Relationship between Plant Roots, Rhizosphere Microorganisms, and Nitrogen and Its Special Focus on Rice
Abstract
1. Introduction
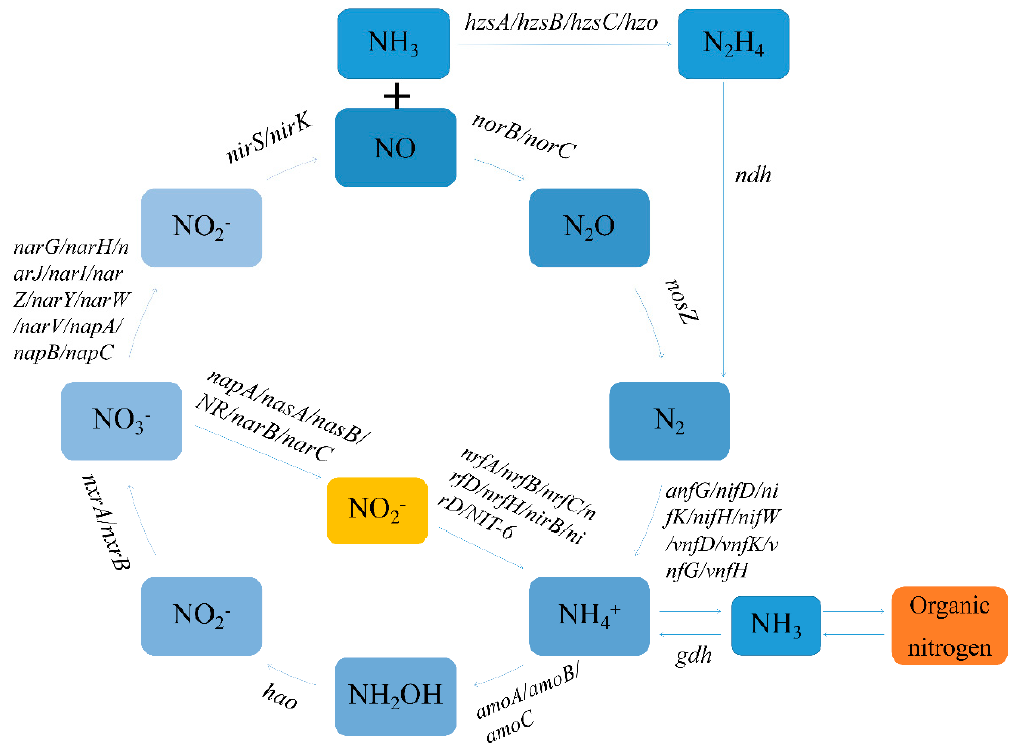
2. Major Nitrogen Cycle Pathways
3. Nitrogen–Root–Rhizosphere Microorganisms Interaction Network
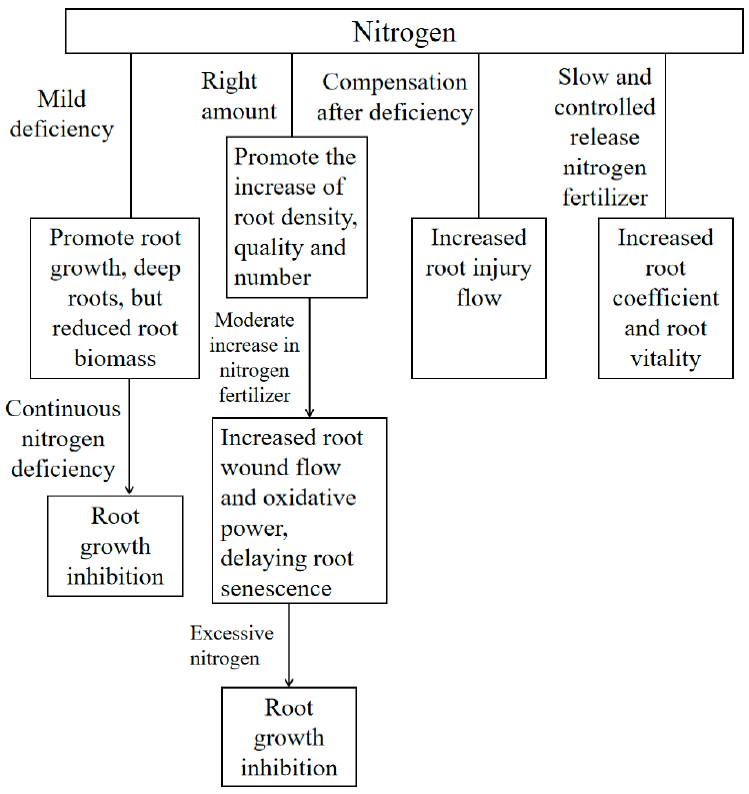
4. Effects of Nitrogen Fertilizer on Plant Root Morphology
5. Effects of N Fertilizer on Plant Root Physiology and Biochemistry
6. Effects of Nitrogen Fertilizer on Soil Enzyme Activity in Plants
7. Effects of Nitrogen Fertilizer on Microorganisms in Rhizosphere Soil of Plants
8. Relationship between Rhizosphere Microorganisms and Crop Yield
9. Relationship between Root Microorganisms and N Utilization Efficiency
10. Effects of Cultivated Habitats on Rhizosphere Microorganisms
11. Summary and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gojon, A. Nitrogen nutrition in plants: Rapid progress and new challenges. J. Exp. Bot. 2017, 68, 2457–2462. [Google Scholar] [CrossRef]
- Xiong, Q.; Tang, G.; Zhong, L.; He, H.; Chen, X. Response to nitrogen deficiency and compensation on physiological characteristics, yield formation, and nitrogen utilization of rice. Front. Plant Sci. 2018, 9, 1075. [Google Scholar] [CrossRef]
- Xiong, Q.; Zhong, L.; Shen, T.; Cao, C. iTRAQ-based quantitative proteomic and physiological analysis of the response to N deficiency and the compensation effect in rice. BMC Genom. 2019, 20, 681. [Google Scholar] [CrossRef]
- Shen, T.; Xiong, Q.; Zhong, L.; Shi, X.; Chen, X. Analysis of main metabolisms during nitrogen deficiency and compensation in rice. Acta Physiol. Plant. 2019, 41, 68. [Google Scholar] [CrossRef]
- Moreau, D.; Bardgett, R.D.; Finlay, R.D.; Jones, D.L.; Philippot, L. A plant perspective on nitrogen cycling in the rhizosphere. Funct. Ecol. 2019, 33, 540–552. [Google Scholar] [CrossRef]
- Courty, P.; Smith, P.M.; Koegel, S.; Redecker, D.; Wipf, D. Inorganic nitrogen uptake and transport in beneficial plant root-microbe interactions. Crit. Rev. Plant Sci. 2015, 34, 4–16. [Google Scholar] [CrossRef]
- White, P.J. Root traits benefitting crop production in environments with limited water and nutrient availability. Ann. Bot. 2019, 124, 883–890. [Google Scholar] [CrossRef]
- Sun, X.; Chen, F.; Yuan, L.; Mi, G. The physiological mechanism underlying root elongation in response to nitrogen deficiency in crop plants. Planta 2020, 251, 84. [Google Scholar] [CrossRef]
- Xin, W.; Zhang, L.; Zhang, W.; Gao, J.; Yi, J.; Zhen, X.; Du, M.; Zhao, Y.; Chen, L. An integrated analysis of the rice transcriptome and metabolome reveals root growth regulation mechanisms in response to nitrogen availability. Int. J. Mol. Sci. 2019, 20, 5893. [Google Scholar] [CrossRef]
- Wang, Q.; Huang, J.; He, F.; Cui, K.; Zeng, J.; Nie, L.; Peng, S. Head rice yield of “super” hybrid rice Liangyoupeijiu grown under different nitrogen rates. Field Crops Res. 2012, 134, 71–79. [Google Scholar] [CrossRef]
- Henneron, L.; Kardol, P.; Wardle, D.A.; Cros, C.; Fontaine, S. Rhizosphere control of soil nitrogen cycling: A key component of plant economic strategies. New Phytol. 2020, 228, 1269–1282. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Caldwell, B.A. Enzyme activities as a component of soil biodiversity: A review. Pedobiologia 2005, 49, 637–644. [Google Scholar]
- Toju, H.; Peay, K.G.; Yamamichi, M.; Narisawa, K.; Hiruma, K.; Naito, K.; Fukuda, S.; Ushio, M.; Nakaoka, S.; Onoda, Y.; et al. Core microbiomes for sustainable agroecosystems. Nat. Plants 2018, 4, 247–257. [Google Scholar] [CrossRef]
- Huang, A.C.; Jiang, T.; Liu, Y.; Bai, Y.; Reed, J.; Qu, B.; Goossens, A.; Nützmann1, H.; Bai, Y.; Osbourn, A. A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science 2019, 364, 6389. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.; Zhang, N.; Hu, B.; Jin, T.; Xu, H.; Qin, Y.; Yan, P.; Zhang, X.; Guo, X.; et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 2019, 37, 676–684. [Google Scholar] [CrossRef]
- Fitzpatrick, C.R.; Salas-González, I.; Conway, J.M.; Finkel, O.M.; Gilbert, S.; Russ, D.; Teixeira, P.J.P.L.; Dang, J.L. The plant microbiome: From ecology to reductionism and beyond. Ann. Rev. Microbiol. 2020, 74, 81–100. [Google Scholar]
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar]
