Cherry Tree Crown Extraction from Natural Orchard Images with Complex Backgrounds
Abstract
1. Introduction
2. Materials Acquisition
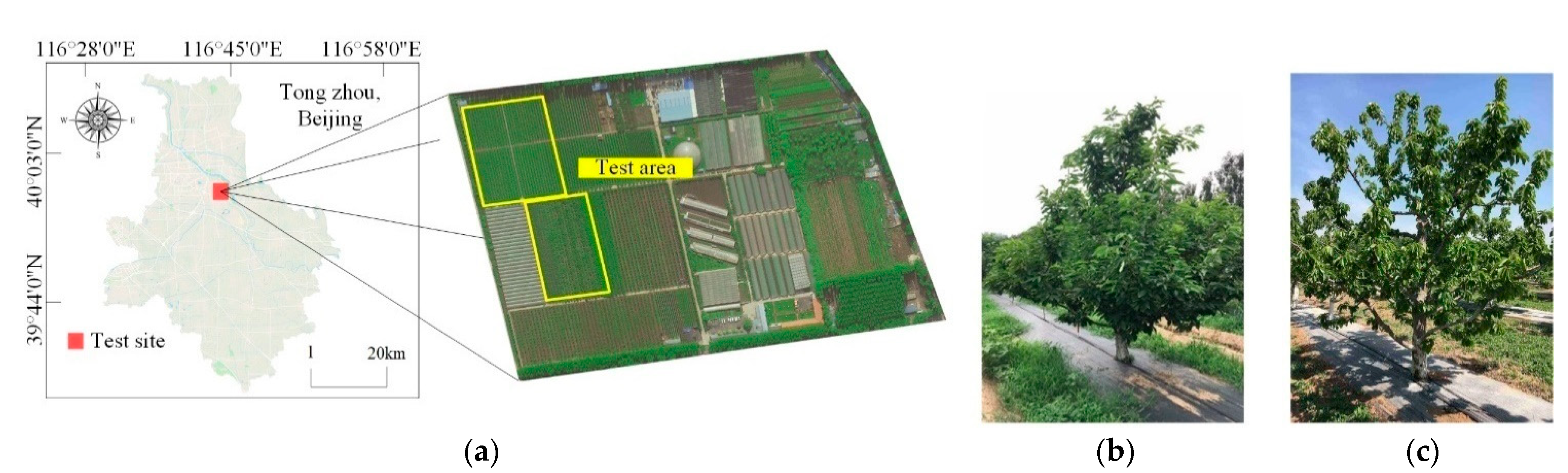
2.1. Test Site and Image Acquisition
Template and Ground Truth Generation
3. Methodology
3.1. Feature Construction of Tree Crown
3.1.1. Color Feature Extraction
3.1.2. Brightness and Height Feature Crossing
3.2. Mahalanobis Distance Computation
- (a)
- Computing mean vectors and covariance matrix: The mean vectors are the average value of the feature, commonly referring to the centroid of data distribution. The feature is a four-dimensional vector (H, S, Y and V) which is extracted from the template and sample images. The mean vectors are calculated as follows:where H, S and V are the hue, saturation and brightness components of HSV color space, respectively; Y is pixel height; n is the number of pixels; i = 1,2, 3, n; and f is a vector composed of H, S, V and Y.The covariance matrix is a square and symmetric matrix containing the variances and covariances associated with components of feature f (H, S, V and Y). The formula to compute the covariance between two variables is as follows:where f is a pair of variables with the four components (H, S, V, Y); µ is the mean vectors obtained by Equation (5); n is the number of pixels.
- (b)
- Computing the Mahalanobis distance: Mahalanobis distance will divide each pixel into two groups described by different mean vectors and covariances. Its formula Equation (8) is as follows:where f is four-dimensional vectors containing the H, S, V and Y values of each pixel; μ is the mean vectors calculated by Equation (5); the Cov is the covariance matrix calculated by Equation (7).
