The Effects of Microbial Inoculants on Bacterial Communities of the Rhizosphere Soil of Maize
Abstract
1. Introduction
2. Materials and Methods
2.1. Screening and Identification of Bacterial Strains
2.2. Preparation of Three Inoculants for Application of Field Experiment
2.3. Conditions and Treatment Design of Field Experiment
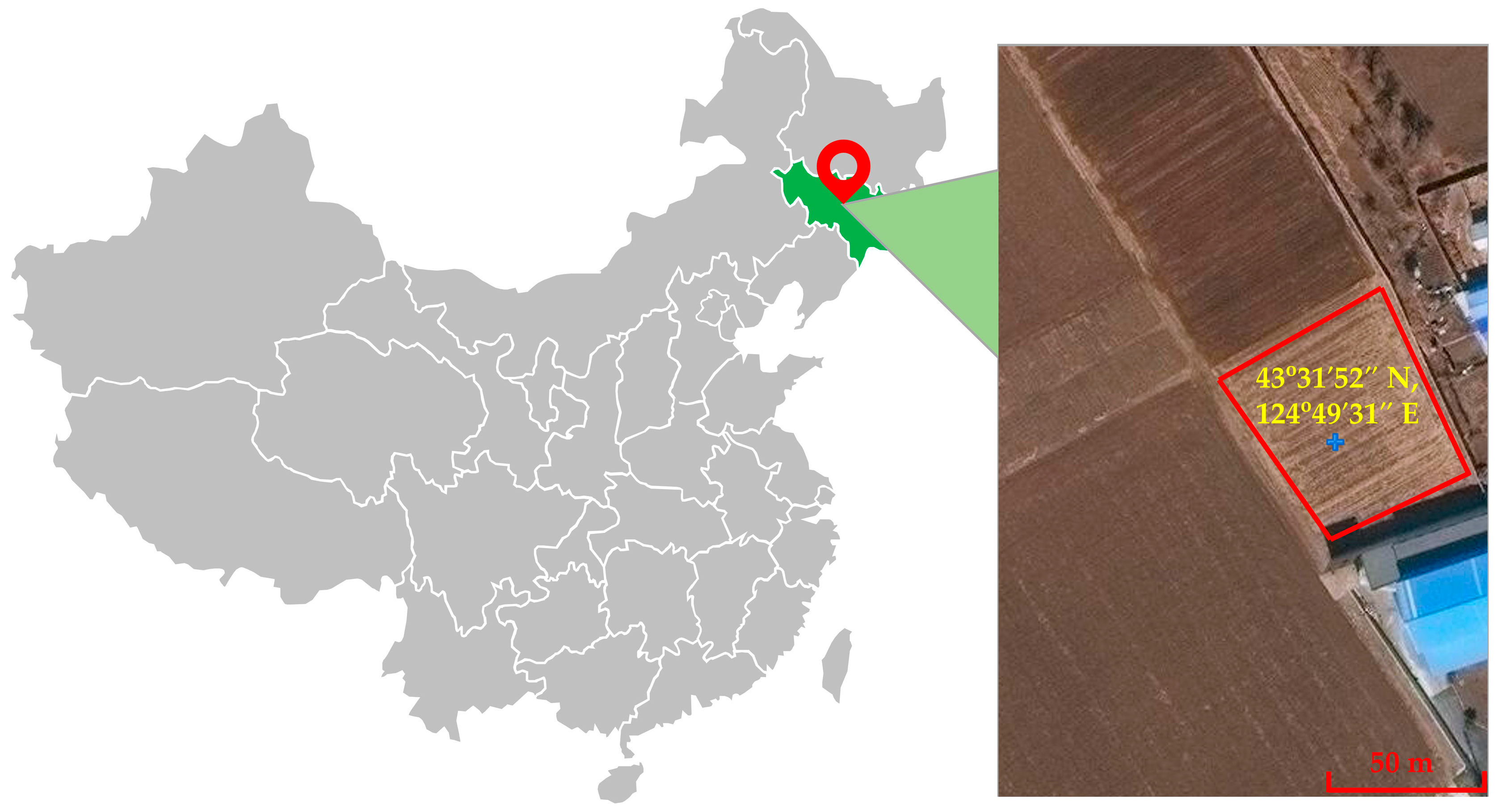
2.4. Sample Collection
2.5. DNA Extraction and Polymerase Chain Reaction (PCR) Amplification
2.6. Library Construction and Sequencing
2.7. Data Processing and OTU Clustering and Annotation
2.8. Bioinformatic Analyses
2.9. Statistical Analysis
3. Results
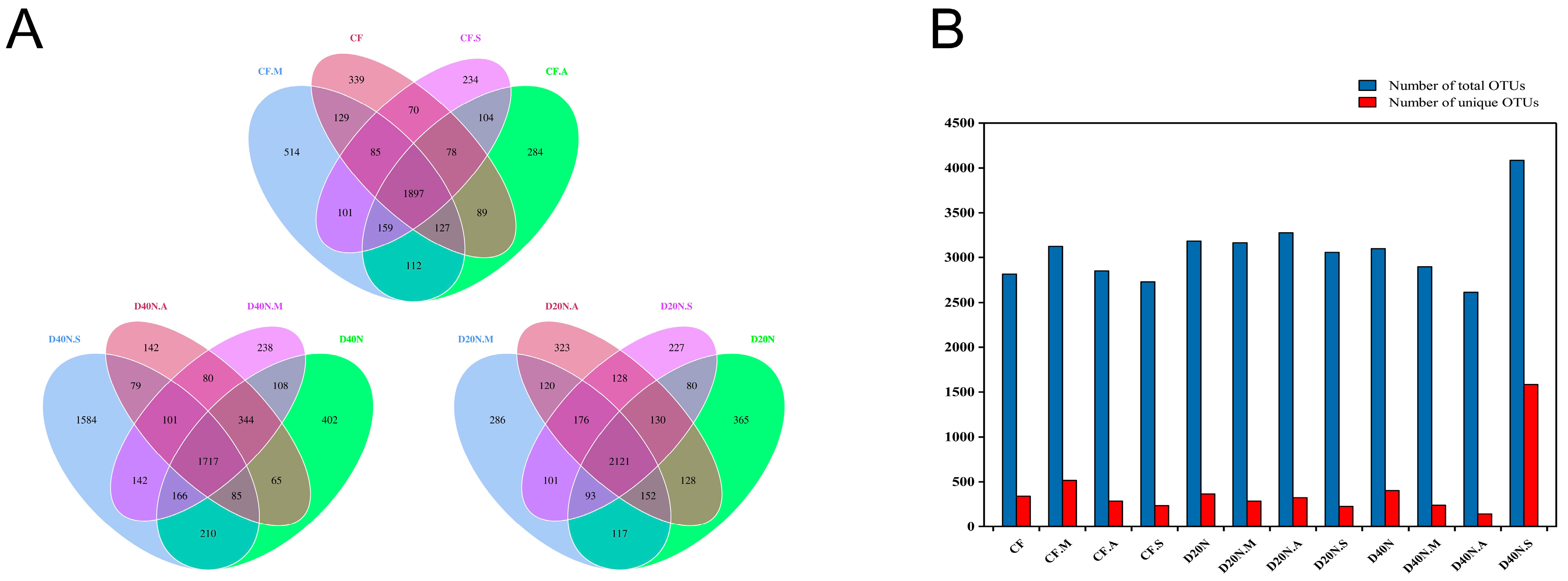
3.1. Sequencing Results
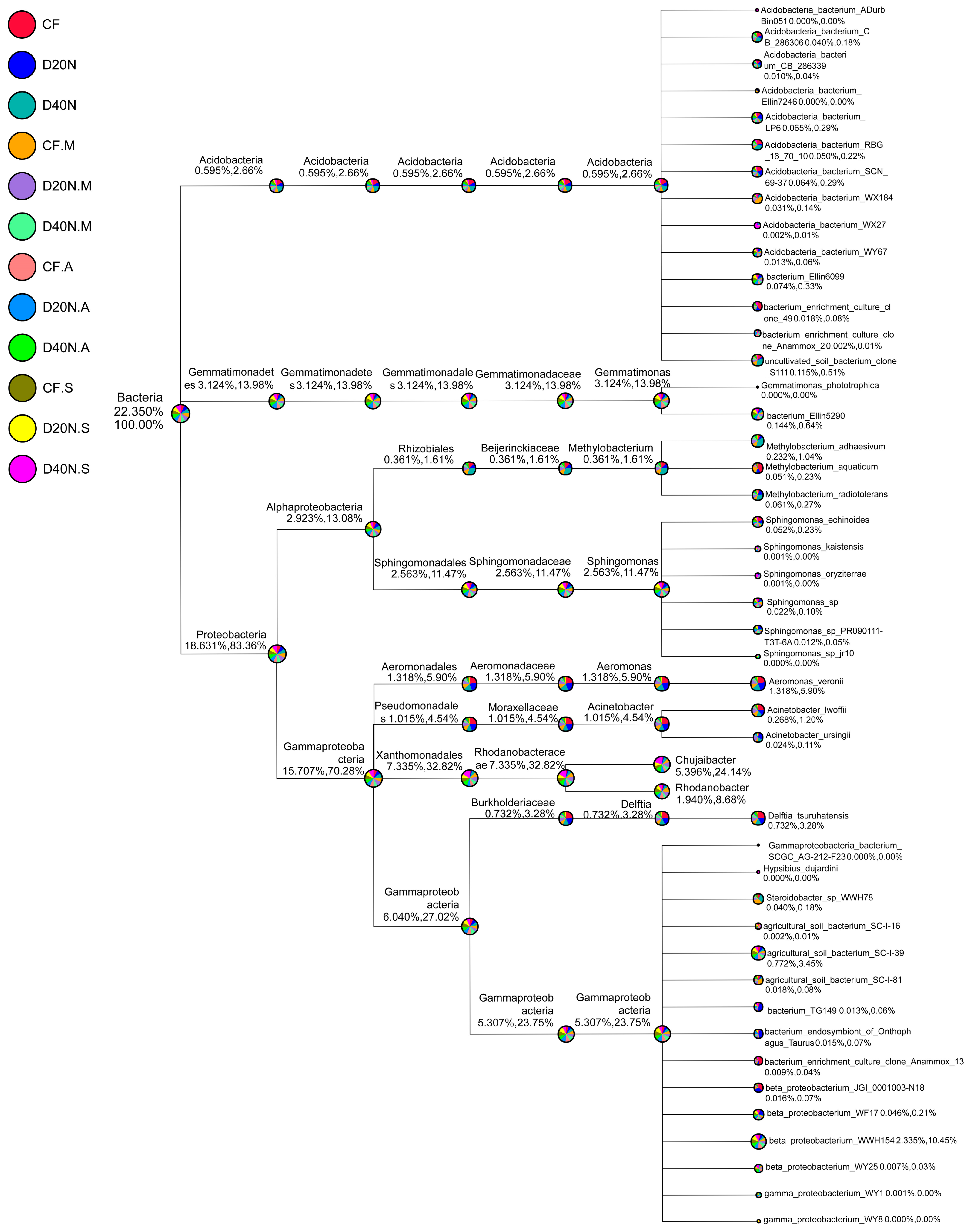
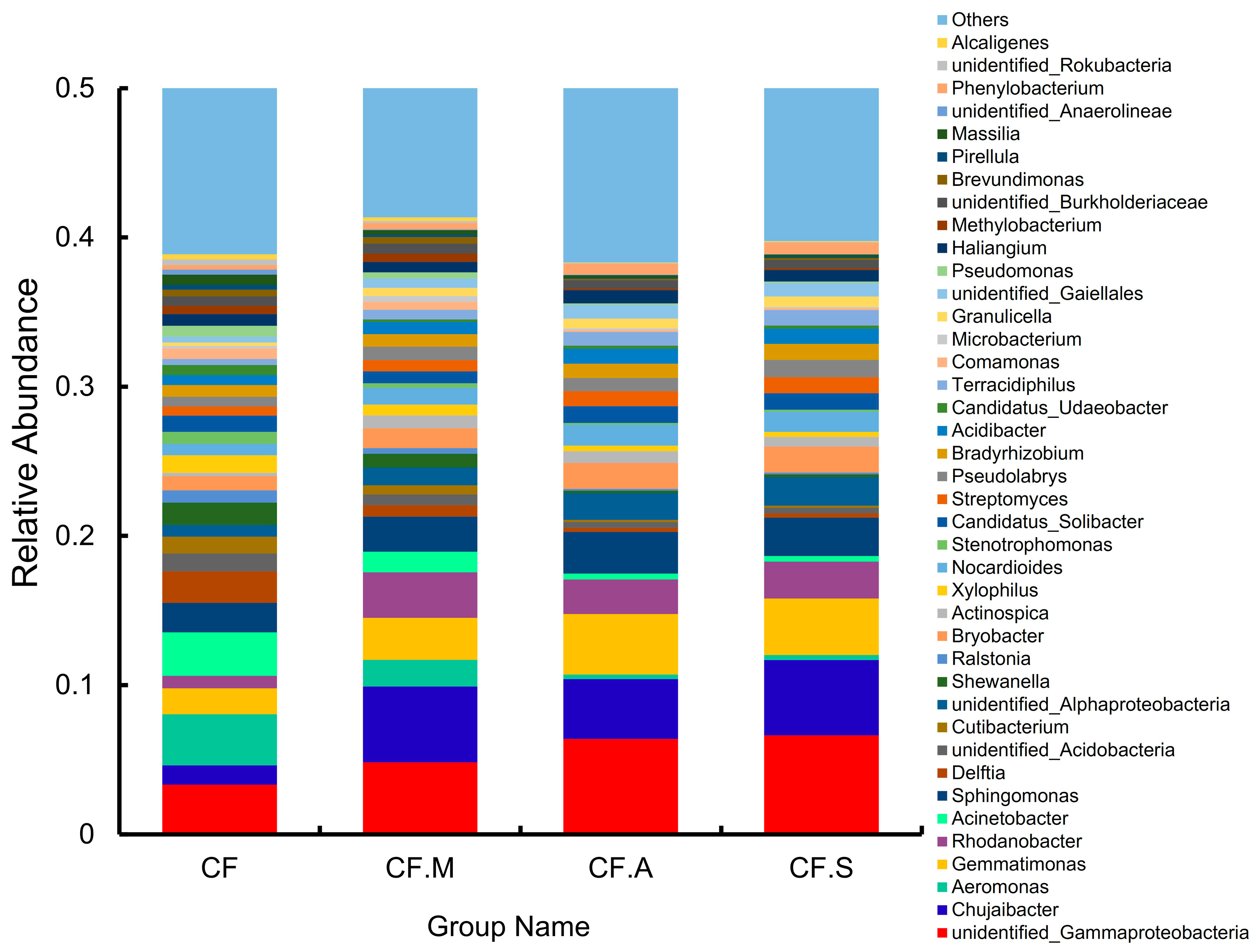
3.2. Overview of Bacterial Taxonomic Composition
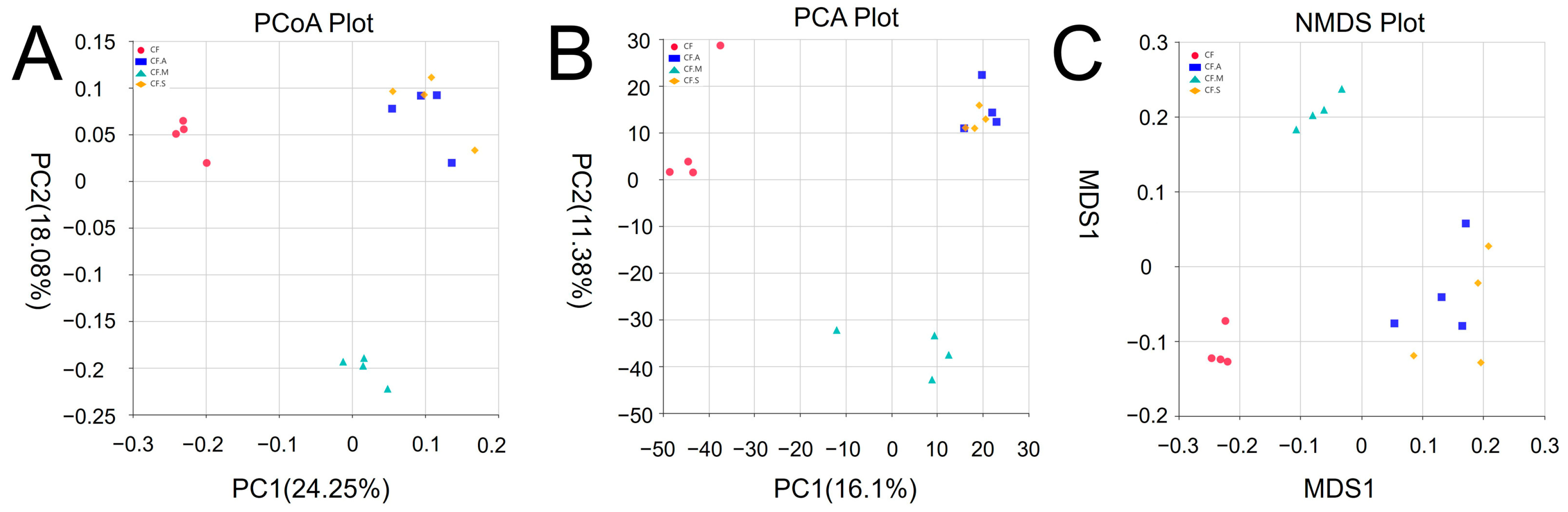
3.3. Dissimilarity of Bacterial Communities in Different Treatments
3.4. Alpha Diversity
3.5. Beta Diversity
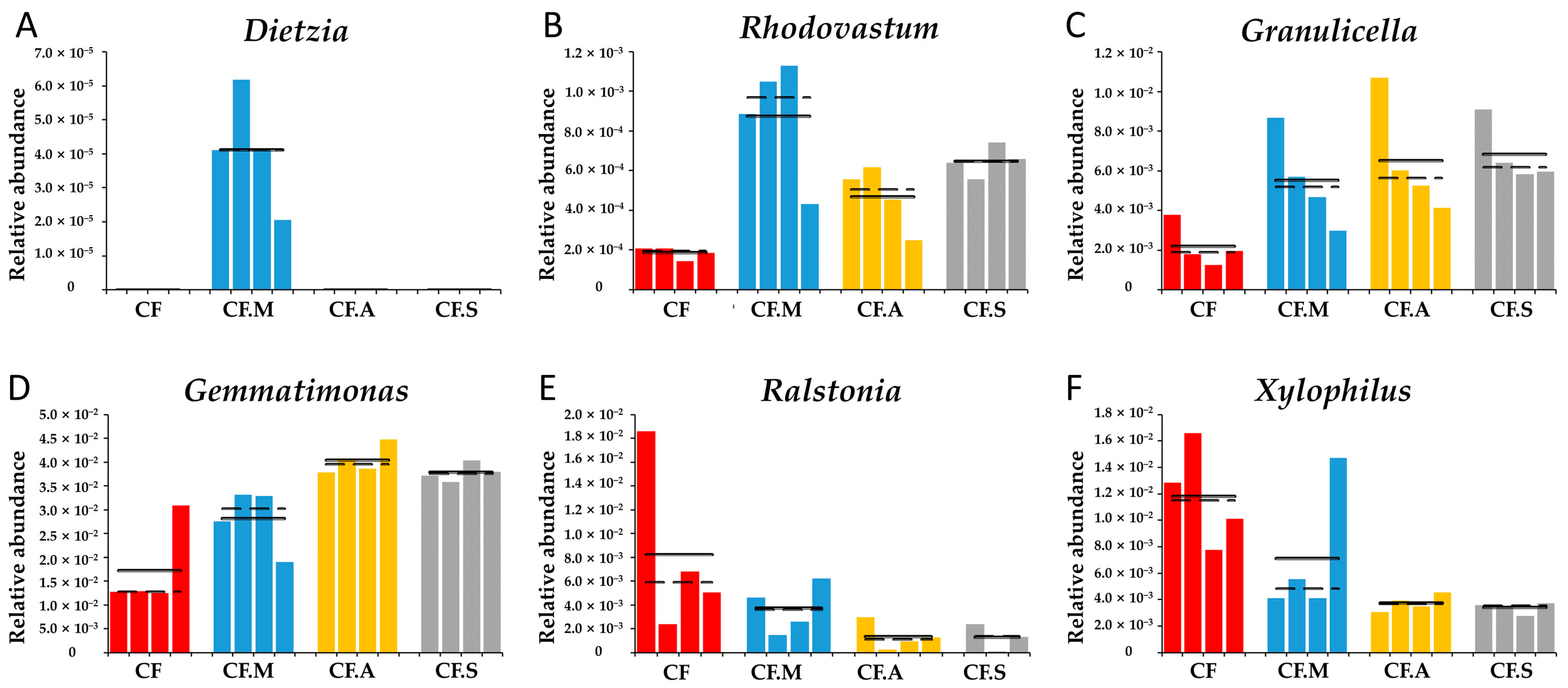
3.6. Inoculant M Mediated the Key Phylotypes of the Rhizosphere Microbiome of Maize
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Busby, P.E.; Soman, C.; Wagner, M.R.; Friesen, M.L.; Kremer, J.; Bennett, A.; Morsy, M.; Eisen, J.A.; Leach, J.E.; Dangl, J.L. Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol. 2017, 15, e2001793. [Google Scholar] [CrossRef]