- Ke, J.; Wang, B.; Yoshikuni, Y. Microbiome engineering: Synthetic biology of plant-associated microbiomes in sustainable agriculture. Trends Biotechnol. 2020. [Google Scholar] [CrossRef]
- Rout, M.E.; Southworth, D. The root microbiome influences scales from molecules to ecosystems: The unseen majority. Am. J. Bot. 2013, 100, 1689–1691. [Google Scholar] [CrossRef]
- Lakshmanan, V.; Ray, P.; Craven, K.D. Toward a resilient, functional microbiome: Drought tolerance-alleviating microbes for sustainable agriculture. Methods Mol. Biol. 2017, 1631, 69–84. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Genre, A.; Lanfranco, L.; Perotto, S.; Bonfante, P. Unique and common traits in mycorrhizal symbioses. Nat. Rev. Microbiol. 2020, 18, 649–660. [Google Scholar] [CrossRef]
- Smith, S.E.; Smith, F.A. Roles of arbuscular mycorrhizas in plant nutrition and growth: New paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 2011, 62, 227–250. [Google Scholar] [CrossRef]
- Spence, C.; Alff, E.; Johnson, C.; Ramos, C.; Donofrio, N.; Sundaresan, V.; Bais, H. Natural rice rhizospheric microbes suppress rice blast infections. BMC Plant Biol. 2014, 14, 130. [Google Scholar] [CrossRef]
- Hu, J.; Wei, Z.; Kowalchuk, G.A.; Xu, Y.; Shen, Q.; Jousset, A. Rhizosphere microbiome functional diversity and pathogen invasion resistance build up during plant development. Environ. Microbiol. 2020, 22, 5005–5018. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef]
- Smith, P.A. Foliage friendships microbial communities on a plant’s roots, stems and leaves may improve crop growth. Sci. Am. 2014, 311, 24–25. [Google Scholar]
- de Vries, F.T.; Griffiths, R.I.; Knight, C.G.; Nicolitch, O.; Williams, A. Harnessing rhizosphere microbiomes for drought-resilient crop production. Science 2020, 368, 270–274. [Google Scholar] [CrossRef]
- Nelson, M.B.; Martiny, A.C.; Martiny, J.B.H. Global biogeography of microbial nitrogen-cycling traits in soil. Proc. Natl. Acad. Sci. USA 2016, 113, 8033–8040. [Google Scholar] [CrossRef]
- Stokstad, E. The nitrogen fix. Science 2016, 353, 1225–1227. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Leyser, O. A plant’s diet, surviving in a variable nutrient environment. Science 2020, 368, eaba0196. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, X.; Yu, M.; Shen, A. Research progress on effects of straw returning on nitrogen cycling microbes and functional genes in paddy soil. Acta Agric. Zhejiangensis 2019, 31, 333–342. [Google Scholar] [CrossRef]
- Xu, J. Effects of soil moisture and nitrogen and phosphorus nutrition on plant root growth. Soil Water Conserv. Sci. Technol. Shanxi 2010, 3, 12–15. [Google Scholar]
- Edwards, J.; Santos-Medellín, C.; Nguyen, B.; Kilmer, J.; Liechty, Z.; Veliz, E.; Ni, J.; Phillips, G.; Sundaresan, V. Soil domestication by rice cultivation results in plant-soil feedback through shifts in soil microbiota. Genome biol. 2019, 20, 221. [Google Scholar] [CrossRef]
- Zhong, Y.; Hu, J.; Xia, Q.; Zhang, S.; Li, X.; Pan, X.; Zhao, R.; Wang, R.; Yan, W.; Shangguan, Z.; et al. Soil microbial mechanisms promoting ultrahigh rice yield. Soil Biol. Biochem. 2020, 143, 107741. [Google Scholar] [CrossRef]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef]
- Chen, S.; Waghmode, T.R.; Sun, R.; Kuramae, E.E.; Hu, C.; Liu, B. Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 2019, 7, 136. [Google Scholar] [CrossRef]
- Fan, K.; Delgado-Baquerizo, M.; Guo, X.; Wang, D.; Zhu, Y.G.; Chu, H. Biodiversity of key-stone phylotypes determines crop production in a four-decade fertilization experiment. ISME J. 2020. [Google Scholar] [CrossRef]
- Müller, D.; Vogel, C.; Bai, Y.; Vorholt, J.A. The plant microbiota: Systems-level insights and perspectives. Ann. Rev. Genet. 2016, 50, 211–234. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Lanfranco, L.; Fiorilli, V.; Gutjahr, C. Partner communication and role of nutrients in the arbuscular mycorrhizal symbiosis. New Phytol. 2018, 220, 1031–1046. [Google Scholar] [CrossRef]
- Nannipieri, P.; Ascher, J.; Ceccherini, M.T.; Landi, L.; Renella, G. Microbial diversity and soil functions. Eur. J. Soil Sci. 2003, 54, 655–670. [Google Scholar] [CrossRef]
- Fan, K.; Delgado-Baquerizo, M.; Guo, X.; Wang, D.; Zhu, Y.G.; Chu, H. Microbial resistance promotes plant production in a four-decade nutrient fertilization experiment. Soil Biol. Biochem. 2020, 141, 107679. [Google Scholar] [CrossRef]
