Genome-Wide Association Study Reveals the Genetic Basis of Cold Tolerance in Rice at the Seedling Stage
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Genotyping
2.2. Cold Treatment and Cold Tolerance Evaluation
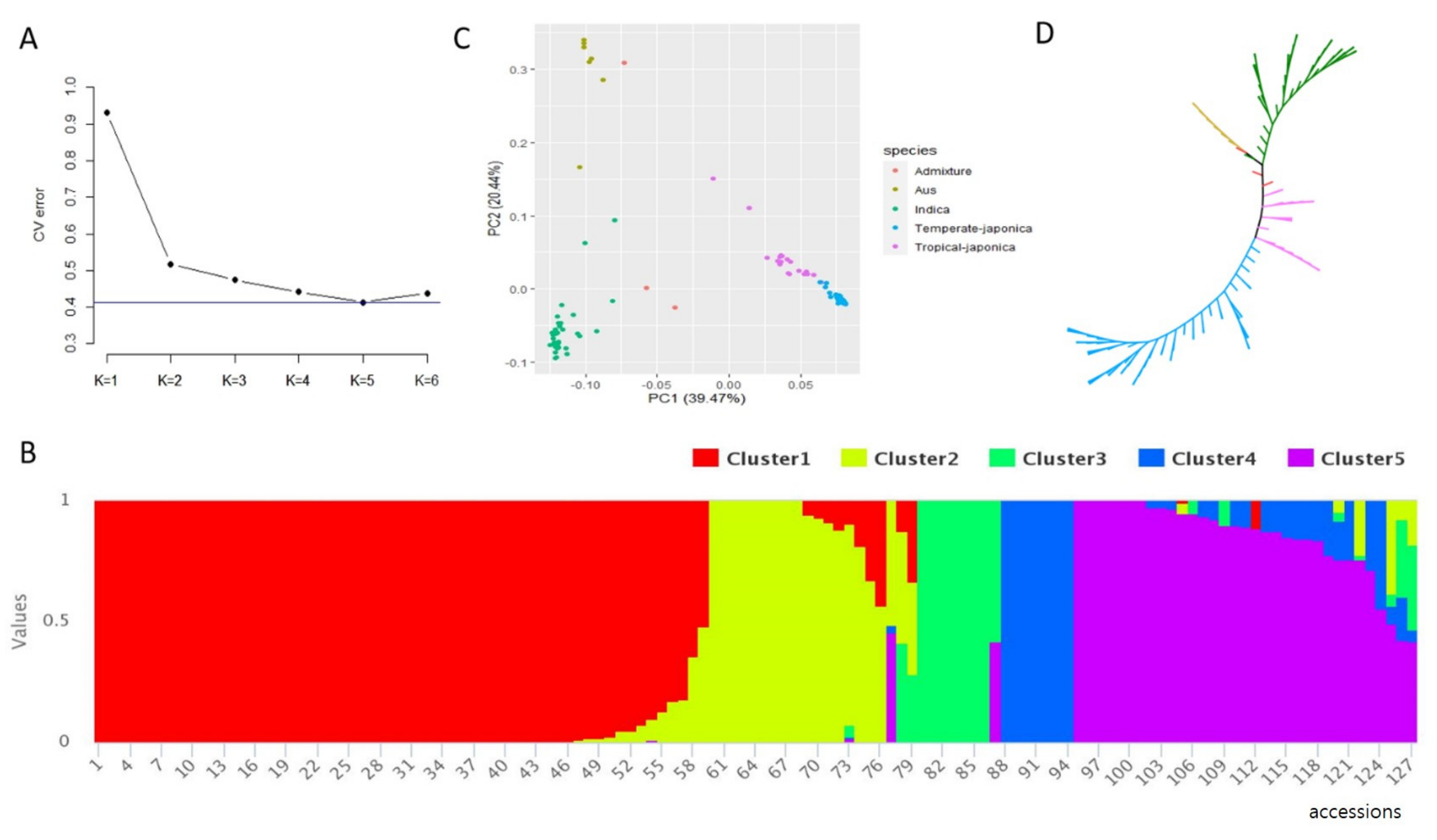
2.3. Population Structure and Linkage Disequilibrium (LD) Decay Analysis
2.4. GWAS
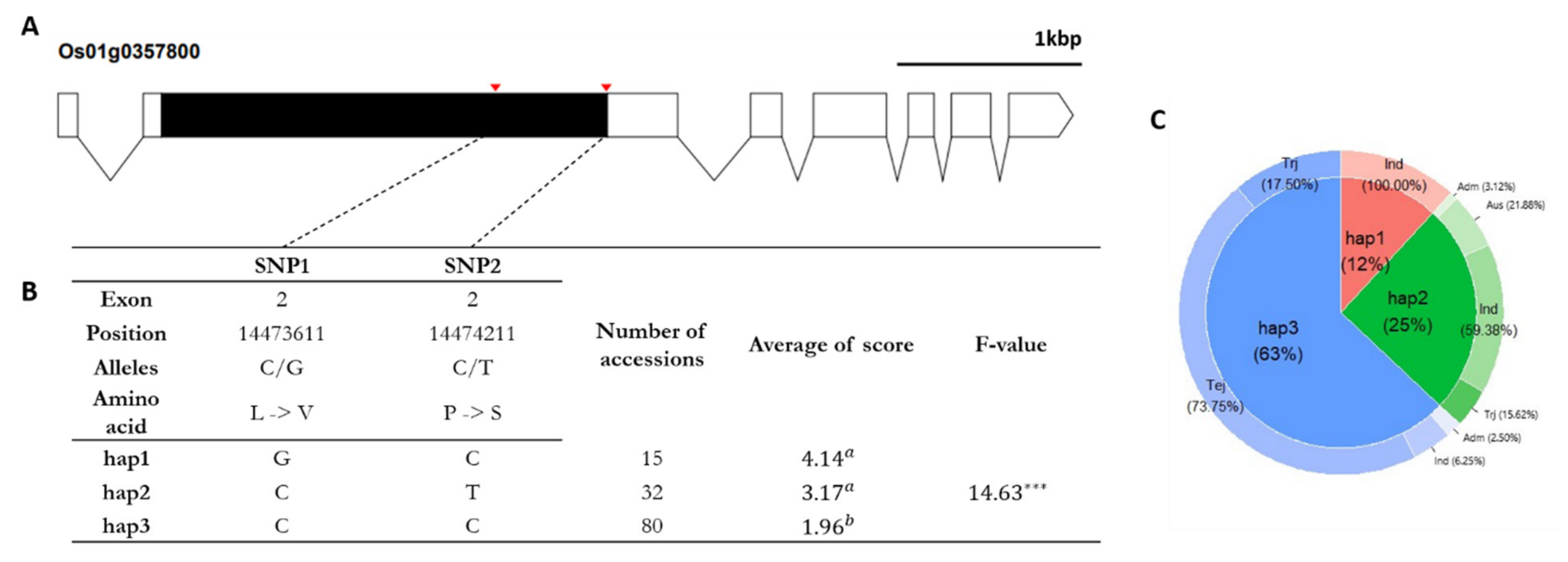
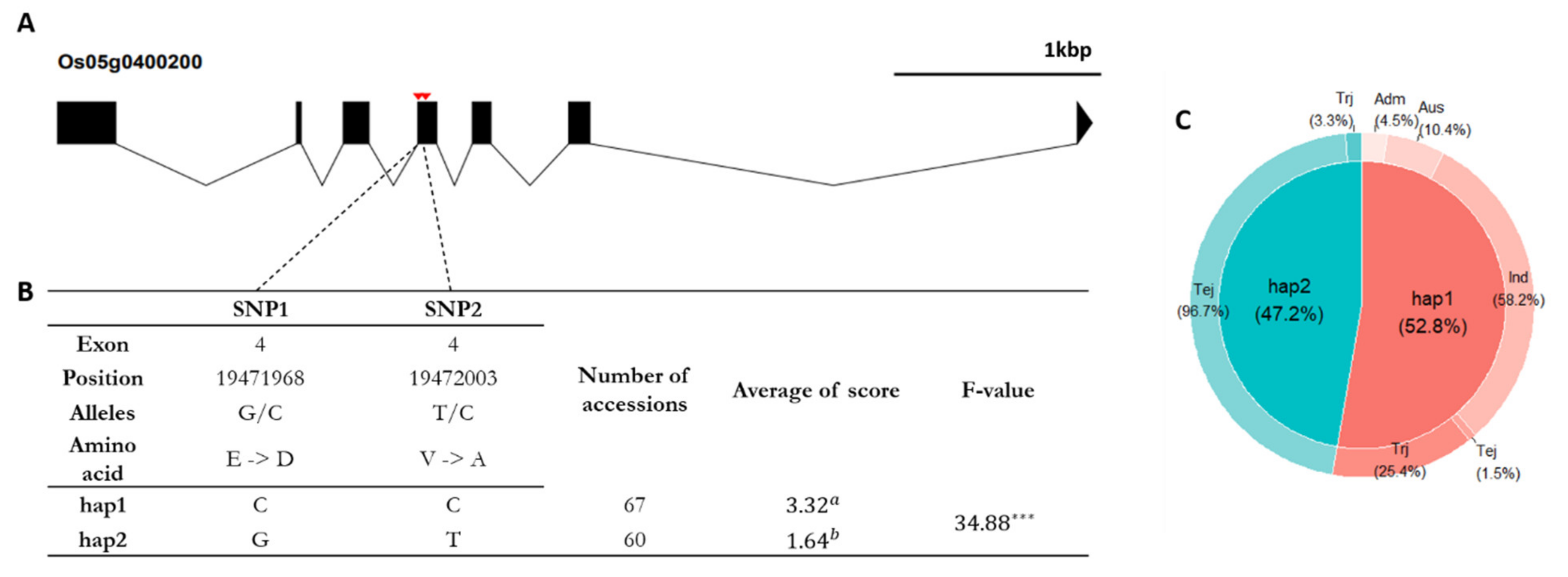
2.5. Candidate Gene Prediction and Haplotype Analysis
3. Results
3.1. Cold Tolerance in Rice at the Seedling Stage
3.2. Population Structure and LD Decay
3.3. GWAS of Cold Stress Tolerance in Rice Seedlings
3.4. Haplotype Analysis to Identify Candidate Genes Underlying Cold Tolerance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Almansouri, M.; Kinet, J.M.; Lutts, S. Effect of salt and osmotic stresses on germination in durum wheat (triticum durum desf.). Plant Soil 2001, 231, 243–254. [Google Scholar] [CrossRef]
- Nakagahra, M.; Okuno, K.; Vaughan, D. Rice genetic resources: History, conservation, investigative characterization and use in japan. Plant Mol. Biol. 1997, 35, 69–77. [Google Scholar] [CrossRef]
- Jiang, L.; Liu, S.J.; Hou, M.Y.; Tang, J.Y.; Chen, L.M.; Zhai, H.Q.; Wan, J.M. Analysis of qtls for seed low temperature germinability and anoxia germinability in rice (Oryza sativa L.). Field Crop. Res. 2006, 98, 68–75. [Google Scholar] [CrossRef]
- Suh, J.P.; Jeung, J.U.; Lee, J.I.; Choi, Y.H.; Yea, J.D.; Virk, P.S.; Mackill, D.J.; Jena, K.K. Identification and analysis of qtls controlling cold tolerance at the reproductive stage and validation of effective qtls in cold-tolerant genotypes of rice (Oryza sativa L.). Theor. Appl. Genet. 2010, 120, 985–995. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, H.; Zhang, D.; Li, J.; Xiong, H.; Yu, J.; Li, J.; Rashid, M.A.R.; Li, G.; Ma, X.; et al. Genetic analysis of cold tolerance at the germination and booting stages in rice by association mapping. PLoS ONE 2015, 10, e0120590. [Google Scholar] [CrossRef]
- Zhao, X.Q.; Wang, W.S.; Zhang, F.; Zhang, T.; Zhao, W.; Fu, B.Y.; Li, Z.K. Temporal profiling of primary metabolites under chilling stress and its association with seedling chilling tolerance of rice (Oryza sativa L.). Rice 2013, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mao, D.H.; Yu, L.; Chen, D.Z.; Li, L.Y.; Zhu, Y.X.; Xiao, Y.Q.; Zhang, D.C.; Chen, C.Y. Multiple cold resistance loci confer the high cold tolerance adaptation of dongxiang wild rice (Oryza rufipogon) to its high-latitude habitat. Theor. Appl. Genet. 2015, 128, 1359–1371. [Google Scholar] [CrossRef]
