean grain: A review. J. Sci. Food Agric. 2010, 90, 2541–2547. [Google Scholar] [CrossRef] [PubMed]
- Benlhabib, O.; Jacobsen, S.-E.; Jellen, E.N.; Maughan, P.J.; Choukr-Allah, R. Status of quinoa production and research in Morocco. In State of the Art Report on Quinoa around the World in 2013; Bazile, D., Bertero, H.D., Nieto, C., Eds.; Food and Agriculture Organization of the United Nations Romz: Quebec City, QC, Canada, 2015; pp. 178–491. [Google Scholar]
- Jacobsen, S.-E.; Mujica, A.; Jensen, C. The resistance of quinoa (Chenopodium quinoa Willd.) to adverse abiotic factors. Food Rev. Int. 2003, 19, 99–109. [Google Scholar] [CrossRef]
- Nowak, V.; Du, J.; Charrondiere, U.R. Assessment of the nutritional composition of quinoa (Chenopodium quinoa Willd.). Food Chem. 2016, 193, 47–54. [Google Scholar] [CrossRef]
- Jacobsen, S.-E. The worldwide potential for quinoa (Chenopodium quinoa Willd.). Food Rev. Int. 2003, 19, 167–177. [Google Scholar] [CrossRef]
- Filho, A.M.M.; Pirozi, M.R.; Borges, J.T.D.S.; Pinheiro Sant’Ana, H.M.; Chaves, J.B.P.; Coimbra, J.S.D.R. Quinoa: Nutritional, functional, and antinutritional aspects. Crit. Rev. Food Sci. Nutr. 2017, 57, 1618–1630. [Google Scholar] [CrossRef]
- Hirich, A.; Choukr-Allah, R.; Jacobsen, S.-E. Quinoa in Morocco—Effect of Sowing Dates on Development and Yield. J. Agron. Crop Sci. 2014, 200. [Google Scholar] [CrossRef]
- Araújo, W.; Lima, A.D.S.; Azevedo, J.; Marcon, J.; Sobral, J.; Lacava, P. Manual: Isolamento de microrganismos endofíticos. Piracicaba Calq 2002, 1, 86. [Google Scholar]
- González-Teuber, M.; Vilo, C.; Bascuñán-Godoy, L. Molecular characterization of endophytic fungi associated with the roots of Chenopodium quinoa inhabiting the Atacama Desert, Chile. Genom. Data 2017, 11, 109–112. [Google Scholar] [CrossRef]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Oteino, N.; Lally, R.D.; Kiwanuka, S.; Lloyd, A.; Ryan, D.; Germaine, K.J.; Dowling, D.N. Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 2015, 6, 745. [Google Scholar] [CrossRef]
- Hill, J.E.; Hemmingsen, S.M.; Town, J.R. Strong PCR Primers and Primer Cocktails. U.S. Patent 7507535B2, 24 March 2009. [Google Scholar]
- Oliver, F. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 2017. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; Team, T.U. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Patel, K.S.; Naik, J.H.; Chaudhari, S.; Amaresan, N. Characterization of culturable bacteria isolated from hot springs for plant growth promoting traits and effect on tomato (Lycopersicon esculentum) seedling. Comptes Rendus Biol. 2017, 340, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Thant, S.; Aung, N.; Aye, O. Phosphate solubilization of Bacillus megaterium isolated from non-saline soils under salt stressed conditions. J. Bacteriol. Mycol. Open Access 2018, 6, 335–341. [Google Scholar] [CrossRef]
- Biswas, J.; Banerjee, A.; Rai, M.; Naidu, R.; Biswas, B.; Vithanage, M.; Chandra Dash, M.; Sarkar, S.; Meers, E. Potential application of selected metal resistant phosphate solubilizing bacteria isolated from the gut of earthworm (Metaphire posthuma) in plant growth promotion. Geoderma 2018, 330, 117–124. [Google Scholar] [CrossRef]