- Wang, Q.; Zhu, Y.; Zou, X.; Li, F.; Zhang, J.; Kang, Z.; Li, X.; Yin, C.; Lin, Y. Nitrogen deficiency-induced decrease in cytokinins content promotes rice seminal root growth by promoting root meristem cell proliferation and cell elongation. Cells 2020, 9, 916. [Google Scholar] [CrossRef]
- Jiang, W.; Ma, Z.; Hu, Q.; Ma, H.; Ren, G.; Zhu, Y.; Liu, G.; Zhang, H.; Wei, H. Effects of slow and controlled release nitrogen fertilizer on rice growth and nitrogen utilization. Jiangsu J. Agric. Sci. 2020, 36, 777–784. [Google Scholar] [CrossRef]
- Zhu, S.; Vivanco, J.M.; Manter, D.K. Nitrogen fertilizer rate affects root exudation, the rhizosphere microbiome and nitrogen-use-efficiency of maize. Appl. Soil Ecol. 2016, 107, 324–333. [Google Scholar] [CrossRef]
- Tang, S.; Yang, S.; Chen, J.; Xu, P.; Zhang, F.; Ai, S.; Huang, X. Studies on the mechanism of single basal application of controlled-release fertilizers for increasing yield of rice (Oryza safiva L.). Agric. Sci. China 2007, 6, 586–596. [Google Scholar] [CrossRef]
- Jia, Z.; Giehl, R.F.H.; Meyer, R.C.; Altmann, T.; von Wirén, N. Natural variation of BSK3 tunes brassinosteroid signaling to regulate root foraging under low nitrogen. Nat. Commun. 2019, 10, 2378. [Google Scholar] [CrossRef]
- Gao, K.; Chen, F.; Yuan, L.; Zhang, F.; Mi, G. A comprehensive analysis of root morphological changes and nitrogen allocation in maize in response to low nitrogen stress. Plant Cell Environ. 2015, 38, 740–750. [Google Scholar] [CrossRef]
- Sun, H.; Tao, J.; Liu, S.; Huang, S.; Chen, S.; Xie, X.; Koichi, Y.; Zhang, Y.; Xu, G. Strigolactones are involved in phosphate-and nitrate-deficiency-induced root development and auxin transport in rice. J. Exp. Bot. 2014, 65, 6735–6746. [Google Scholar] [CrossRef]
- Sun, H.; Bi, Y.; Tao, J.; Huang, S.; Hou, M.; Xue, R.; Liang, Z.; Gu, P.; Koichi, Y.; Xie, X.; et al. Strigolactones are required for nitric oxide to induce root elongation in response to nitrogen and phosphate deficiencies in rice. Plant Cell Environ. 2016, 39, 1473–1484. [Google Scholar] [CrossRef]
- Sun, H.; Feng, F.; Liu, J.; Zhao, Q. Nitric oxide affects rice root growth by regulating auxin transport under nitrate supply. Front. Plant Sci. 2018, 9, 659. [Google Scholar] [CrossRef]
- Ren, Y.; Qian, Y.; Xu, Y.; Zou, C.; Liu, D.; Zhao, X.; Zhang, A.; Tong, Y. Characterization of QTLs for root traits of wheat grown under different nitrogen and phosphorus supply levels. Front. Plant Sci. 2017, 8, 2096. [Google Scholar] [CrossRef]
- Ren, Y.; Yue, H.; Li, L.; Xu, Y.; Wang, Z.; Xin, Z.; Lin, T. Identification and characterization of circRNAs involved in the regulation of low nitrogen-promoted root growth in hexaploid wheat. Biol. Res. 2018, 51, 43. [Google Scholar] [CrossRef]
- Xu, Y.; Ren, Y.; Lin, T.; Cui, D. Identification and characterization of circRNAs involved in the regulation of wheat root length. Biol. Res. 2019, 52, 19. [Google Scholar] [CrossRef]
- Xu, Y.; Ren, Y.; Li, J.; Li, L.; Chen, S.; Wang, Z.; Xin, Z.; Chen, F.; Lin, T.; Cui, D.; et al. Comparative proteomic analysis provides new insights into low nitrogen-promoted primary root growth in hexaploid wheat. Front. Plant Sci. 2019, 10, 151. [Google Scholar] [CrossRef]
- Baskin, T.I. Patterns of root growth acclimation: Constant processes, changing boundaries. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 65–73. [Google Scholar] [CrossRef]
- Sun, W.; Liu, S.; Feng, J.; Wang, P.; Ma, D.; Xie, Y.; Lu, H.; Wang, C. Effects of water and nitrogen source types on soil enzyme activity and nitrogen utilization efficiency of wheat. Chin. J. Appl. Ecol. 2020, 31, 2583–2592. [Google Scholar] [CrossRef]
- Takatsuka, H.; Umeda, M. Hormonal control of cell division and elongation along differentiation trajectories in roots. J. Exp. Bot. 2014, 65, 2633–2643. [Google Scholar] [CrossRef]
- Pacifici, E.; Polverari, L.; Sabatini, S. Plant hormone cross-talk: The pivot of root growth. J. Exp. Bot. 2015, 66, 1113–1121. [Google Scholar]
- Steffens, B.; Rasmussen, A. The physiology of adventitious roots. Plant Physiol. 2016, 170, 603–617. [Google Scholar] [CrossRef]
- Zhang, H.; Xue, Y.; Wang, Z.; Yang, J.; Zhang, J. Morphological and physiological traits of roots and their relationships with shoot growth in “super” rice. Field Crops Res. 2009, 113, 31–40. [Google Scholar] [CrossRef]