- Elser, J.J.; Bracken, M.E.; Cleland, E.E.; Gruner, D.S.; Harpole, W.S.; Hillebrand, H.; Ngai, J.T.; Seabloom, E.W.; Shurin, J.B.; Smith, J.E. Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol. Lett. 2007, 10, 1135–1142. [Google Scholar] [CrossRef]
- Ling, N.; Zhu, C.; Xue, C.; Chen, H.; Duan, Y.; Peng, C.; Guo, S.; Shen, Q. Insight into how organic amendments can shape the soil microbiome in long-term field experiments as revealed by network analysis. Soil Biol. Biochem. 2016, 99, 137–149. [Google Scholar] [CrossRef]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef]
- Carlson, R.; Tugizimana, F.; Steenkamp, P.A.; Dubery, I.A.; Hassen, A.I.; Labuschagne, N. Rhizobacteria-induced systemic tolerance against drought stress in Sorghum bicolor (L.) Moench. Microbiol. Res. 2020, 232, 126388. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.M.H.; Soares, H.; Soares, E.V. Promising bacterial genera for agricultural practices: An insight on plant growth-promoting properties and microbial safety aspects. Sci. Total Environ. 2019, 682, 779–799. [Google Scholar] [CrossRef]
- Chakraborty, S.; Newton, A.C. Climate change, plant diseases and food security: An overview. Plant Pathol. 2011, 60, 2–14. [Google Scholar] [CrossRef]