- Jensen, H. Nonsymbiotic nitrogen fixation. Soil Nitrogen 1965, 10, 436–480. [Google Scholar] [CrossRef]
- Ehmann, A. The van URK-Salkowski reagent—A sensitive and specific chromogenic reagent for silica gel thin-layer chromatographic detection and identification of indole derivatives. J. Chromatogr. A 1977, 132, 267–276. [Google Scholar] [CrossRef]
- Leveau, J.H.; Lindow, S.E. Utilization of the plant hormone indole-3-acetic acid for growth by Pseudomonas putida strain 1290. Appl. Environ. Microbiol. 2005, 71, 2365–2371. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, A.; Muralidharan, G. Assessment of zinc solubilizing potentiality of Acinetobacter sp. isolated from rice rhizosphere. Eur. J. Soil Biol. 2016, 76, 1–8. [Google Scholar] [CrossRef]
- Khanghahi, M.Y.; Ricciuti, P.; Allegretta, I.; Terzano, R.; Crecchio, C. Solubilization of insoluble zinc compounds by zinc solubilizing bacteria (ZSB) and optimization of their growth conditions. Environ. Sci. Pollut. Res. 2018, 25, 25862–25868. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Cappuccino, J.G.; Sherman, N. Microbiology: A Laboratory ManualA Laboratory Manual, 3rd ed.; Benjamin/Cumming Pub. Co.: New York, NY, USA, 1992. [Google Scholar]
- Chrouqi, L.; Lahcen, O.; Jadrane, I.; Koussa, T.; Alfeddy, M.N. Screening of soil rhizobacteria isolated from wheat plants grown in the Marrakech region (Morocco, North Africa) for plant growth promoting activities. JMES 2017, 8, 3382–3390. [Google Scholar]
- Kasana, R.C.; Salwan, R.; Dhar, H.; Dutt, S.; Gulati, A. A rapid and easy method for the detection of microbial cellulases on agar plates using gram’s iodine. Curr. Microbiol. 2008, 57, 503–507. [Google Scholar] [CrossRef]
- Kavitha, T.; Nelson, R.; Jesi, S.J. Screening of rhizobacteria for plant growth promoting traits and antifungal activity against charcoal rot pathogen Macrophomina phaseolina. Int. J. Pharma Bio Sci. 2013, 4, B-177–B-186. [Google Scholar]
- Smibert, R. Phenotypic characterization. Methods Gen. Mol. Bacteriol. 1994, 16, 3–11. [Google Scholar]
- Armalytė, J.; Skerniškytė, J.; Bakienė, E.; Krasauskas, R.; Šiugždinienė, R.; Kareivienė, V.; Kerzienė, S.; Klimienė, I.; Sužiedėlienė, E.; Ružauskas, M. Microbial Diversity and Antimicrobial Resistance Profile in Microbiota From Soils of Conventional and Organic Farming Systems. Front. Microbiol. 2019, 10, 892. [Google Scholar] [CrossRef]
- Syal, K.; Mo, M.; Yu, H.; Iriya, R.; Jing, W.; Guodong, S.; Wang, S.; Grys, T.E.; Haydel, S.E.; Tao, N. Current and emerging techniques for antibiotic susceptibility tests. Theranostics 2017, 7, 1795–1805. [Google Scholar] [CrossRef]
- Taylor, R.G.; Walker, D.C.; McInnes, R.R. E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res. 1993, 21, 1677–1678. [Google Scholar] [CrossRef]
- Bauer, A. Antibiotic susceptibility testing by a standardized single disc method. Am. J. Clin. Pathol. 1966, 45, 149–158. [Google Scholar] [CrossRef]
- Ryan, K.J.; Schoenknecht, F.D.; Kirby, W.M. Disc sensitivity testing. Hosp. Pract. 1970, 5, 91–100. [Google Scholar] [CrossRef]