- Liu, Z.; Zhao, Y.; Guo, S.; Cheng, S.; Guan, Y.; Cai, H.; Mi, G.; Yuan, L.; Chen, F. Enhanced crown root number and length confers potential for yield improvement and fertilizer reduction in nitrogen-efficient maize cultivars. Field Crops Res. 2019, 241, 107562. [Google Scholar] [CrossRef]
- Zhang, H.; Fu, X.; Wang, X.; Gui, H.; Dong, Q.; Pang, N.; Wang, Z.; Zhang, X.; Song, M. Identification and screening of nitrogen-efficient cotton genotypes under low and normal nitrogen environments at the seedling stage. J. Cotton Res. 2018, 1, 6. [Google Scholar] [CrossRef]
- Cassman, K.G.; Dobermann, A.; Walters, D.T. Agroecosystems, nitrogen-use efficiency and nitrogen management. Ambio 2002, 31, 132–140. [Google Scholar]
- Wei, H.; Zhang, H.; Zhang, S.; Hang, J.; Dai, Q.; Huo, Z.; Xu, K.; Ma, Q.; Zhang, Q.; Liu, Y. Root morphological and physiological characteristics in rice genotypes with different N use efficiencies. Acta Agron. Sin. 2008, 34, 429–436. [Google Scholar] [CrossRef]
- Li, X. Effects of N Application on the Roots Characteristics and Its Relation with Aerial Part Growth and Yield Formation of the Machine-Trasplanted; Guizhou University: Guiyang, China, 2016. [Google Scholar]
- Li, H.; Sun, Y.; Qu, J.; Wei, C.; Sun, G.; Zhao, Y.; Chai, Y. Influence of nitrogen levels on morphological and physiological characteristics of root system in japonica rice in northeast China. Chin. J. Rice Sci. 2012, 26, 723–730. [Google Scholar]
- Zhao, R.; Chen, H.; Ou, X.; Liu, J.; Jia, G.; Yao, S.; Liu, M.; Huang, L. Effect of nitrogen levels on oxidase activities in leaves and roots of winter wheat. Chin. Agric. Sci. Bull. 2020, 36, 1–7. [Google Scholar]
- Sharma, N.; Sinha, V.B.; Prem Kumar, N.A.; Subrahmanyam, D.; Neeraja, C.N.; Kuchi, S.; Jha, A.; Parsad, R.; Sitaramam, V.; Raghuram, N. Nitrogen use efficiency phenotype and associated genes: Roles of germination, flowering, root/shoot length and biomass. Front. Plant Sci. 2021, 11, 587464. [Google Scholar] [CrossRef]
- Gu, X.; Cai, H.; Du, Y.; Li, Y. Effects of film mulching and nitrogen fertilization on rhizosphere soil environment, root growth and nutrient uptake of winter oilseed rape in northwest China. Soil Tillage Res. 2019, 187, 194–203. [Google Scholar] [CrossRef]
- Chen, Z.; Hu, Y.; Han, Y. Response of nitrogen uptake and root traits of rapeseed to the low nitrogen stress. Hubei Agric. Sci. 2020, 59, 57–62. [Google Scholar] [CrossRef]
- Liang, Z.; Guo, C.; Wang, C.; Li, Y.; Zhang, J. Synergistic effect of combined application of nitrogen and zinc on construction of good morphology and high physiological activities of wheat root. J. Plant Nutr. Fertil. 2020, 26, 826–839. [Google Scholar] [CrossRef]
- Rekha, K.; Ramasamy, M.; Usha, B. Root exudation of organic acids as affected by plant growth-promoting rhizobacteria Bacillus subtilis RR4 in rice. J. Crop Improv. 2020, 1–16. [Google Scholar] [CrossRef]
- Canarini, A.; Kaiser, C.; Merchant, A.; Richter, A.; Wanek, W. Root exudation of primary metabolites: Mechanisms and their roles in plant responses to environmental stimuli. Front. Plant Sci. 2019, 10, 157. [Google Scholar] [CrossRef]
- Wang, X.; Whalley, W.R.; Miller, A.J.; White, P.; Zhang, F.; Shen, J. Sustainable cropping requires adaptation to a heterogeneous rhizosphere. Trends Plant Sci. 2020, 25, 1194–1202. [Google Scholar] [CrossRef]
- Wen, T.; Zhao, M.; Yuan, J.; Kowalchuk, G.A.; Shen, Q. Root exudates mediate plant defense against foliar pathogens by recruiting beneficial microbes. Soil Ecol. Lett. 2020. [Google Scholar] [CrossRef]
- Vives-Peris, V.; de Ollas, C.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Root exudates: From plant to rhizosphere and beyond. Plant Cell Rep. 2020, 39, 3–17. [Google Scholar] [CrossRef]
- Eichmann, R.; Richards, L.; Schfer, P. Hormones as go-betweens in plant microbiome assembly. Plant J. 2020. [Google Scholar] [CrossRef]
- Brzezińska, M. Significance of soil enzymes in nutrient transformations. Acta Agrophys. 2020, 2002, 5–23. [Google Scholar]
- Skujins, J. History of Abiontic Soil Enzyme Research; Academic Press: London, UK, 1978; pp. 1–49. [Google Scholar]
- Zhang, X.; Dippold, M.A.; Kuzyakov, Y.; Razavi, B.S. Spatial pattern of enzyme activities depends on root exudate composition. Soil Biol. Biochem. 2019, 133, 83–93. [Google Scholar] [CrossRef]
- Sherene, T. Role of soil enzymes in nutrient transformation: A review. Bio. Bull. 2017, 3, 109–131. [Google Scholar]