- Ghorbanpour, M.; Omidvari, M.; Abbaszadeh-Dahaji, P.; Omidvar, R.; Kariman, K. Mechanisms underlying the protective effects of beneficial fungi against plant diseases. Biol. Control 2018, 117, 147–157. [Google Scholar] [CrossRef]
- Khoshnevisan, B.; Rafiee, S.; Pan, J.; Zhang, Y.; Liu, H. A multi-criteria evolutionary-based algorithm as a regional scale decision support system to optimize nitrogen consumption rate; A case study in North China plain. J. Clean Prod. 2020, 256, 120213. [Google Scholar] [CrossRef]
- Xun, W.; Zhao, J.; Xue, C.; Zhang, G.; Ran, W.; Wang, B.; Shen, Q.; Zhang, R. Significant alteration of soil bacterial communities and organic carbon decomposition by different long-term fertilization management conditions of extremely low-productivity arable soil in South China. Environ. Microbiol. 2016, 18, 1907–1917. [Google Scholar] [CrossRef]
- Ahmed, M.; Rauf, M.; Mukhtar, Z.; Saeed, N.A. Excessive use of nitrogenous fertilizers: An unawareness causing serious threats to environment and human health. Environ. Sci. Pollut. Res. 2017, 24, 26983–26987. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Zhu, Z.; Jiang, Y. Long-Term impact of fertilization on soil pH and fertility in an apple production system. J. Soil Sci. Plant Nutr. 2018, 18, 282–293. [Google Scholar] [CrossRef]