- Ranawake, A.L.; Manangkil, O.E.; Yoshida, S.; Ishii, T.; Mori, N.; Nakamura, C. Mapping qtls for cold tolerance at germination and the early seedling stage in rice (Oryza sativa L.). Biotechnol. Biotec. Eq. 2014, 28, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.M.; Huang, D.Q.; Tang, W.Q.; Zheng, Y.; Liang, K.J.; Cutler, A.J.; Wu, W.R. Mapping of quantitative trait loci underlying cold tolerance in rice seedlings via high-throughput sequencing of pooled extremes. PLoS ONE 2013, 8, e68433. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.J.; Chen, K.; Mi, X.F.; Chen, T.X.; Ali, J.; Ye, G.Y.; Xu, J.L.; Li, Z.K. Identification and fine mapping of a stably expressed qtl for cold tolerance at the booting stage using an interconnected breeding population in rice. PLoS ONE 2015, 10, e0145704. [Google Scholar] [CrossRef] [PubMed]
- Xiao, N.; Huang, W.N.; Zhang, X.X.; Gao, Y.; Li, A.H.; Dai, Y.; Yu, L.; Liu, G.Q.; Pan, C.H.; Li, Y.H.; et al. Fine mapping of qrc10-2, a quantitative trait locus for cold tolerance of rice roots at seedling and mature stages. PLoS ONE 2014, 9, e96046. [Google Scholar] [CrossRef] [PubMed]
- Cruz, R.P.d.; Milach, S.C.K. Cold tolerance at the germination stage of rice: Methods of evaluation and characterization of genotypes. Sci. Agric. 2004, 61, 1–8. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, Q.H.; Wang, S.L.; Hong, Y.H.; Wang, Z.L. Rice and cold stress: Methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice 2014, 7, 24. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ma, X.F.; Gao, Y.M.; Hao, X.B.; Li, Z.K. Genome-wide response to selection and genetic basis of cold tolerance in rice (Oryza sativa l.). BMC Genet. 2014, 15, 55. [Google Scholar] [CrossRef] [PubMed]
- Fujino, K.; Sekiguchi, H.; Matsuda, Y.; Sugimoto, K.; Ono, K.; Yano, M. Molecular identification of a major quantitative trait locus, qltg3-1, controlling low-temperature germinability in rice. Proc. Natl. Acad. Sci. USA 2008, 105, 12623–12628. [Google Scholar] [CrossRef]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. Cold1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef]