3.3. Conditional Random Field for Image Segmentation
3.3.1. Energy Function Construction
- (1)
- Model construction: establishing the mapping relationship between X and Y through the conditional probability distribution P(Y|X). In the fully connected conditional random field model, P(Y|X) is expressed in the form of Gibbs distribution:where X indicates the feature set f, and Y corresponds to the class labels, Y∈{L1, L2}. L1 represents the tree crown, and L2 is the background. Z is a normalization term that ensures the distribution P sums to 1 and is defined as follows:where Ε(Y|X) denotes the Energy function.
- (2)
- The Energy function minimization: CRF aims to find the output Y with the maximum conditional probability P(Y|X). According to Equation (9), the problem of conditional probability maximization is the problem of energy minimization, which can be expressed as follows:where y* is the minimization of Energy function Ε(Y|X) The Ε(Y|X) consists of two types of potential energy: unary potentials and pairwise potentials:where ψu(yi) is the unary potential for the probability of pixel i taking the label yi, denoting the pixel’s local information; ψp(yi, yj) is the pairwise potential, representing the label class similarity relationship between nearby pixels i and j, including inter-pixel global information; and i, j∈{1, 2,3, N} are the pixel indices.
- (3)
- The unary potential construction: The unary potential is the probability that a pixel obtains the corresponding label, indicating the category information of the current observation point. The study employed the Mahalanobis distance classifier results described in Section 3.2 to construct the unary potential energy. The unary potential takes the negative logarithm to provide a framework that unifies energy minimization:
- (4)
- The pairwise potential computation: The pairwise potential pixels are constraints of the final label assignment. Its goal is to assign adjacent labeled pixels with similar characteristics to the same category. The punishment strength is positively correlated to the feature difference between adjacent pixels under the same label, thereby restricting the classifier’s misclassification behavior. The general form of the paired potential function is a linear combination of Gaussian kernel functions:where u (yi, yj) is a constant symmetric label compatibility function between the labels yi, and yj to punish the similar pixels with different class labels. When the classifier assigns different labels to adjacent pixels, the greater the difference between pixel features, the smaller the penalty is, which is consistent with Gibbs energy minimization. Moreover, ω (m) is the coefficient weight of the given kernels; m = (1, 2,3, N) is the number of kernel K (m); K (m) (fj, fj) is the kernel potential function on feature vectors; and fj is feature vectors of pixels i, while fj is feature vectors of pixels j.
3.3.2. CRF Inference
3.4. Evaluation Indices and Competing Segmentation Methods for Segmentation Performance
3.4.1. Competing Segmentation Methods
3.4.2. Evaluation Indices
4. Results and Discussion
4.1. Segmentation Results of the Four Competing Methods
4.2. Performance Results under Different Overlapping Conditions and at Different Day Times
4.3. Segmentation Results in Different Years and Seasons
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cisternas, I.; Velásquez, I.; Caro, A.; Rodríguez, A. Systematic literature review of implementations of precision agriculture. Comput. Electron. Agric. 2020, 176, 105626. [Google Scholar] [CrossRef]