- Cong, P.; Ouyang, Z.; Hou, R.; Han, D. Effects of application of microbial fertilizer on aggregation and aggregate-associated carbon in saline soils. Soil Tillage Res. 2017, 168, 33–41. [Google Scholar] [CrossRef]
- Wichern, J.; Wichern, F.; Joergensen, R.G. Impact of salinity on soil microbial communities and the decomposition of maize in acidic soils. Geoderma 2006, 137, 100–108. [Google Scholar] [CrossRef]
- Kandula, D.R.; Jones, E.E.; Stewart, A.; McLean, K.L.; Hampton, J.G. Trichoderma species for biocontrol of soil-borne plant pathogens of pasture species. Biocontrol Sci. Technol. 2015, 25, 1052–1069. [Google Scholar] [CrossRef]
- Vurukonda, S.S.K.P.; Giovanardi, D.; Stefani, E. Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci. 2018, 19, 952. [Google Scholar] [CrossRef]
- Harman, G.E.; Howell, C.R.; Viterbo, A.; Chet, I.; Lorito, M. Trichoderma species—Opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2004, 2, 43–56. [Google Scholar] [CrossRef]
- Afzal, A.; Bano, A. Rhizobium and phosphate solubilizing bacteria improve the yield and phosphorus uptake in wheat (Triticum aestivum). Int. J. Agric. Biol. 2008, 10, 85–88. [Google Scholar]
- Malusá, E.; Sas-Paszt, L.; Ciesielska, J. Technologies for beneficial microorganisms inocula used as biofertilizers. Sci. World J. 2012, 491206. [Google Scholar] [CrossRef]