- Fuchs, P.C.; Barry, A.L.; Brown, S.D. Selection of zone size interpretive criteria for disk diffusion susceptibility tests of three antibiotics against Streptococcus pneumoniae, using the New Guidelines of the National Committee for Clinical Laboratory Standards. Antimicrob. Agents Chemother. 2002, 46, 398–401. [Google Scholar] [CrossRef]
- Cervantes-Vega, C.; Chavez, J.; Córdova, N.; Amador, J.V. Resistance to metals by Pseudomonas aeruginosa clinical isolates. Microbios 1986, 48, 159–163. [Google Scholar]
- Khan, N.; Zandi, P.; Ali, S.; Mehmood, A.; Adnan Shahid, M.; Yang, J. Impact of salicylic acid and PGPR on the drought tolerance and phytoremediation potential of Helianthus annus. Front. Microbiol. 2018, 9, 2507. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.; Ghosh, P.K.; Pramanik, K.; Mitra, S.; Soren, T.; Pandey, S.; Mondal, M.H.; Maiti, T.K. A halotolerant Enterobacter sp. displaying ACC deaminase activity promotes rice seedling growth under salt stress. Res. Microbiol. 2018, 169, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.H.; Gururani, M.A.; Chun, S.C. Isolation and characterization of plant growth promoting endophytic diazotrophic bacteria from Korean rice cultivars. Microbiol. Res. 2014, 169, 83–98. [Google Scholar] [CrossRef] [PubMed]
- Bybordi, A. The influence of salt stress on seed germination, growth and yield of canola cultivars. Not. Bot. Horti Agrobot. Cluj-Napoca 2010, 38, 128–133. [Google Scholar] [CrossRef]
- Islam, S.; Mannan Akanda, A.; Prova, A.; Islam, T.; Hossain, M. Isolation and Identification of Plant Growth Promoting Rhizobacteria from Cucumber Rhizosphere and Their Effect on Plant Growth Promotion and Disease Suppression. Front. Microbiol. 2016, 6, 1360. [Google Scholar] [CrossRef]
- Usha, S.; Padmavathi, T. Phosphate solubilizers from the rhizosphere of Piper nigrum L. in Karnataka, India. Chil. J. Agric. Res. 2012, 72, 397–403. [Google Scholar] [CrossRef]
- Welbaum, G.E.; Sturz, A.V.; Dong, Z.; Nowak, J. Managing soil microorganisms to improve productivity of agro-ecosystems. Crit. Rev. Plant Sci. 2004, 23, 175–193. [Google Scholar] [CrossRef]
- Kshetri, L.; Naseem, F.; Pandey, P. Role of Serratia sp. as Biocontrol Agent and Plant Growth Stimulator, with Prospects of Biotic Stress Management in Plant. In Plant Growth Promoting Rhizobacteria for Sustainable Stress Management; Springer: Berlin/Heidelberg, Germany, 2019; pp. 169–200. [Google Scholar] [CrossRef]
- Alraey, D.A.; Haroun, S.A.; Omar, M.N.; Abd-ElGawad, A.M.; El-Shobaky, A.M.; Mowafy, A.M. Fluctuation of essential oil constituents in Origanum syriacum subsp. sinaicum in response to plant growth promoting bacteria. J. Essent. Oil Bear. Plants 2019, 22, 1022–1033. [Google Scholar] [CrossRef]
- Sabu, R.; Aswani, R.; Jishma, P.; Jasim, B.; Mathew, J.; Radhakrishnan, E.K. Plant Growth Promoting Endophytic Serratia sp. ZoB14 Protecting Ginger from Fungal Pathogens. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2019, 89, 213–220. [Google Scholar] [CrossRef]
- Lee, S.; Flores-Encarnacion, M.; Contreras-Zentella, M.; Garcia-Flores, L.; Escamilla, J.; Kennedy, C. Indole-3-acetic acid biosynthesis is deficient in Gluconacetobacter diazotrophicus strains with mutations in cytochrome c biogenesis genes. J. Bacteriol. 2004, 186, 5384–5391. [Google Scholar] [CrossRef]