- Miles, C. The combine will tell the truth: On precision agriculture and algorithmic rationality. Big Data Soc. 2019, 6, 1–12. [Google Scholar] [CrossRef]
- Shafi, U.; Mumtaz, R.; García-Nieto, J.; Hassan, S.A.; Zaidi, S.A.R.; Iqbal, N. Precision agriculture techniques and practices: From considerations to applications. Sensors 2019, 19, 3796. [Google Scholar] [CrossRef] [PubMed]
- Xiao, K.; Ma, Y.; Gao, G. An intelligent precision orchard pesticide spray technique based on the depth-of-field extraction algorithm. Comput. Electron. Agric. 2017, 133, 30–36. [Google Scholar] [CrossRef]
- Solanelles, F.; Escolà, A.; Planas, S.; Rosell, J.R.; Camp, F.; Gràcia, F. An electronic control system for pesticide application proportional to the canopy width of tree crops. Biosyst. Eng. 2006, 95, 473–481. [Google Scholar] [CrossRef]
- Miranda-Fuentes, A.; Llorens, J.; Rodríguez-Lizana, A.; Cuenca, A.; Gil, E.; Blanco-Roldán, G.L.; Gil-Ribes, J.A. Assessing the optimal liquid volume to be sprayed on isolated olive trees according to their canopy volumes. Sci. Total Environ. 2016, 568, 269–305. [Google Scholar] [CrossRef] [PubMed]
- Tona, E.; Calcante, A.; Oberti, R. The profitability of precision spraying on specialty crops: A technical-economic analysis of protection equipment at increasing technological levels. Precis. Agric. 2018, 19, 606–629. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, X.; Lai, Q.; Zhang, Z. Review of variable-rate sprayer applications based on real-time sensor technologies. In Automation in Agriculture—Securing Food Supplies for Future Generations; Hussmann, S., Ed.; London, UK, 2018; pp. 53–79. [Google Scholar] [CrossRef]
- Pallottino, F.; Antonucci, F.; Costa, C.; Bisaglia, C.; Figorilli, S.; Menesatti, P. Optoelectronic proximal sensing vehicle-mounted technologies in precision agriculture: A review. Comput. Electron. Agric. 2019, 162, 859–873. [Google Scholar] [CrossRef]
- Virlet, N.; Gomez-Candon, D.; Lebourgeois, V.; Martinez, S.; Jolivot, A.; Lauri, P.E.; Costes, E.; Labbe, S.; Regnard, J.L. Contribution of high-resolution remotely sensed thermal-infrared imagery to high-throughput field phenotyping of an apple progeny submitted to water constraints. Acta Hortic. 2016, 1127, 243–250. [Google Scholar] [CrossRef]
- Jurado, J.M.; Ortega, L.; Cubillas, J.J.; Feito, F.R. Multispectral mapping on 3D models and multi-temporal monitoring for individual characterization of olive trees. Remote Sens. 2020, 12, 1106. [Google Scholar] [CrossRef]
- Ma, X.; Meng, Q.; Zhang, L.; Liu, G.; Zhou, W. Image mosaics reconstruction of canopy organ morphology of apple trees. Nongye Gongcheng Xuebao Trans. Chin. Soc. Agric. Eng. 2014, 30, 154–162. [Google Scholar]
- Dong, W.; Isler, V. Tree morphology for phenotyping from semantics-based mapping in orchard environments. arXiv 2018, arXiv:1804.05905. [Google Scholar]
- Xu, H.; Ying, Y. Detecting citrus in a tree canopy using infrared thermal imaging. In Monitoring Food Safety, Agriculture, and Plant Health; International Society for Optics and Photonics: San Diego, CA, USA, 2004; Volume 5271, pp. 321–327. [Google Scholar] [CrossRef]
- Coupel-Ledru, A.; Pallas, B.; Delalande, M.; Boudon, F.; Carrié, E.; Martinez, S.; Regnard, J.L.; Costes, E. Multi-scale high-throughput phenotyping of apple architectural and functional traits in orchard reveals genotypic variability under contrasted watering regimes. Hortic. Res. 2019, 6, 52. [Google Scholar] [CrossRef]