- Reddy, C.A.; Saravanan, R.S. Polymicrobial multi-functional approach for enhancement of crop productivity. Adv. Appl. Microbiol. 2013, 82, 53–113. [Google Scholar] [CrossRef]
- Cepeda, V. Effects of Microbial Inoculants on Biocontrol and Plant Growth Promotion. Master’s Thesis, Ohio State University, Columbus, OH, USA, 2012. [Google Scholar]
- Ye, L.; Zhao, X.; Bao, E.; Li, J.; Zou, Z.; Cao, K. Bio-Organic fertilizer with reduced rates of chemical fertilization improves soil fertility and enhances tomato yield and quality. Sci. Rep. 2020, 10, 177. [Google Scholar] [CrossRef] [PubMed]
- Good, A.G.; Beatty, P.H. Fertilizing nature: A tragedy of excess in the commons. PLoS. Biol. 2011, 9, e1001124. [Google Scholar] [CrossRef]
- Chen, L.H.; Huang, X.Q.; Zhang, F.G.; Zhao, D.K.; Yang, X.M.; Shen, Q.R. Application of Trichoderma harzianum SQR-T037 bio-organic fertiliser significantly controls Fusarium wilt and affects the microbial communities of continuously cropped soil of cucumber. J. Sci. Food Agric. 2012, 92, 2465–2470. [Google Scholar] [CrossRef]
- Kandil, E.E.; Abdelsalam, N.R.; Mansour, M.A.; Ali, H.M.; Siddiqui, M.H. Potentials of organic manure and potassium forms on maize (Zea mays L.) growth and production. Sci. Rep. 2020, 10, 8752. [Google Scholar] [CrossRef]
- Faostat, F.J.Q. Available online: http://www.fao.org/faostat/en/#data.2017 (accessed on 11 November 2020).
