Abstract
As one of the world’s most popular fruits, watermelon (Citrus lanatus) is cultivated in more than 3 million hectares across the globe, with a yearly yield of more than 100 million tons. According to ‘97103’ genome version 1, a previous study has shown that the watermelon genome consists of 11 PIN genes. However, the higher quality ‘97103’ genome version 2 was recently assembled by using PacBio long reads with the benefit of fast development sequencing technology. Using this new assembly, we conducted a new genome-wide search for PIN genes in watermelon and compared it with cucumber and melon genomes. We identified nine, nine, and eight PINs in watermelon, cucumber, and melon, respectively. Phylogenetic analysis revealed a distinct evolutionary history of PIN proteins in watermelon, which is shown by the orphan PIN6 in watermelon. We further conducted an expression analysis of the watermelon PIN genes in silico and by qRT-PCR. ClaPIN6 might not play an essential role during shoot regeneration, which is closely related to species-specific evolution. However, the up-regulated expression patterns of ClaPIN1-1 and ClaPIN1-3 indicated their important roles during shoot regeneration. The result of this research will benefit future studies to understand the regulating mechanisms of auxin in watermelon shoot regeneration.
1. Introduction
As one of the most important hormones, auxin plays a vital role in the growth and development of plants [1]. Auxin is transported from its biosynthesis tissues to the sink organs where it acts through the phloem stream of photosynthetic assimilates in a fast manner [2]. Local auxin transport depends on polar auxin transport (PAT) between neighboring cells, which is slow but essential for generating auxin maxima in developing tissues [3]. Among the gene families related to auxin transport, the PIN-FORMED (PIN) family has been thoroughly characterized in Arabidopsis. [4]. Among the eight Arabidopsis PIN proteins, seven contribute to cell-to-cell PAT (AtPIN1–4, 7) and local auxin homeostasis in-cell (AtPIN5, 8) [4,5,6]. These functions are determined by their subcellular localizations. Besides, AtPIN6 is located both at the plasma membrane and endoplasmic reticulum, which indicates its combined functions of cell-to-cell PAT and intracellular auxin homeostasis [7]. Arabidopsis PIN proteins are involved in regulating diverse growth and developmental processes, such as AtPIN1–3, 7 in gravitropic response [8,9,10,11], AtPIN1, 3, 4, 7 in phototropic responses [12,13], AtPIN1, 8 in flower development [14,15], AtPIN3, 4, 7 in apical hook development [13,16], AtPIN2–6 in root formation and development [6,7,17,18,19], and AtPIN2, 5, 7 in embryogenesis [1,6,20].
Watermelon (Citrullus lanatus) is an important fruit crop, which is cultivated in over 3 million hectares around the world, with a yearly production of over 100 million tons [21]. Watermelon contains a series of healthy nutritional compounds, such as lycopene, citrulline, arginine, and glutathione [22]. Consequently, it is of great importance to improve the fruit’s quality traits [23]. Newly invented CRISPR/Cas9 mediated gene editing technology is an efficient method for genetic improvement of crops, which has long been successfully applied to watermelon [24,25]. However, there is limited knowledge of the mechanisms of shoot regeneration, which is a major impediment for genetic transformation of watermelon. Zhang et al. have clarified the importance of the WOX and SAUR gene families during shoot regeneration of watermelon [26,27]. In Arabidopsis, a complex gene regulatory network for shoot regeneration has been identified [28]. Among the genes involved, AtPINs play essential roles in callus formation and shoot initiation by regulating PAT [29,30]. However, it still remains unknown exactly how the PIN gene family regulates shoot regeneration in watermelon. Using the watermelon 97103 genome (version 1), a previous study has identified the PIN gene family and its expression patterns in response to auxin and extreme stress conditions [31]. Recently, a new high-quality 97103 genome (version 2), which has been assembled using PacBio long reads combined with BioNano optical and Hi-C maps, has been released [23]. In the present study, we have identified 9 PIN genes in the new genome. Two previous PIN genes were reassembled into a single gene. A phylogenetic analysis of PIN proteins was carried out to investigate the evolutionary pattern of PIN genes in Arabidopsis, watermelon, cucumber (Cucumis sativus), and melon (Cucumis melo). We further examined their expression patterns in watermelon tissues and during shoot regeneration. The results presented here will benefit future studies on auxin-mediated regulation mechanisms of shoot regeneration in watermelon.