- Zelaya-Molina, L.X.; Hernández-Soto, L.M.; Guerra-Camacho, J.E.; Monterrubio-López, R.; Patiño-Siciliano, A.; Villa-Tanaca, L.; Hernández-Rodríguez, C. Ammonia-Oligotrophic and Diazotrophic Heavy Metal-Resistant Serratia liquefaciens Strains from Pioneer Plants and Mine Tailings. Microb. Ecol. 2016, 72, 324–346. [Google Scholar] [CrossRef]
- Yang, R.; Li, J.; Wei-Xie, L.; Shao, L. Oligotrophic Nitrification and Denitrification Bacterial Communities in a Constructed Sewage Treatment Ecosystem and Nitrogen Removal of Delftia tsuruhatensis NF4. Pol. J. Microbiol. 2020, 69, 99–108. [Google Scholar] [CrossRef]
- Gorzala, G.; Jablonska-Gorzala, D.; Chojnicki, J.; Gozdowski, D.; Russel, S. Influence of soil pH modification on the number of oligonitrophilic yeast. Rocz. Glebozn. 2003, 54, 35–42. [Google Scholar]
- Jamil, A.; Riaz, S.; Ashraf, M.; Foolad, M.R. Gene expression profiling of plants under salt stress. Crit. Rev. Plant Sci. 2011, 30, 435–458. [Google Scholar] [CrossRef]
- Zhang, H.; Sun, Y.; Xie, X.; Kim, M.S.; Dowd, S.E.; Paré, P.W. A soil bacterium regulates plant acquisition of iron via deficiency-inducible mechanisms. Plant J. 2009, 58, 568–577. [Google Scholar] [CrossRef] [PubMed]
- Barriuso, J.; Solano, B.R.; Gutiérrez Mañero, F. Protection against pathogen and salt stress by four plant growth-promoting rhizobacteria isolated from Pinus sp. on Arabidopsis thaliana. Phytopathology 2008, 98, 666–672. [Google Scholar] [CrossRef]
- Mayak, S.; Tirosh, T.; Glick, B.R. Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol. Biochem. 2004, 42, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.; Khalil, S.; Ayub, N.; Alam, S.; Latif, F. Organic acids production and phosphate solubilization by phosphate solubilizing microorganisms (PSM) under in vitro conditions. Pak. J. Biol. Sci. 2004, 7, 187–196. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Yandigeri, M.S.; Kashyap, S.; Alagawadi, A.R. Effect of salt on survival and P-solubilization potential of phosphate solubilizing microorganisms from salt affected soils. Saudi J. Biol. Sci. 2012, 19, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Demiİr, Y.; Kocaçalişkan, İ. Effects of NaCl and Proline on Polyphenol Oxidase Activity in Bean Seedlings. Biol. Plant. 2001, 44, 607–609. [Google Scholar] [CrossRef]
- Qin, S.; Zhang, Y.-J.; Yuan, B.; Xu, P.-Y.; Xing, K.; Wang, J.; Jiang, J.-H. Isolation of ACC deaminase-producing habitat-adapted symbiotic bacteria associated with halophyte Limonium sinense (Girard) Kuntze and evaluating their plant growth-promoting activity under salt stress. Plant Soil 2014, 374, 753–766. [Google Scholar] [CrossRef]
- Gujral, M.S.; Agrawal, P.; Khetmalas, M.B.; Pandey, R. Colonization and plant growth promotion of Sorghum seedlings by endorhizospheric Serratia sp. Acta Biol. Indica 2013, 2, 343–352. [Google Scholar]
- Patten, C.L.; Glick, B.R. Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 2002, 68, 3795–3801. [Google Scholar] [CrossRef]
- Sharafzadeh, S. Effects of PGPR on growth and nutrients uptake of tomato. Int. J. Adv. Eng. Technol. 2012, 2, 27. [Google Scholar]