- Calvo, P.; Nelson, L.; Kloepper, J.W. Agricultural uses of plant biostimulants. Plant Soil 2014, 383, 3–41. [Google Scholar] [CrossRef]
- Timmusk, S.; Behers, L.; Muthoni, J.; Muraya, A.; Aronsson, A.C. Perspectives and Challenges of Microbial Application for Crop Improvement. Front. Plant Sci. 2017, 8, 49. [Google Scholar] [CrossRef]
- Barampuram, S.; Allen, G.; Krasnyanski, S. Effect of various sterilization procedures on the in vitro germination of cotton seeds. Plant Cell Tissue Organ Cult. 2014, 118, 179–185. [Google Scholar] [CrossRef]
- Aliye, N.; Fininsa, C.; Hiskias, Y. Evaluation of rhizosphere bacterial antagonists for their potential to bioprotect potato (Solanum tuberosum) against bacterial wilt (Ralstonia solanacearum). Biol. Control 2008, 47, 282–288. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, H.; Liu, X.; Chen, Y.; Lu, Y.; Shen, M.; Dang, K.; Zhao, Y.; Dong, Y.; Li, Q.; et al. Organic fertilizer enhances rice growth in severe saline-alkali soil by increasing soil bacterial diversity. Soil Use Manag. 2021. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.S.; Mcgarell, D.M.; Marsh, T.; Garrity, G.M.; et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009, 37, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.L.; Cao, M.; Liu, X.M.; Du, Y.M.; Shen, G.M.; Zhang, Z.F.; Li, J.H.; Zhang, P. Composition and Genetic Diversity of the Nicotiana tabacum Microbiome in Different Topographic Areas and Growth Periods. Int. J. Mol. Sci. 2018, 19, 3421. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Hou, X.-D.; Yan, N.; Du, Y.-M.; Liang, H.; Zhang, Z.-F.; Yuan, X.-L. Consumption of Wild Rice (Zizania latifolia) Prevents Metabolic Associated Fatty Liver Disease through the Modulation of the Gut Microbiota in Mice Model. Int. J. Mol. Sci. 2020, 21, 5375. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Zhang, Y.; Zuo, Q.; Du, B.; Chen, W.; Wei, D.; Huang, Q. Contrasting responses of bacterial and fungal communities to aggregate-size fractions and long-term fertilizations in soils of northeastern China. Sci. Total Environ. 2018, 635, 784–792. [Google Scholar] [CrossRef]
- Qin, Y.; Shang, Q.; Zhang, Y.; Li, P.; Chai, Y. Bacillus amyloliquefaciens L-S60 Reforms the Rhizosphere Bacterial Community and Improves Growth Conditions in Cucumber Plug Seedling. Front. Microbiol. 2017, 8, 2620. [Google Scholar] [CrossRef]
- Peralta, A.L.; Matthews, J.W.; Kent, A.D. Microbial community structure and denitrification in a wetland mitigation bank. Appl. Environ. Microbiol. 2010, 76, 4207–4215. [Google Scholar] [CrossRef]
- Kerry, B.R. Rhizosphere Interactions and the Exploitation of Microbial Agents for the Biological Control of Plant-Parasitic Nematodes. Annu. Rev. Phytopathol. 2000, 38, 423–441. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, G.; Xue, S.; Wang, G. Soil bacterial community dynamics reflect changes in plant community and soil properties during the secondary succession of abandoned farmland in the Loess Plateau. Soil Biol. Biochem. 2016, 97, 40–49. [Google Scholar] [CrossRef]
- Gustave, W.; Yuan, Z.-F.; Sekar, R.; Ren, Y.-X.; Chang, H.-C.; Liu, J.-Y.; Chen, Z. The change in biotic and abiotic soil components influenced by paddy soil microbial fuel cells loaded with various resistances. J. Soils Sediments 2019, 19, 106–115. [Google Scholar] [CrossRef]
- Bennett, J.A.; Klironomos, J. Mechanisms of plant–soil feedback: Interactions among biotic and abiotic drivers. New Phytol. 2019, 222, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Roman, K.K.; Konieczna, A. Evaluation of a different fertilisation in technology of corn for silage, sugar beet and meadow grasses production and their impact on the environment in Poland. Afr. J. Agric. Res. 2015, 10, 1351–1358. [Google Scholar]
- Eo, J.; Park, K.-C. Long-term effects of imbalanced fertilization on the composition and diversity of soil bacterial community. Agric. Ecosyst. Environ. 2016, 231, 176–182. [Google Scholar] [CrossRef]
- Wagner, B.D.; Grunwald, G.K.; Zerbe, G.O.; Mikulich-Gilbertson, S.K.; Robertson, C.E.; Zemanick, E.T.; Harris, J.K. On the use of diversity measures in longitudinal sequencing studies of microbial communities. Front. Microbiol. 2018, 9, 1037. [Google Scholar] [CrossRef] [PubMed]