- Mina, L.B.; Saha, S.; Kumar, N.; Srivastva, A.K.; Gupta, H.S. Changes in soil nutrient content and enzymatic activity under conventional and zero-tillage practices in an Indian sandy clay loam soil. Nutr. Cycl. Agroecosyst. 2008, 82, 273–281. [Google Scholar] [CrossRef]
- Su, J.; Li, X.; Bao, J. Effects of nitrogen addition on soil physico-chemical properties and enzyme activities in desertified steppe. Chin. J. Appl. Ecol. 2014, 25, 664–670. [Google Scholar]
- Song, D.; Chen, L.; Zhang, S.; Zheng, Q.; Wang, X. Combined biochar and nitrogen fertilizer change soil enzyme and microbial activities in a 2-year field trial. Eur. J. Soil Biol. 2020, 99, 103212. [Google Scholar] [CrossRef]
- Pokharel, P.; Ma, Z.; Chang, S.X. Biochar increases soil microbial biomass with changes in extra-and intracellular enzyme activities: A global meta-analysis. Biochar 2020, 2, 65–79. [Google Scholar]
- Li, S.; Wang, S.; Fan, M.; Wu, Y.; Shangguan, Z. Interactions between biochar and nitrogen impact soil carbon mineralization and the microbial community. Soil Tillage Res. 2020, 196, 104437. [Google Scholar] [CrossRef]
- Zhao, J.; Ni, T.; Li, Y.; Xiong, W.; Ran, W.; Shen, B.; Shen, Q.; Zhang, R. Responses of bacterial communities in arable soils in a rice-wheat cropping system to different fertilizer regimes and sampling times. PLoS ONE 2014, 9, e85301. [Google Scholar] [CrossRef]
- Qin, J.; Xiong, H.; Ma, H.; Li, Z. Effects of different fertilizers on residues of oxytetracycline and microbial activity in soil. Environ. Sci. Pollut. Res. 2019, 26, 161–170. [Google Scholar] [CrossRef]
- Yang, D.; Chen, H.; You, Q.; Wang, Z.; Xie, H.; Zhuo, C.; Xie, H. Ecological effect of different nitrogen application modes on rhizosphere microbes in soil of rice root. J. Northwest A F Univ. (Nat. Sci. Ed.) 2008, 36, 88–94. [Google Scholar]
- Macias-Benitez, S.; Garcia-Martinez, A.M.; Caballero, J.P.; Gonzalez, J.M.; Moral, T.; Rubio, J.P. Rhizospheric organic acids as biostimulants: Monitoring feedbacks on soil microorganisms and biochemical properties. Front. Plant Sci. 2020, 11, 633. [Google Scholar] [CrossRef]
- Brucker, E.; Kernchen, S.; Spohn, M. Release of phosphorus and silicon from minerals by soil microorganisms depends on the availability of organic carbon. Soil Biol. Biochem. 2020, 143, 107737. [Google Scholar] [CrossRef]
- Ahn, J.; Song, J.; Kim, B.; Kim, M.; Joa, J.; Weon, H. Characterization of the bacterial and archaeal communities in rice field soils subjected to long-term fertilization practices. J. Microbiol. 2012, 50, 754–765. [Google Scholar] [CrossRef]
- Liu, K.L.; Li, Y.Z.; Zhou, L.J.; Chen, Y.; Huang, Q.; Yu, X.; Li, D. Comparison of crop productivity and soil microbial activity among different fertilization patterns in red upland and paddy soils. Acta Ecol. Sin. 2018, 38, 262–267. [Google Scholar] [CrossRef]
- Chaudhry, A.N.; Jilani, G.; Khan, M.A.; Iqbal, T. Improved processing of poultry litter reduces nitrate leaching and enhances its fertilizer quality. Asian J. Chem. 2009, 21, 4997–5003. [Google Scholar]
- Ren, N.; Wang, Y.; Ye, Y.; Zhao, Y.; Chu, X. Effects of continuous nitrogen fertilizer application on the diversity and composition of rhizosphere soil bacteria. Front. Microbiol. 2020, 11, 1948. [Google Scholar] [CrossRef]
- Wang, C.; Lu, X.; Mori, T.; Mao, Q.; Zhou, K.; Zhou, G.; Nie, Y.; Mo, J. Responses of soil microbial community to continuous experimental nitrogen additions for 13 years in a nitrogen-rich tropical forest. Soil Biol. Biochem. 2018, 121, 103–112. [Google Scholar] [CrossRef]
- Wang, C.; Liu, D.W.; Bai, E. Decreasing soil microbial diversity is associated with decreasing microbial biomass under nitrogen addition. Soil Biol. Biochem. 2018, 120, 126–133. [Google Scholar] [CrossRef]
- Chen, J.; Luo, Y.; Van Groenigen, K.J.; Hungate, B.A.; Cao, J.; Zhou, X.; Wang, R.W. A keystone microbial enzyme for nitrogen control of soil carbon storage. Sci. Adv. 2018, 4, eaaq1689. [Google Scholar] [CrossRef]
- Luo, R.; Kuzyakov, Y.; Liu, D.; Fan, J.; Luo, J.; Lindsey, S.; He, J.; Ding, W. Nutrient addition reduces carbon sequestration in a Tibetan grassland soil: Disentangling microbial and physical controls. Soil Biol. Biochem. 2020, 144, 107746. [Google Scholar] [CrossRef]