- Gyaneshwar, P.; Parekh, L.; Archana, G.; Poole, P.; Collins, M.; Hutson, R.; Kumar, G.N. Involvement of a phosphate starvation inducible glucose dehydrogenase in soil phosphate solubilization by Enterobacter asburiae. FEMS Microbiol. Lett. 1999, 171, 223–229. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Siddikee, M.A.; Chauhan, P.; Anandham, R.; Han, G.-H.; Sa, T. Isolation, characterization, and use for plant growth promotion under salt stress, of ACC deaminase-producing halotolerant bacteria derived from coastal soil. J. Microbiol. Biotechnol. 2010, 20, 1577–1584. [Google Scholar] [CrossRef] [PubMed]
- Jha, B.; Gontia, I.; Hartmann, A. The roots of the halophyte Salicornia brachiata are a source of new halotolerant diazotrophic bacteria with plant growth-promoting potential. Plant Soil 2012, 356, 265–277. [Google Scholar] [CrossRef]
- Berg, G.; Alavi, M.; Schmidt, C.S.; Zachow, C.; Egamberdieva, D.; Kamilova, F.; Lugtenberg, B. Biocontrol and osmoprotection for plants under salinated conditions. Mol. Microb. Ecol. Rhizosphere 2013, 1, 561–573. [Google Scholar]
- Dodd, I.C.; Pérez-Alfocea, F. Microbial amelioration of crop salinity stress. J. Exp. Bot. 2012, 63, 3415–3428. [Google Scholar] [CrossRef] [PubMed]
- Havenaar, R.; Ten Brink, B.; Huis, J.H. Selection of strains for probiotic use. In Probiotics; Springer: Berlin/Heidelberg, Germany, 1992; pp. 209–224. [Google Scholar] [CrossRef]
- Ali, H.; Khan, E.; Sajad, M.A. Phytoremediation of heavy metals—Concepts and applications. Chemosphere 2013, 91, 869–881. [Google Scholar] [CrossRef]
- Oves, M.; Khan, M.S.; Qari, H.A. Ensifer adhaerens for heavy metal bioaccumulation, biosorption, and phosphate solubilization under metal stress condition. J. Taiwan Inst. Chem. Eng. 2017, 80, 540–552. [Google Scholar] [CrossRef]
- Campos, V.; Moraga, R.; Yánez, J.; Zaror, C.; Mondaca, M. Chromate reduction by Serratia marcescens isolated from tannery effluent. Bull. Environ. Contam. Toxicol. 2005, 75, 400. [Google Scholar] [CrossRef] [PubMed]
- Halder, U.; Banerjee, A.; Biswas, R.; Sharma, A.; Pal, S.; Adhikary, A.; Bandopadhyay, R. Production of prodigiosin by a drug-resistant Serratia rubidaea HB01 isolated from sewage. Environ. Sustain. 2020, 3, 279–287. [Google Scholar] [CrossRef]
- Jalal, K.; UT, N.F.; Mardiana, M.; Shahbudin, S.; Omar, M.N. Antibiotic resistance microbes in tropical mangrove sediments in east coast peninsular, Malaysia. Afr. J. Microbiol. Res. 2010, 4, 640–645. [Google Scholar] [CrossRef]
- Ibrahim, M.K.; Galal, A.-M.M.; Al-Turk, I.M.; Al-Zhrany, K.D. Antibiotic resistance in Gram-negative pathogenic bacteria in hospitals’ drain in Al-Madina Al-Munnawara. J. Taibah Univ. Sci. 2010, 3, 14–22. [Google Scholar] [CrossRef][Green Version]
- Gentille, D.; Pérez, M.; Centelles, M.J. [Bacteremia by a Serratia rubidaea with an atypical quinolones resistance phenotype]. Rev. Chil. Infectol 2014, 31, 351–352. [Google Scholar] [CrossRef] [PubMed]
- Ursua, P.R.; Unzaga, M.J.; Melero, P.; Iturburu, I.; Ezpeleta, C.; Cisterna, R. Serratia rubidaea as an invasive pathogen. J. Clin. Microbiol. 1996, 34, 216–217. [Google Scholar] [CrossRef]
- Litterio, M.R.; Arazi, S.; Hernández, C.; Lopardo, H. Isolation of Serratia rubidaea from a mixed infection after a horse bite. Rev. Argent. De Microbiol. 2012, 44, 272–274. [Google Scholar]