- Pusdá-Chulde, M.R.; Salazar-Fierro, F.A.; Sandoval-Pillajo, L.; Herrera-Granda, E.P.; García-Santillán, I.D.; de Giusti, A. Image analysis based on heterogeneous architectures for precision agriculture: A systematic literature review. Adv. Intell. Syst. Comput. 2020, 1078, 51–70. [Google Scholar] [CrossRef]
- Moreno, W.F.; Tangarife, H.I.; Escobar Díaz, A. Image analysis aplications in precision agriculture. Visión Electrónica 2018, 11, 200–210. [Google Scholar] [CrossRef]
- Delgado-Vera, C.; Mite-Baidal, K.; Gomez-Chabla, R.; Solís-Avilés, E.; Merchán-Benavides, S.; Rodríguez, A. Use of technologies of image recognition in agriculture: Systematic review of literature. Commun. Comput. Inform. Sci. 2018, 883, 15–29. [Google Scholar] [CrossRef]
- Hernández-Hernández, J.L.; García-Mateos, G.; González-Esquiva, J.M.; Escarabajal-Henarejos, D.; Ruiz-Canales, A.; Molina-Martínez, J.M. Optimal color space selection method for plant/soil segmentation in agriculture. Comput. Electron. Agric. 2016, 122, 124–132. [Google Scholar] [CrossRef]
- Hamuda, E.; Glavin, M.; Jones, E. A survey of image processing techniques for plant extraction and segmentation in the field. Comput. Electron. Agric. 2016, 125, 184–199. [Google Scholar] [CrossRef]
- Xue, J.; Su, B. Significant remote sensing vegetation indices: A review of developments and applications. J. Sens. 2017, 1, 1–17. [Google Scholar] [CrossRef]
- Woebbecke, D.M.; Meyer, G.E.; von Bargen, K.; Mortensen, D.A. Color indices for weed identification under various soil, residue, and lighting conditions. Trans. Am. Soc. Agric. Eng. 1995, 38, 259–269. [Google Scholar] [CrossRef]
- Meyer, G.E.; Neto, J.C.; Jones, D.D.; Hindman, T.W. Intensified fuzzy clusters for classifying plant, soil, and residue regions of interest from color images. Comput. Electron. Agric. 2004, 42, 161–180. [Google Scholar] [CrossRef]
- Weier, J.; Herring, D. Measuring Vegetation (NDVI & EVI); Normalized Difference Vegetation Index (NDVI); Nasa Earth Observatory: Washington, DC, USA, 2011. [Google Scholar]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Hassaan, O.; Nasir, A.K.; Roth, H.; Khan, M.F. Precision forestry: Trees counting in urban areas using visible imagery based on an unmanned aerial vehicle. IFAC PapersOnLine 2016, 49, 16–21. [Google Scholar] [CrossRef]
- Wang, A.; Zhang, W.; Wei, X. A review on weed detection using ground-based machine vision and image processing techniques. Comput. Electron. Agric. 2019, 158, 226–240. [Google Scholar] [CrossRef]
- Mu, Y.; Fujii, Y.; Takata, D.; Zheng, B.; Noshita, K.; Honda, K.; Ninomiya, S.; Guo, W. Characterization of peach tree crown by using high-resolution images from an unmanned aerial vehicle. Hortic. Res. 2018, 5, 74. [Google Scholar] [CrossRef]
- Dong, X.; Zhang, Z.; Yu, R.; Tian, Q.; Zhu, X. Extraction of information about individual trees from high-spatial-resolution UAV-acquired images of an orchard. Remote Sens. 2020, 12, 133. [Google Scholar] [CrossRef]
- Chen, Y.R.; Chao, K.; Kim, M.S. Machine vision technology for agricultural applications. Comput. Electron. Agric. 2002, 36, 173–191. [Google Scholar] [CrossRef]
- Patrício, D.I.; Rieder, R. Computer vision and artificial intelligence in precision agriculture for grain crops: A systematic review. Comput. Electron. Agric. 2018, 153, 69–81. [Google Scholar] [CrossRef]
- Sridharan, M.; Gowda, P. Application of statistical machine learning algorithms in precision agriculture. In Proceedings of the 7th Asian-Australasian Conference on Precision Agriculture, Hamilton, New Zealand, 16–18 October 2017. [Google Scholar] [CrossRef]