- Fujino, K.; Obara, M.; Shimizu, T.; Koyanagi, K.O.; Ikegaya, T. Genome-wide association mapping focusing on a rice population derived from rice breeding programs in a region. Breeding Sci. 2015, 65, 403–410. [Google Scholar] [CrossRef]
- Sales, E.; Viruel, J.; Domingo, C.; Marques, L. Genome wide association analysis of cold tolerance at germination in temperate japonica rice (Oryza sativa L.) varieties. PLoS ONE 2017, 12, e0183416. [Google Scholar] [CrossRef] [PubMed]
- Shakiba, E.; Edwards, J.D.; Jodari, F.; Duke, S.E.; Baldo, A.M.; Korniliev, P.; McCouch, S.R.; Eizenga, G.C. Genetic architecture of cold tolerance in rice (Oryza sativa) determined through high resolution genome-wide analysis. PLoS ONE 2017, 12, e0172133. [Google Scholar] [CrossRef] [PubMed]
- de Freitas, G.M.; Thomas, J.; Liyanage, R.; Lay, J.O.; Basu, S.; Ramegowda, V.; do Amaral, M.N.; Benitez, L.C.; Bolacel Braga, E.J.; Pereira, A. Cold tolerance response mechanisms revealed through comparative analysis of gene and protein expression in multiple rice genotypes. PLoS ONE 2019, 14, e0218019. [Google Scholar]
- Yadav, S.K. Cold stress tolerance mechanisms in plants. A review. Agron Sustain. Dev. 2010, 30, 515–527. [Google Scholar] [CrossRef]
- Welti, R.; Li, W.; Li, M.; Sang, Y.; Biesiada, H.; Zhou, H.E.; Rajashekar, C.B.; Williams, T.D.; Wang, X. Profiling membrane lipids in plant stress responses. Role of phospholipase d alpha in freezing-induced lipid changes in arabidopsis. J. Biol. Chem. 2002, 277, 31994–32002. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K.; Seki, M. Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant. Biol. 2003, 6, 410–417. [Google Scholar] [CrossRef]
- Maruyama, K.; Sakuma, Y.; Kasuga, M.; Ito, Y.; Seki, M.; Goda, H.; Shimada, Y.; Yoshida, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Identification of cold-inducible downstream genes of the arabidopsis dreb1a/cbf3 transcriptional factor using two microarray systems. Plant. J. 2004, 38, 982–993. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Pan, Y.; Li, J.; Zhou, L.; Shi, H.; Zeng, Y.; Guo, H.; Yang, S.; Zheng, W.; et al. Natural variation in ctb4a enhances rice adaptation to cold habitats. Nat. Commun. 2017, 8, 14788. [Google Scholar] [CrossRef]