- Vázquez, M.M.; César, S.; Azcón, R.; Barea, J.M. Interactions between arbuscular mycorrhizal fungi and other microbial inoculants (Azospirillum, Pseudomonas, Trichoderma) and their effects on microbial population and enzyme activities in the rhizosphere of maize plants. Appl. Soil Ecol. 2000, 15, 261–272. [Google Scholar] [CrossRef]
- Li, L.; Wang, S.; Li, X.; Li, T.; He, X.; Tao, Y. Effects of Pseudomonas chenduensis and biochar on cadmium availability and microbial community in the paddy soil. Sci. Total Environ. 2018, 640–641, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Win, K.T.; Okazaki, K.; Ohkama-Ohtsu, N.; Yokoyama, T.; Ohwaki, Y. Short-Term effects of biochar and Bacillus pumilus TUAT-1 on the growth of forage rice and its associated soil microbial community and soil properties. Biol. Fertil. Soils 2020, 1–17. [Google Scholar] [CrossRef]
- Zhong, Y.; Yang, Y.; Liu, P.; Xu, R.; Rensing, C.; Fu, X.; Liao, H. Genotype and rhizobium inoculation modulate the assembly of soybean rhizobacterial communities. Plant Cell Environ. 2019, 42, 2028–2044. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Vargas, L.; de Carvalho, T.L.G.; Ferreira, P.C.G.; Baldani, V.L.D.; Baldani, J.I.; Hemerly, A.S. Early responses of rice (Oryza sativa L.) seedlings to inoculation with beneficial diazotrophic bacteria are dependent on plant and bacterial genotypes. Plant Soil 2012, 356, 127–137. [Google Scholar] [CrossRef]
- Paulson, J.N.; Stine, O.C.; Bravo, H.C.; Pop, M. Differential abundance analysis for microbial marker-gene surveys. Nat. Methods 2013, 10, 1200–1202. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef]
- Rodriguez-Sanchez, A.; Leyva-Diaz, J.C.; Gonzalez-Martinez, A.; Poyatos, J.M. Linkage of microbial kinetics and bacterial community structure of M BR and hybrid M BBR–MBR systems to treat salinity-amended urban wastewater. Biotechnol. Prog. 2017, 33, 1483–1495. [Google Scholar] [CrossRef]
- Kämpfer, P.; Young, C.-C.; Arun, A.; Shen, F.-T.; Jäckel, U.; Rossello-Mora, R.; Lai, W.-A.; Rekha, P. Pseudolabrys taiwanensis gen. nov., sp. nov., An alphaproteobacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 2006, 56, 2469–2472. [Google Scholar] [CrossRef]
- García-Fraile, P.; Benada, O.; Cajthaml, T.; Baldrian, P.; Lladó, S. Terracidiphilus gabretensis gen. nov., sp. nov., An abundant and active forest soil acidobacterium important in organic matter transformation. Appl. Environ. Microbiol. 2016, 82, 560–569. [Google Scholar] [CrossRef]
- Khan, I.U.; Hussain, F.; Habib, N.; Wadaan, M.A.M.; Ahmed, I.; Im, W.T.; Hozzein, W.N.; Zhi, X.Y.; Li, W.J. Phenylobacterium deserti sp. nov., isolated from desert soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 4722–4727. [Google Scholar] [CrossRef]
- Rawat, S.R.; Mannisto, M.K.; Starovoytov, V.; Goodwin, L.; Nolan, M.; Hauser, L.J.; Land, M.; Davenport, K.W.; Woyke, T.; Haggblom, M.M. Complete genome sequence of Granulicella mallensis type strain MP5ACTX8(T), an acidobacterium from tundra soil. Stand. Genomic Sci. 2013, 9, 71–82. [Google Scholar] [CrossRef]
- Park, D.; Kim, H.; Yoon, S. Nitrous Oxide Reduction by an Obligate Aerobic Bacterium, Gemmatimonas aurantiaca Strain T-27. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Van Den Heuvel, R.; Van Der Biezen, E.; Jetten, M.; Hefting, M.; Kartal, B. Denitrification at pH 4 by a soil-derived Rhodanobacter-dominated community. Environ. Microbiol. 2010, 12, 3264–3271. [Google Scholar] [CrossRef]
- Salanoubat, M.; Genin, S.; Artiguenave, F.; Gouzy, J.; Mangenot, S.; Arlat, M.; Billault, A.; Brottier, P.; Camus, J.C.; Cattolico, L.; et al. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 2002, 415, 497–502. [Google Scholar] [CrossRef]
- Kikuchi, T.; Cotton, J.A.; Dalzell, J.J.; Hasegawa, K.; Kanzaki, N.; McVeigh, P.; Takanashi, T.; Tsai, I.J.; Assefa, S.A.; Cock, P.J. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS. Pathog. 2011, 7, e1002219. [Google Scholar] [CrossRef]
- Wu, Y.; Zaiden, N.; Cao, B. The Core- and Pan-Genomic Analyses of the Genus Comamonas: From Environmental Adaptation to Potential Virulence. Front. Microbiol. 2018, 9, 3096. [Google Scholar] [CrossRef]