- Alteio, L.V.; Schulz, F.; Seshadri, R.; Varghese, N.; Rodriguez-Reillo, W.; Ryan, E.; Goudeau, D.; Eichorst, S.A.; Malmstrom, R.R.; Bowers, R.M.; et al. Complementary metagenomic approaches improve reconstruction of microbial diversity in a forest soil. Msystems 2020, 5, e00768-19. [Google Scholar] [CrossRef]
- Emmett, B.D.; Buckley, D.H.; Drinkwater, L.E. Plant growth rate and nitrogen uptake shape rhizosphere bacterial community composition and activity in an agricultural field. New Phytol. 2020, 225, 960–973. [Google Scholar] [CrossRef]
- Zhang, B.H.; Hong, J.P.; Zhang, Q.; Jin, D.S.; Gao, C.H. Contrast in soil microbial metabolic functional diversity to fertilization and crop rotation under rhizosphere and non-rhizosphere in the coal gangue landfill reclamation area of Loess Hills. PLoS ONE 2020, 15, e0229341. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, 911–920. [Google Scholar] [CrossRef]
- Barber, N.A.; Adler, L.S.; Theis, N.; Hazzard, R.V.; Kiers, E.T. Herbivory reduces plant interactions with above- and belowground antagonists and mutualists. Ecology 2012, 93, 1560–1570. [Google Scholar] [CrossRef]
- Li, H.; Su, J.Q.; Yang, X.R.; Zhu, Y.G. Distinct rhizosphere effect on active and total bacterial communities in paddy soils. Sci. Total Environ. 2019, 649, 422–430. [Google Scholar] [CrossRef]
- Maarastawi, S.A.; Frindte, K.; Geer, R.; Kröber, E.; Knief, C. Temporal dynamics and compartment specific rice straw degradation in bulk soil and the rhizosphere of maize. Soil Biol. Biochem. 2018, 127, 200–212. [Google Scholar] [CrossRef]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loqué, D.; Bowen, B.P.; et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef]
- Bernaola, L.; Cosme, M.; Schneider, R.W.; Stout, M. Belowground inoculation with arbuscular mycorrhizal fungi increases local and systemic susceptibility of rice plants to different pest organisms. Front. Plant Sci. 2018, 9, 747. [Google Scholar] [CrossRef]
- Richardson, A.E.; José-Miguel, B.; Mcneill, A.M.; Prigent-Combaret, C. Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 2009, 321, 305–339. [Google Scholar] [CrossRef]
- Yasunori, I.; Yasuhiro, D.; Amiu, S.; Tomoko, S.; Arisa, S.; Kie, K.; Fumiaki, F.; Kenji, W.; Kohei, Y.; Akio, U.; et al. Multi-omics analysis on an agroecosystem reveals the significant role of organic nitrogen to increase agricultural crop yield. Proc. Natl. Acad. Sci. USA 2020, 117, 14552–14560. [Google Scholar] [CrossRef]
- Narayan, O.P.; Verma, N.; Jogawat, A.; Dua, M.; Johri, A.K. Sulfur transfer from the endophytic fungus Serendipita indica improves maize growth and requires the sulfate transporter SiSulT. Plant Cell 2021, koab006. [Google Scholar] [CrossRef]
- Paul, K.; Saha, C.; Nag, M.; Mandal, D.; Naiya, H.; Sen, D.; Mitra, S.; Kumar, M.; Bose, D.; Mukherjee, G.; et al. A tripartite interaction among the basidiomycete Rhodotorula mucilaginosa, N2-fixing endobacteria, and rice improves plant nitrogen nutrition. Plant Cell 2020, 32, 486–507. [Google Scholar] [CrossRef]
- Abdelaziz, M.E.; Abdelsattar, M.; Abdeldaym, E.A.; Omar, M.A.; Hirt, H. Piriformospora indica alters Na+/K+ homeostasis, antioxidant enzymes and lenhx1 expression of greenhouse tomato grown under salt stress. Sci. Hortic. 2019, 256, 108532. [Google Scholar] [CrossRef]
- Pennisi, E. Salty soil is no problem for these tomatoes, thanks to some microbial helpers. Science 2019. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, E. Effect of various rotation systems on yield of Angelica sinensis and microbial populations in its rhizosphere. Chin. Tradit. Herbal Drugs 2008, 39, 267–269. [Google Scholar]
- Yong, T.W.; Yang, W.Y.; Xiang, D.B.; Chen, X.R. Effects of different cropping modes on crop root growth, yield, and rhizosphere soil microbes’ number. Chin. J. Appl. Ecol. 2012, 23, 125–132. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, H.; He, N.; Wu, F.; Tang, B.; Huang, R.; Li, P.; Li, C. Effects of agronomic measures and water-retaining agent on yield and rhizospheric biological activity of summer maize. Chin. Agric. Sci. Bull. 2016, 32, 43–48. [Google Scholar]
- Guo, L.; Ma, J.; Huang, G. Effect of rhizobia inoculation on pea yield and microbial number in pea rhizosphere. Soil Res. Agric. Mod. 2010, 31, 630–633. [Google Scholar]