- Dai, L.Y.C.; Yu, T.; Xu, F. Studies on cold tolerance of rice, Oryza sativa L. I. Description on types of cold injury and classifications of evaluation methods on cold tolerance in rice. Southwest China J. Agrc. Sci. 2002, 15, 5. [Google Scholar]
- Zhao, W.G.; Cho, G.T.; Ma, K.H.; Chung, J.W.; Gwag, J.G.; Park, Y.J. Development of an allele-mining set in rice using a heuristic algorithm and ssr genotype data with least redundancy for the post-genomic era. Mol. Breed. 2010, 26, 639–651. [Google Scholar] [CrossRef]
- Kim, K.W.; Chung, H.K.; Cho, G.T.; Ma, K.H.; Chandrabalan, D.; Gwag, J.G.; Kim, T.S.; Cho, E.G.; Park, Y.J. Powercore: A program applying the advanced m strategy with a heuristic search for establishing core sets. Bioinformatics 2007, 23, 2155–2162. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.S.; He, Q.; Kim, K.W.; Yoon, M.Y.; Ra, W.H.; Li, F.P.; Tong, W.; Yu, J.; Oo, W.H.; Choi, B.; et al. Genome-wide resequencing of krice_core reveals their potential for future breeding, as well as functional and evolutionary studies in the post-genomic era. BMC Genom. 2016, 17, 408. [Google Scholar] [CrossRef]
- Kim, J.; Zhang, Y.W.; Pan, W. Powerful and adaptive testing for multi-trait and multi-snp associations with gwas and sequencing data. Genet. Epidemiol. 2016, 40, 646. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. Plink: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, L.; Harper, J.; Uno, H.; Gaito, G.; Audette-Arruda, J.; Kurata, S.; Berman, C.; Primka, R.; Pikounis, B. The effects of finasteride (proscar) on hair growth, hair cycle stage, and serum testosterone and dihydrotestosterone in adult male and female stumptail macaques (macaca arctoides). J. Clin. Endocrinol. Metab. 1994, 79, 991–996. [Google Scholar] [PubMed]
- IRRI. Standard Evaluation System(ses), 5th ed.; IRRI: Laguna, Philippines, 2013; p. 35. [Google Scholar]
- Alexander, D.H.; Lange, K. Enhancements to the admixture algorithm for individual ancestry estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef] [PubMed]
- Francis, R.M. Pophelper: An r package and web app to analyse and visualize population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega x: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (itol) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.-S.; Xu, J.-Y.; He, W.-M.; Yang, T.-L. Poplddecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Kang, H.M.; Zaitlen, N.A.; Wade, C.M.; Kirby, A.; Heckerman, D.; Daly, M.J.; Eskin, E. Efficient control of population structure in model organism association mapping. Genetics 2008, 178, 1709–1723. [Google Scholar] [CrossRef]