- Bharti, N.; Pandey, S.S.; Barnawal, D.; Patel, V.K.; Kalra, A. Plant growth promoting rhizobacteria Dietzia natronolimnaea modulates the expression of stress responsive genes providing protection of wheat from salinity stress. Sci. Rep. 2016, 6, 34768. [Google Scholar] [CrossRef]
- Okamura, K.; Hisada, T.; Kanbe, T.; Hiraishi, A. Rhodovastum atsumiense gen. nov., sp. nov., A phototrophic alphaproteobacterium isolated from paddy soil. J. Gen. Appl. Microbiol. 2009, 55, 43–50. [Google Scholar] [CrossRef]



| No. | Treatments | Instructions |
|---|---|---|
| 1 | CF | Urea, 600.00 kg ha−1; Calcium Superphosphate, 1000.00 kg ha−1; Potassium Sulfate, 240.00 kg ha−1 |
| 2 | D20N | Urea, 480.00 kg ha−1; Calcium Superphosphate, 1000.00 kg ha−1; Potassium Sulfate, 240.00 kg ha−1 |
| 3 | D40N | Urea, 360.00 kg ha−1; Calcium Superphosphate, 1000.00 kg ha−1; Potassium Sulfate, 240.00 kg ha−1 |
| 4 | CF + M | Inoculant M, 75.00 dm3 ha−1 |
| 5 | D20N + M | |
| 6 | D40N + M | |
| 7 | CF + A | Inoculant A, 75.00 dm3 ha−1 |
| 8 | D20N + A | |
| 9 | D40N + A | |
| 10 | CF + S | Inoculant S, 75.00 dm3 ha−1 |
| 11 | D20N + S | |
| 12 | D40N + S |
| Sample Name | Observed Species | Shannon | Simpson | Chao1 | ACE | Goods Coverage | PD Whole Tree |
|---|---|---|---|---|---|---|---|
| CF | 1953.75 ± 213.81 b,c,d | 8.87 ± 0.31 | 0.99 ± 0.00 b | 2104.32 ± 217.44 b,c,d | 2235.96 ± 223.64 b,c,d | 0.99 ± 0.00 d,e | 182.44 ± 15.05 c,d,e |
| CF.M | 2076.50 ± 125.05 c,d | 8.88 ± 0.11 | 0.99 ± 0.00 b | 2293.59 ± 99.03 d | 2474.94 ± 127.62 d | 0.99 ± 0.00 b,c | 186.97 ± 9.66 d,e |
| CF.A | 1820.25 ± 57.16 a,b,c | 8.73 ± 0.09 | 0.99 ± 0.00 b | 1973.39 ± 62.99 a,b,c | 2149.73 ± 73.83 bc | 0.99 ± 0.00 d,e | 158.04 ± 4.42 ab |
| CF.S | 1705.75 ± 82.42 a,b | 8.57 ± 0.06 | 0.99 ± 0.00 b | 1854.81 ± 96.38 a,b | 2009.90 ± 117.01 a,b | 0.99 ± 0.00 d,e | 152.14 ± 4.74 a |
| D20N | 2104.00 ± 299.20 d | 8.98 ± 0.45 | 0.99 ± 0.00 b | 2237.29 ± 310.51 c,d | 2373.10 ± 302.13 c,d | 0.99 ± 0.00 d,e | 196.57 ± 22.35 e |
| D20N.M | 2089.50 ± 124.42 c,d | 8.90 ± 0.61 | 0.99 ± 0.01 a,b | 2252.00 ± 94.90 d | 2421.19 ± 47.15 c,d | 0.99 ± 0.00 c,d | 192.34 ± 14.26 de |
| D20N.A | 2009.50 ± 170.35 c,d | 8.84 ± 0.14 | 0.99 ± 0.00 b | 2186.80 ± 173.65 c,d | 2374.97 ± 177.58 c,d | 0.99 ± 0.00 c,d | 174.18 ± 16.00 b,c,d |
| D20N.S | 1821.00 ± 154.70 a,b,c | 8.57 ± 0.12 | 0.99 ± 0.00 a,b | 1964.32 ± 162.53 a,b,c | 2130.63 ± 174.49 b,c | 0.99 ± 0.00 d,e | 162.33 ± 15.15 a,b,c |
| D40N | 1988.75 ± 308.07 c,d | 8.78 ± 0.34 | 0.99 ± 0.00 b | 2274.14 ± 331.58 d | 2494.88 ± 378.75 d | 0.99 ± 0.00 b | 182.94 ± 22.65 c,d,e |
| D40N.M | 1814.75 ± 52.69 a,b,c | 8.74 ± 0.14 | 0.99 ± 0.00 b | 1968.25 ± 53.76 a,b,c | 2130.81 ± 75.93 b,c | 0.99 ± 0.00 d,e | 159.83 ± 5.75 a,b |
| D40N.A | 1587.75 ± 86.92 a | 8.47 ± 0.12 | 0.99 ± 0.00 a,b | 1713.85 ± 102.57 a | 1838.79 ± 113.34 a | 0.99 ± 0.00 e | 147.05 ± 6.84 a |
| D40N.S | 2449.25 ± 135.71 e | 8.43 ± 0.64 | 0.98 ± 0.02 a | 2803.00 ± 38.66 e | 3184.54 ± 67.32 e | 0.98 ± 0.00 a | 202.65 ± 7.91 e |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, M.; Li, J.; Dong, Y.; Zhang, Z.; Zhao, Y.; Li, Q.; Dang, K.; Peng, J.; Liu, H. The Effects of Microbial Inoculants on Bacterial Communities of the Rhizosphere Soil of Maize. Agriculture 2021, 11, 389. https://doi.org/10.3390/agriculture11050389
Shen M, Li J, Dong Y, Zhang Z, Zhao Y, Li Q, Dang K, Peng J, Liu H. The Effects of Microbial Inoculants on Bacterial Communities of the Rhizosphere Soil of Maize. Agriculture. 2021; 11(5):389. https://doi.org/10.3390/agriculture11050389
Chicago/Turabian StyleShen, Minchong, Jiangang Li, Yuanhua Dong, Zhengkun Zhang, Yu Zhao, Qiyun Li, Keke Dang, Junwei Peng, and Hong Liu. 2021. "The Effects of Microbial Inoculants on Bacterial Communities of the Rhizosphere Soil of Maize" Agriculture 11, no. 5: 389. https://doi.org/10.3390/agriculture11050389
APA StyleShen, M., Li, J., Dong, Y., Zhang, Z., Zhao, Y., Li, Q., Dang, K., Peng, J., & Liu, H. (2021). The Effects of Microbial Inoculants on Bacterial Communities of the Rhizosphere Soil of Maize. Agriculture, 11(5), 389. https://doi.org/10.3390/agriculture11050389