- Liu, W.; Zhong, T.; Song, Y. Prediction of trees diameter at breast height based on unmanned aerial vehicle image analysis. Nongye Gongcheng Xuebao Trans. Chin. Soc. Agric. Eng. 2017, 33, 99–104. [Google Scholar]
- Qi, L.; Cheng, Y.; Cheng, Z.; Yang, Z.; Wu, Y.; Ge, L. Estimation of upper and lower canopy volume ratio of fruit trees based on M-K clustering. Nongye Jixie Xuebao Trans. Chin. Soc. Agric. Mach. 2018, 49, 57–64. [Google Scholar]
- Abdalla, A.; Cen, H.; El-Manawy, A.; He, Y. Infield oilseed rape images segmentation via improved unsupervised learning models combined with supreme color features. Comput. Electron. Agric. 2019, 162, 1057–1068. [Google Scholar] [CrossRef]
- Rehman, T.U.; Mahmud, M.S.; Chang, Y.K.; Jin, J.; Shin, J. Current and future applications of statistical machine learning algorithms for agricultural machine vision systems. Comput. Electron. Agric. 2019, 156, 585–605. [Google Scholar] [CrossRef]
- Elavarasan, D.; Vincent, D.R.; Sharma, V.; Zomaya, A.Y.; Srinivasan, K. Forecasting yield by integrating agrarian factors and machine learning models: A survey. Comput. Electron. Agric. 2018, 155, 257–282. [Google Scholar] [CrossRef]
- Valente, J.; Doldersum, M.; Roers, C.; Kooistra, L. Detecting rumex obtusifolius weed plants in grasslands from UAV RGB imagery using deep learning. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2019, 4, 179–185. [Google Scholar] [CrossRef]
- Chen, Y.; Hou, C.; Tang, Y.; Zhuang, J.; Lin, J.; He, Y.; Guo, Q.; Zhong, Z.; Lei, H.; Luo, S. Citrus tree segmentation from UAV images based on monocular machine vision in a natural orchard environment. Sensors 2019, 19, 5558. [Google Scholar] [CrossRef] [PubMed]
- Zortea, M.; Macedo, M.M.G.; Mattos, A.B.; Ruga, B.C.; Gemignani, B.H. Automatic citrus tree detection from UAV images based on convolutional neural networks. Drones 2018, 11, 1–7. [Google Scholar]
- Wu, J.; Yang, G.; Yang, H.; Zhu, Y.; Li, Z.; Lei, L.; Zhao, C. Extracting apple tree crown information from remote imagery using deep learning. Comput. Electron. Agric. 2020, 174, 105504. [Google Scholar] [CrossRef]
- Lu, Y.; Young, S. A survey of public datasets for computer vision tasks in precision agriculture. Comput. Electron. Agric. 2020, 178, 105760. [Google Scholar] [CrossRef]
- Cheng, Z.; Qi, L.; Cheng, Y.; Wu, Y.; Zhang, H. Interlacing orchard canopy separation and assessment using UAV images. Remote Sens. 2001, 34, 2259–2281. [Google Scholar] [CrossRef]
- Cheng, H.D.; Jiang, X.H.; Sun, Y.; Wang, J. Color image segmentation: Advances and prospects. Pattern Recognit. 2001, 34, 2259–2281. [Google Scholar] [CrossRef]
- Hamuda, E.; Ginley, B.M.; Glavin, M.; Jones, E. Automatic crop detection under field conditions using the HSV colour space and morphological operations. Comput. Electron. Agric. 2017, 133, 97–107. [Google Scholar] [CrossRef]
- Kozlowski, T.T.; Pallardy, S.G. Environmental regulation of vegetative growth. In Growth Control in Woody Plants; Academic Press: Cambridge, MA, USA, 1997; pp. 195–322. [Google Scholar]
- García-Santillán, I.D.; Pajares, G. On-line crop/weed discrimination through the Mahalanobis distance from images in maize fields. Biosyst. Eng. 2018, 166, 28–43. [Google Scholar] [CrossRef]
- Lafferty, J.; Andrew, M.; Fernando, C.N.P. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. In Proceedings of the 18th International Conference on Machine Learning, Williamstown, MA, USA, 28 June–1 July 2001; pp. 282–289. [Google Scholar] [CrossRef]