- Yang, W.; Guo, Z.; Huang, C.; Duan, L.; Chen, G.; Jiang, N.; Fang, W.; Feng, H.; Xie, W.; Lian, X. Combining high-throughput phenotyping and genome-wide association studies to reveal natural genetic variation in rice. Nat. Commun. 2014, 5, 5087. [Google Scholar] [CrossRef]
- Karmanova, I.G.; Aristakesian, E.A.; Shilling, N.V. Neurophysiologic analysis of hypothalamic mechanisms of primary sleep and hypobiosis. Dokl. Akad. Nauk SSSR 1987, 294, 245–248. [Google Scholar]
- Andaya, V.C.; Tai, T.H. Fine mapping of the qcts4 locus associated with seedling cold tolerance in rice (Oryza sativa L.). Mol. Breed. 2007, 20, 349–358. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Qu, X.S.; Wan, S.; Chen, L.H.; Zhu, Y.G. Comparison of qtl controlling seedling vigour under different temperature conditions using recombinant inbred lines in rice (Oryza sativa). Ann. Bot. 2005, 95, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Kamoshita, A.; Wade, J.; Ali, L.; Pathan, S.; Zhang, J.; Sarkarung, S.; Nguyen, T. Mapping qtls for root morphology of a rice population adapted to rainfed lowland conditions. Theor. Appl. Genet. 2002, 104, 880–893. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, K.; Kobayashi, N.; Ono, K.; Yano, M.; Ohsugi, R. Are contents of rubisco, soluble protein and nitrogen in flag leaves of rice controlled by the same genetics? J. Exp. Bot. 2001, 52, 1827–1833. [Google Scholar] [CrossRef] [PubMed]
- Ming, F.; Zheng, X.; Mi, G.; Zhu, L.; Zhang, F. Detection and verification of quantitative trait loci affecting tolerance to low phosphorus in rice. J. Plant Nutr. 2001, 24, 1399–1408. [Google Scholar]
- Huang, Z.; Yu, T.; Su, L.; Yu, S.B.; Zhang, Z.H.; Zhu, Y.G. Identification of chromosome regions associated with seedling vigor in rice. Acta Genet. Sin. 2004, 31, 596–603. [Google Scholar]
- Mackill, D.J.; Lei, X. Genetic variation for traits related to temperate adaptation of rice cultivars. Crop. Sci. 1997, 37, 1340–1346. [Google Scholar] [CrossRef]
- Ni, J.; Pujar, A.; Youens-Clark, K.; Yap, I.; Jaiswal, P.; Tecle, I.; Tung, C.-W.; Ren, L.; Spooner, W.; Wei, X. Gramene qtl database: Development, content and applications. Database 2009, 2009, bap005. [Google Scholar] [CrossRef]