- Xue, Y.; Sun, Q.; Liu, W.; Xiuyun, Y.; Guangqian, P.; Wang, Y.; An, X.; Zhiqiang, M.; Yaping, L.; Yigang, T. Complete genome sequence of Serratia rubidaea isolated in China. Genome Announc. 2016, 4. [Google Scholar] [CrossRef]
- Yasmin, F.; Othman, R.; Sijam, K.; Saad, M.S. Characterization of beneficial properties of plant growth-promoting rhizobacteria isolated from sweet potato rhizosphere. Afr. J. Microbiol. Res. 2009, 3, 815–821. [Google Scholar] [CrossRef]
- Lim, Y.-L.; Yong, D.; Ee, R.; Krishnan, T.; Tee, K.-K.; Yin, W.-F.; Chan, K.-G. Complete genome sequence of Serratia fonticola DSM 4576T, a potential plant growth promoting bacterium. J. Biotechnol. 2015, 214, 43–44. [Google Scholar] [CrossRef]
- Aylward, F.O.; Tremmel, D.M.; Starrett, G.J.; Bruce, D.C.; Chain, P.; Chen, A.; Davenport, K.W.; Detter, C.; Han, C.S.; Han, J. Complete genome of Serratia sp. strain FGI 94, a strain associated with leaf-cutter ant fungus gardens. Genome Announc. 2013, 1. [Google Scholar] [CrossRef]
- Kluepfel, D.A. The behavior and tracking of bacteria in the rhizosphere. Annu. Rev. Phytopathol. 1993, 31, 441–472. [Google Scholar] [CrossRef]
- Trivedi, P.; Pandey, A.; Palni, L.M.S.; Bag, N.; Tamang, M. Colonization of Rhizosphere of Tea by Growth Promoting Bacteria. 2004. Available online: http://hdl.handle.net/2263/8370 (accessed on 8 March 2021).
- Ramakrishna, W.; Yadav, R.; Li, K. Plant growth promoting bacteria in agriculture: Two sides of a coin. Appl. Soil Ecol. 2019, 138, 10–18. [Google Scholar] [CrossRef]
- Kumar, A.; Munder, A.; Aravind, R.; Eapen, S.; Tümmler, B.; Raaijmakers, J. Friend or foe: Genetic and functional characterization of plant endophytic P seudomonas aeruginosa. Environ. Microbiol. 2013, 15, 764–779. [Google Scholar] [CrossRef]
- Wu, L.; Wang, H.; Zhang, Z.; Lin, R.; Zhang, Z.; Lin, W. Comparative metaproteomic analysis on consecutively Rehmannia glutinosa-monocultured rhizosphere soil. PLoS ONE 2011, 6, e20611. [Google Scholar] [CrossRef]
- Farmer, J.; Davis, B.R.; Hickman-Brenner, F.; McWhorter, A.; Huntley-Carter, G.; Asbury, M.; Riddle, C.; Wathen-Grady, H.; Elias, C.; Fanning, G. Biochemical identification of new species and biogroups of Enterobacteriaceae isolated from clinical specimens. J. Clin. Microbiol. 1985, 21, 46–76. [Google Scholar] [CrossRef]
- Nalini, S.; Parthasarathi, R. Optimization of rhamnolipid biosurfactant production from Serratia rubidaea SNAU02 under solid-state fermentation and its biocontrol efficacy against Fusarium wilt of eggplant. Ann. Agrar. Sci. 2018, 16, 108–115. [Google Scholar] [CrossRef]
- Kalbe, C.; Marten, P.; Berg, G. Strains of the genus Serratia as beneficial rhizobacteria of oilseed rape with antifungal properties. Microbiol. Res. 1996, 151, 433–439. [Google Scholar] [CrossRef]
- Al-Mijalli, S.H.S. Isolation and characterization of plant and human pathogenic bacteria from green pepper (Capsicum annum L.) in Riyadh, Saudi Arabia. 3 Biotech 2014, 4, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Pence, N.S.; Larsen, P.B.; Ebbs, S.D.; Letham, D.L.; Lasat, M.M.; Garvin, D.F.; Eide, D.; Kochian, L.V. The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc. Natl. Acad. Sci. USA 2000, 97, 4956–4960. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).