- Di Salvo, L.P.; Cellucci, G.C.; Carlino, M.E.; de García, S.; Inés, E. Plant growth-promoting rhizobacteria inoculation and nitrogen fertilization increase maize (Zea mays L.) grain yield and modified rhizosphere microbial communities. Appl. Soil Ecol. 2018, 126, 113–120. [Google Scholar] [CrossRef]
- Golubkina, N.; Zamana, S.; Seredin, T.; Poluboyarinov, P.; Sokolov, S.; Baranova, H.; Krivenkov, L.; Pietrantonio, L.; Caruso, G. Effect of selenium biofortification and beneficial microorganism inoculation on yield, quality and antioxidant properties of shallot bulbs. Plants 2019, 8, 102. [Google Scholar] [CrossRef]
- Duan, H.; Zhang, N.; Li, J.; Yang, G.; Shi, C. Preliminary study on physiological populations and quantity of rhizosphere main microorganism in super-yield rice fields. Chin. Agric. Sci. Bull. 2007, 23, 285–289. [Google Scholar]
- Pan, L.; Xiao, W.; Dong, Y.; Li, G.; Zhang, N.; Duan, H.; Zhang, S. Microbial species and functional diversity in rice rhizosphere of high-yield special ecological areas. J. Agric. Res. Environ. 2016, 33, 583–590. [Google Scholar] [CrossRef]
- Mommer, L.; Kirkegaard, J.; van Ruijven, J. Root-root interactions: Towards a rhizosphere framework. Trends Plant Sci. 2016, 21, 209–217. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Murray, J.D.; Poole, P.S.; Downie, J.A. The rules of engagement in the legume-rhizobial symbiosis. Ann. Rev. Genet. 2011, 45, 119–144. [Google Scholar] [CrossRef]
- Mbodj, D.; Effa-Effa, B.; Kane, A.; Manneh, B.; Gantet, P.; Laplaze, L.; Diedhiou, A.G.; Grondin, A. Arbuscular mycorrhizal symbiosis in rice: Establishment, environmental control and impact on plant growth and resistance to abiotic stresses. Rhizosphere 2018, 8, 12–26. [Google Scholar] [CrossRef]
- Hu, B.; Wang, W.; Ou, S.; Tang, J.; Li, H.; Che, R.; Zhang, Z.; Chai, X.; Wang, H.; Wang, Y.; et al. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 2015, 47, 834–838. [Google Scholar] [CrossRef]
- Di Benedetto, N.A.; Corbo, M.R.; Campaniello, D.; Cataldi, M.P.; Bevilacqua, A.; Sinigaglia, M.; Flagella, Z. The role of Plant Growth Promoting Bacteria in improving nitrogen use efficiency for sustainable crop production: A focus on wheat. AIMS Microbiol. 2017, 3, 413–434. [Google Scholar] [CrossRef]
- Cheng, W.; Johnson, D.W.; Fu, S. Rhizosphere effects on decomposition. Soil Sci. Soc. Am. J. 2003, 67, 1418–1427. [Google Scholar] [CrossRef]
- Nelson, D.R.; Mele, P.M. The impact of crop residue amendments and lime on microbial community structure and nitrogenfixing bacteria in the wheat rhizosphere. Soil Res. 2006, 44, 319–329. [Google Scholar]
- Venieraki, A.; Dimou, M.; Pergalis, P.; Kefalogianni, I.; Chatzipavlidis, I.; Katinakis, P. The genetic diversity of culturable nitrogen-fixing bacteria in the rhizosphere of wheat. Microb. Ecol. 2011, 61, 277–285. [Google Scholar] [CrossRef]
- Vessey, J.K. Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 2003, 255, 571–586. [Google Scholar] [CrossRef]
- Behl, R.K.; Ruppel, S.; Kothe, E.; Narula, N. Wheat×Azotobacter×VA Mycorrhiza interactions towards plant nutrition and growth-a review. J. Appl. Bot. Food Qual. 2012, 81, 95–109. [Google Scholar] [CrossRef]
- Neiverth, A.; Delai, S.; Garcia, D.M.; Saatkamp, K.; de Souza, E.M.; de Oliveira Pedrosa, F.; Guimarães, V.F.; Santos, M.F.; da Costa, A.C.T.; Vendruscolo, E.C.G.; et al. Performance of different wheat genotypes inoculated with the plant growth promoting bacterium Herbaspirillum seropedicae. Eur. J. Soil Biol. 2014, 64, 1–5. [Google Scholar] [CrossRef]
- Macintosh, K.A.; Doody, D.G.; Withers, P.J.A.; McDowell, R.W.; Smith, D.R.; Johnson, L.T.; Bruulsema, T.W.; O’Flaherty, V.; McGrath, J.W. Transforming soil phosphorus fertility management strategies to support the delivery of multiple ecosystem services from agricultural systems. Sci. Total Environ. 2019, 649, 90–98. [Google Scholar] [CrossRef]
- Lin, X.; Shi, H.; Wu, L.; Cheng, Y.; Cai, S.; Huang, S.; He, S.; Huang, Q.; Zhang, K. Effects of cultivation methods on soil microbial community structure and diversity in red paddy. Ecol. Environ. Sci. 2020, 29, 2206–2214. [Google Scholar] [CrossRef]
- Pang, Z.; Xu, P.; Yu, D. Environmental adaptation of the root microbiome in two rice ecotypes. Microbiol. Res. 2020, 241, 126588. [Google Scholar] [CrossRef]