- Morin, E.; Kohler, A.; Baker, A.R.; Foulongne-Oriol, M.; Lombard, V.; Nagy, L.G.; Ohm, R.A.; Patyshakuliyeva, A.; Brun, A.; Aerts, A.L.; et al. Genome sequence of the button mushroom agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc. Natl. Acad. Sci. USA 2012, 109, 17501–17506. [Google Scholar] [CrossRef]
- Kawahara, Y.; Oono, Y.; Wakimoto, H.; Ogata, J.; Kanamori, H.; Sasaki, H.; Mori, S.; Matsumoto, T.; Itoh, T. Tenor: Database for comprehensive mrna-seq experiments in rice. Plant. Cell Physiol. 2016, 57, e7. [Google Scholar] [CrossRef]
- Xing, H.; Fu, X.; Yang, C.; Tang, X.; Guo, L.; Li, C.; Xu, C.; Luo, K. Genome-wide investigation of pentatricopeptide repeat gene family in poplar and their expression analysis in response to biotic and abiotic stresses. Sci. Rep. 2018, 8, 2817. [Google Scholar]
- Koussevitzky, S.; Nott, A.; Mockler, T.C.; Hong, F.; Sachetto-Martins, G.; Surpin, M.; Lim, J.; Mittler, R.; Chory, J. Signals from chloroplasts converge to regulate nuclear gene expression. Science 2007, 316, 715–719. [Google Scholar]
- Kobayashi, K.; Suzuki, M.; Tang, J.; Nagata, N.; Ohyama, K.; Seki, H.; Kiuchi, R.; Kaneko, Y.; Nakazawa, M.; Matsui, M. Lovastatin insensitive 1, a novel pentatricopeptide repeat protein, is a potential regulatory factor of isoprenoid biosynthesis in arabidopsis. Plant. Cell Physiol. 2007, 48, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Kobayashi, K.; Suzuki, M.; Matsumoto, S.; Muranaka, T. The mitochondrial ppr protein lovastatin insensitive 1 plays regulatory roles in cytosolic and plastidial isoprenoid biosynthesis through rna editing. Plant. J. 2010, 61, 456–466. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Liu, D. Functional disruption of the pentatricopeptide protein slg1 affects mitochondrial rna editing, plant development, and responses to abiotic stresses in arabidopsis. Plant. J. 2012, 70, 432–444. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Bennetzen, J.L. Plant retrotransposons. Annu. Rev. Genet. 1999, 33, 479–532. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Sugimoto, K.; Otsuki, H.; Hirochika, H. A 13-bp cis-regulatory element in the ltr promoter of the tobacco retrotransposon tto1 is involved in responsiveness to tissue culture, wounding, methyl jasmonate and fungal elicitors. Plant. J. 1999, 18, 383–393. [Google Scholar] [CrossRef]