- Krähenbühl, P.; Koltun, V. Efficient inference in fully connected crfs with Gaussian edge potentials. In Proceedings of the Advances in Neural Information Processing Systems 24: 25th Annual Conference on Neural Information Processing Systems, Granada, Spain, 12–14 December 2011; pp. 109–117. [Google Scholar]
- Liu, T.; Huang, X.; Ma, J. Conditional random fields for image labeling. Math. Probl. Eng. 2016, 2016, 1–15. [Google Scholar] [CrossRef]
- Cheng, Z.; Qi, L.; Cheng, Y.; Wu, Y.; Zhang, H.; Xiao, Y. Fruit tree canopy image segmentation method based on M-LP features weighted clustering. Nongye Jixie Xuebao Trans. Chin. Soc. Agric. Mach. 2020, 51, 191–198. [Google Scholar]
- Liu, H.; Zhu, S.; Shen, Y.; Tang, J. Fast segmentation algorithm of tree trunks based on multi-feature fusion. Nongye Jixie Xuebao Trans. Chin. Soc. Agric. Mach. 2020, 51, 221–229. [Google Scholar]
- Ferreira, M.P.; de Almeida, D.R.A.; de Almeida, D.P.; Minervino, J.B.S.; Veras, H.F.P.; Formighieri, A.; Santos, C.A.N.; Ferreira, M.A.D.; Figueiredo, E.O.; Ferreira, E.J.L. Individual tree detection and species classification of Amazonian palms using UAV images and deep learning. For. Ecol. Manag. 2020, 475, 118397. [Google Scholar] [CrossRef]
- Selim, S.Z.; Ismail, M.A. K-means-type algorithms: A generalized convergence theorem and characterization of local optimality. IEEE Trans. Pattern Anal. Mach. Intell. 1984, 6, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. Lect. Notes Comput. Sci. 2018, 833–8521. [Google Scholar] [CrossRef]
- Rother, C.; Kolmogorov, V.; Blake, A. “GrabCut”—Interactive foreground extraction using iterated graph cuts. ACM Trans. Graph. 2004, 23, 309–314. [Google Scholar] [CrossRef]
- WHO. Statement on the Second Meeting of the International Health Regulations (2005) Emergency Committee Regarding the Outbreak of Novel Coronavirus (2019-nCoV); WHO: Geneva, Switzerland, 2020. [Google Scholar]












| Method | Segmentation Evaluation Index | Computational Cost Assessment Index | ||||
|---|---|---|---|---|---|---|
| Average P (%) | Average R (%) | Average F1 (%) | Average Time (s) | The Number of Labeled Images | Training Time-Consuming | |
| K-means | 58.1 | 79.7 | 68.9 | 0.366 | - | - |
| DeaplabV3+ | 82.4 | 73.8 | 78.1 | 0.554 | 500 | 8 h |
| Grabcut | 86.3 | 80.3 | 83.8 | 0.978 | 200 | - |
| Proposed Algorithm | 92.1 | 94.5 | 93.3 | 0.736 | 2 | - |
| Indices | Overlapping Degrees | Image Shooting Day Times | In Total | ||||
|---|---|---|---|---|---|---|---|
| Slightly | Partly | Heavily | Morning | Noon | Evening | ||
| Average P/% | 94.9 | 92.9 | 90.8 | 94.1 | 93.9 | 93.1 | 93.2 |
| Average R/% | 95.3 | 94.9 | 93.8 | 94.9 | 90.4 | 92.1 | 93.5 |
| Average F1/% | 95.1 | 93.9 | 92.3 | 94.5 | 92.1 | 92.6 | 93.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, Z.; Qi, L.; Cheng, Y. Cherry Tree Crown Extraction from Natural Orchard Images with Complex Backgrounds. Agriculture 2021, 11, 431. https://doi.org/10.3390/agriculture11050431
Cheng Z, Qi L, Cheng Y. Cherry Tree Crown Extraction from Natural Orchard Images with Complex Backgrounds. Agriculture. 2021; 11(5):431. https://doi.org/10.3390/agriculture11050431
Chicago/Turabian StyleCheng, Zhenzhen, Lijun Qi, and Yifan Cheng. 2021. "Cherry Tree Crown Extraction from Natural Orchard Images with Complex Backgrounds" Agriculture 11, no. 5: 431. https://doi.org/10.3390/agriculture11050431
APA StyleCheng, Z., Qi, L., & Cheng, Y. (2021). Cherry Tree Crown Extraction from Natural Orchard Images with Complex Backgrounds. Agriculture, 11(5), 431. https://doi.org/10.3390/agriculture11050431