- Pang, Z.; Zhao, Y.; Xu, P.; Yu, D. Microbial diversity of upland rice roots and their influence on rice growth and drought tolerance. Microorganisms 2020, 8, 1329. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, J.; Qiu, C.; Bao, Y.; Feng, Y.; Lin, X. Study on community assembly processes under paddy-upland rotation. Soils 2020, 52, 710–717. [Google Scholar] [CrossRef]
- Bao, Q.; Wang, F.; Bao, W.; Huang, Y. Effects of rice straw addition on methanogenic archaea and bacteria in two paddy soils. Environ. Sci. 2019, 40, 4202–4212. [Google Scholar] [CrossRef]
- Vaksmaa, A.; van Alen, T.A.; Ettwig, K.F.; Lupotto, E.; Valè, G.; Jetten, M.S.M.; Lüke, C. Stratification of diversity and activity of methanogenic and methanotrophic microorganisms in a nitrogen-fertilized Italian paddy soil. Front. Microbiol. 2017, 8, 2127. [Google Scholar] [CrossRef]
- Fan, D.J.; Liu, T.Q.; Sheng, F.; Li, S.H.; Cao, C.G.; Li, C.F. Nitrogen deep placement mitigates methane emissions by regulating methanogens and methanotrophs in no-tillage paddy fields. Biol. Fertil. Soils 2020, 56, 711–727. [Google Scholar] [CrossRef]
- Bao, Q.; Huang, Y.; Wang, F.; Nie, S.; Nicol, G.W.; Yao, H.; Ding, L. Effect of nitrogen fertilizer and/or rice straw amendment on methanogenic archaeal communities and methane production from a rice paddy soil. Appl. Microbiol. Biotechnol. 2016, 100, 5989–5998. [Google Scholar] [CrossRef]
- Bai, Y.; Mueller, D.B.; Srinivas, G.; Garrido-Oter, R.; Potthoff, E.; Rott, M.; Dombrowski, N.; Münch, P.C.; Spaepen, S.; Remus-Emsermann, M.; et al. Functional overlap of the arabidopsis leaf and root microbiota. Nature 2015, 528, 364–369. [Google Scholar] [CrossRef]
- Pascual-García, A.; Bonhoeffer, S.; Bell, T. Metabolically cohesive microbial consortia and ecosystem functioning. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190245. [Google Scholar] [CrossRef]
- Fux, C.A.; Shirtliff, M.; Stoodley, P.; Costerton, J.W. Can laboratory reference strains mirror ‘real-world’ pathogenesis? Trends Microbiol. 2005, 13, 58–63. [Google Scholar] [CrossRef]
- Zoetendal, E.G.; Ben-Amor, K.; Akkermans, A.D.; Abee, T.; De-Vos, W.M. DNA isolation protocols affect the detection limit of PCR approaches of bacteria in samples from the human gastrointestinal tract. Syst. Appl. Microbiol. 2001, 24, 405–410. [Google Scholar] [CrossRef]
- Zhou, J.; He, Z.; Yang, Y.; Deng, Y.; Alvarez-Cohen, L. High-throughput metagenomic technologies for complex microbial community analysis: Open and closed formats. MBio 2015, 6, e0228814. [Google Scholar] [CrossRef]
- Song, L.; Xie, K. Engineering CRISPR/Cas9 to mitigate abundant host contamination for 16S rRNA gene-based amplicon sequencing. Microbiome 2020, 8, 80. [Google Scholar] [CrossRef]
- Rubin, B.E.; Diamond, S.; Cress, B.F.; Crits-Christoph, A.; He, C.; Xu, M.; Zhou, Z.; Smock, D.C.; Tang, K.; Owens, T.K.; et al. Targeted genome editing of bacteria within microbial communities. bioRxiv 2020. [Google Scholar] [CrossRef]
- Strecker, J.; Ladha, A.; Gardner, Z.; Schmid-Burgk, J.L.; Makarova, K.S.; Koonin, E.V.; Zhang, F. RNA-guided DNA insertion with CRISPR-associated transposases. Science 2019, 365, 48–53. [Google Scholar] [CrossRef]
- Klompe, S.E.; Vo, P.L.H.; Halpin-Healy, T.S.; Sternberg, S.H. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature 2019, 571, 219–225. [Google Scholar] [CrossRef]
- Petassi, M.T.; Hsieh, S.C.; Peters, J.E. Guide RNA categorization enables target site choice in Tn7-CRISPR-Cas transposons. Cell 2020, 183, 1757–1771. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, Q.; Hu, J.; Wei, H.; Zhang, H.; Zhu, J. Relationship between Plant Roots, Rhizosphere Microorganisms, and Nitrogen and Its Special Focus on Rice. Agriculture 2021, 11, 234. https://doi.org/10.3390/agriculture11030234
Xiong Q, Hu J, Wei H, Zhang H, Zhu J. Relationship between Plant Roots, Rhizosphere Microorganisms, and Nitrogen and Its Special Focus on Rice. Agriculture. 2021; 11(3):234. https://doi.org/10.3390/agriculture11030234
Chicago/Turabian StyleXiong, Qiangqiang, Jinlong Hu, Haiyan Wei, Hongcheng Zhang, and Jinyan Zhu. 2021. "Relationship between Plant Roots, Rhizosphere Microorganisms, and Nitrogen and Its Special Focus on Rice" Agriculture 11, no. 3: 234. https://doi.org/10.3390/agriculture11030234
APA StyleXiong, Q., Hu, J., Wei, H., Zhang, H., & Zhu, J. (2021). Relationship between Plant Roots, Rhizosphere Microorganisms, and Nitrogen and Its Special Focus on Rice. Agriculture, 11(3), 234. https://doi.org/10.3390/agriculture11030234