- Kanazawa, A.; Liu, B.; Kong, F.; Arase, S.; Abe, J. Adaptive evolution involving gene duplication and insertion of a novel ty1/copia-like retrotransposon in soybean. J. Mol. Evol. 2009, 69, 164–175. [Google Scholar] [CrossRef]
- Li, S.; Wei, X.; Ren, Y.; Qiu, J.; Jiao, G.; Guo, X.; Tang, S.; Wan, J.; Hu, P. Osbt1 encodes an adp-glucose transporter involved in starch synthesis and compound granule formation in rice endosperm. Sci. Rep. 2017, 7, 40124. [Google Scholar] [CrossRef]
- Thalmann, M.; Santelia, D. Starch as a determinant of plant fitness under abiotic stress. New Phytol. 2017, 214, 943–951. [Google Scholar] [CrossRef]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef]
- Rook, F.; Hadingham, S.A.; Li, Y.; Bevan, M.W. Sugar and aba response pathways and the control of gene expression. Plant. Cell Environ. 2006, 29, 426–434. [Google Scholar] [CrossRef]




| QTL | Lead SNP | Chr | −log10(P) | Reported QTL | Reference of Previously Reported QTLs | |
|---|---|---|---|---|---|---|
| QTL Accession ID | Related Trait | |||||
| qCTS1 | 14,648,882 | 1 | 6.64 | AQAL060 | Root thickness | [44] |
| qCTS4 | 3,829,001 | 4 | 6.34 | CQAA8 | Cold tolerance | [42,46] |
| qCTS5-1 | 4,437,944 | 5 | 6.40 | CQE39 | Rubisco content | [45] |
| qCTS5-2 | 19,300,152 | 5 | 6.97 | AQFR028 | Seedling vigor | [47] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ham, T.-H.; Kwon, Y.; Lee, Y.; Choi, J.; Lee, J. Genome-Wide Association Study Reveals the Genetic Basis of Cold Tolerance in Rice at the Seedling Stage. Agriculture 2021, 11, 318. https://doi.org/10.3390/agriculture11040318
Ham T-H, Kwon Y, Lee Y, Choi J, Lee J. Genome-Wide Association Study Reveals the Genetic Basis of Cold Tolerance in Rice at the Seedling Stage. Agriculture. 2021; 11(4):318. https://doi.org/10.3390/agriculture11040318
Chicago/Turabian StyleHam, Tae-Ho, Yebin Kwon, Yoonjung Lee, Jisu Choi, and Joohyun Lee. 2021. "Genome-Wide Association Study Reveals the Genetic Basis of Cold Tolerance in Rice at the Seedling Stage" Agriculture 11, no. 4: 318. https://doi.org/10.3390/agriculture11040318
APA StyleHam, T.-H., Kwon, Y., Lee, Y., Choi, J., & Lee, J. (2021). Genome-Wide Association Study Reveals the Genetic Basis of Cold Tolerance in Rice at the Seedling Stage. Agriculture, 11(4), 318. https://doi.org/10.3390/agriculture11040318