2. Materials and Methods
2.1. Sequence Retrieval and Bioinformatic Analysis
All Arabidopsis PIN proteins were selected as the queries to search against the genomes of watermelon, cucumber, and melon in the Cucurbit Genomics Database (CuGenDB) with the TBLASTN method [3,32,33]. After removing the redundant sequences, PIN genes from the three species were named after their Arabidopsis homologs and chromosome positions. The details of these sequences are listed in Table 1. The ProtParam tool was used to predict the characteristics of proteins, including sequence lengths, molecular weights, and theoretical pI [34]. The CELLO software was selected for the prediction of subcellular localization [35]. Protein transmembrane topology was analyzed by the TMHMM Server [36]. A Neighbor-Joining (NJ) phylogenetic tree was constructed with the bootstrap value of 1000 trails in the MEGA7 software [37]. The conserved domains were analyzed using DNAMAN 7 software [38]. The TBtools software was used to extract the promoter (2Kb upstream sequence) and genomic sequences of the PIN genes [39]. The PlantCARE software was selected to identify putative cis-elements related to growth, development, and stress responses [40]. The images of chromosome position, cis-element, and intron/exon were displayed in TBtools [39].

Table 1.
The IDs, CuGenDB accessions, sequence lengths, molecular weights, pI, and CELLO localizations of PIN genes in watermelon, cucumber, and melon.
2.2. Plant Materials and RNA Extraction
The diploid watermelon inbred line A7 was selected for shoot regeneration, as previously reported [26]. Decoated seeds were cultured on Murashige and Skoog (MS) medium after sterilizing with NaClO solution (10%) for 5 min. After 1-week-germination, cotyledons were collected and cut into segments (0.5 × 0.5 mm), which were incubated on MS medium (1.0 mg/L 6-BA and 1.0 mg/L NAA, Sigma-Aldrich, Saint Louis, MO, USA) at 25 ± 2 °C and a photoperiod of 16/8 h (light/dark). The samples were collected 0, 7, 14, 21, and 28 days after incubation (DAI). Samples at each time point were collected from three different plates as biological replicates, frozen in liquid nitrogen, and ground into a powder for RNA isolation. Total RNA extraction was carried out with a Tiangen RNA prep Pure Plant Kit (Tiangen Biomart, Beijing, China), following the manufacturer’s instructions. All RNA samples were stored at −80 °C until expression analysis.
2.3. Expression Analysis
In silico expression data were calculated using the Cucurbit Expression Atlas Database of CuGenDB and the RPKM values were downloaded with the accession of each watermelon PIN gene [32]. For qRT-PCR expression analysis, RNA samples were used for cDNA synthesis with the GoScript Reverse Transcription System (Promega, Madison, WI, USA). The 20 μL mixture for qRT-PCR reaction contained 10 μL TransStart Tip Green qPCR Supermix (Transgen Biotech, Beijing, China), 0.5 μL gene-specific primers (10 μM), 0.4 μL Passive ReferenceDye (50×) (Transgen Biotech, Beijing, China), 1 μL cDNA template, and 7.6 μL ddH2O. A QuantStudio 6 Flex Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) was used for qRT-PCR reaction. The thermal cycles were 94 °C for 30 s, 40 cycles of 94 °C for 5 s, and 60 °C for 30 s, followed with a dissociation stage. Three reactions were taken for each sample as the technical repeat. Specific primers for watermelon PIN genes were designed using the online tool Primer 3 (Table 2). [41]. Watermelon GAPDH gene was used as a reference gene [26]. The relative expression levels were calculated by the ΔΔCt method [42].

Table 2.
Primers of watermelon 9 PIN genes and GAPDH gene (as the endogenous control) used for qRT-PCR.
3. Results
3.1. Genome-Wide Identification and Phylogenetic Analysis of PINs in Cucurbitaceae Species
After the whole genome survey, we identified nine PINs in watermelon (ClaPINs), nine in cucumber (CsaPINs), and eight in melon (CmePINs) (Table 1). The CELLO results indicated that all the Cucurbitaceae PINs were located at the plasma membrane, implying their potential function in auxin transport. Most ClaPINs contained 4–5 transmembrane helices at N- or C-termini (Figure S1).
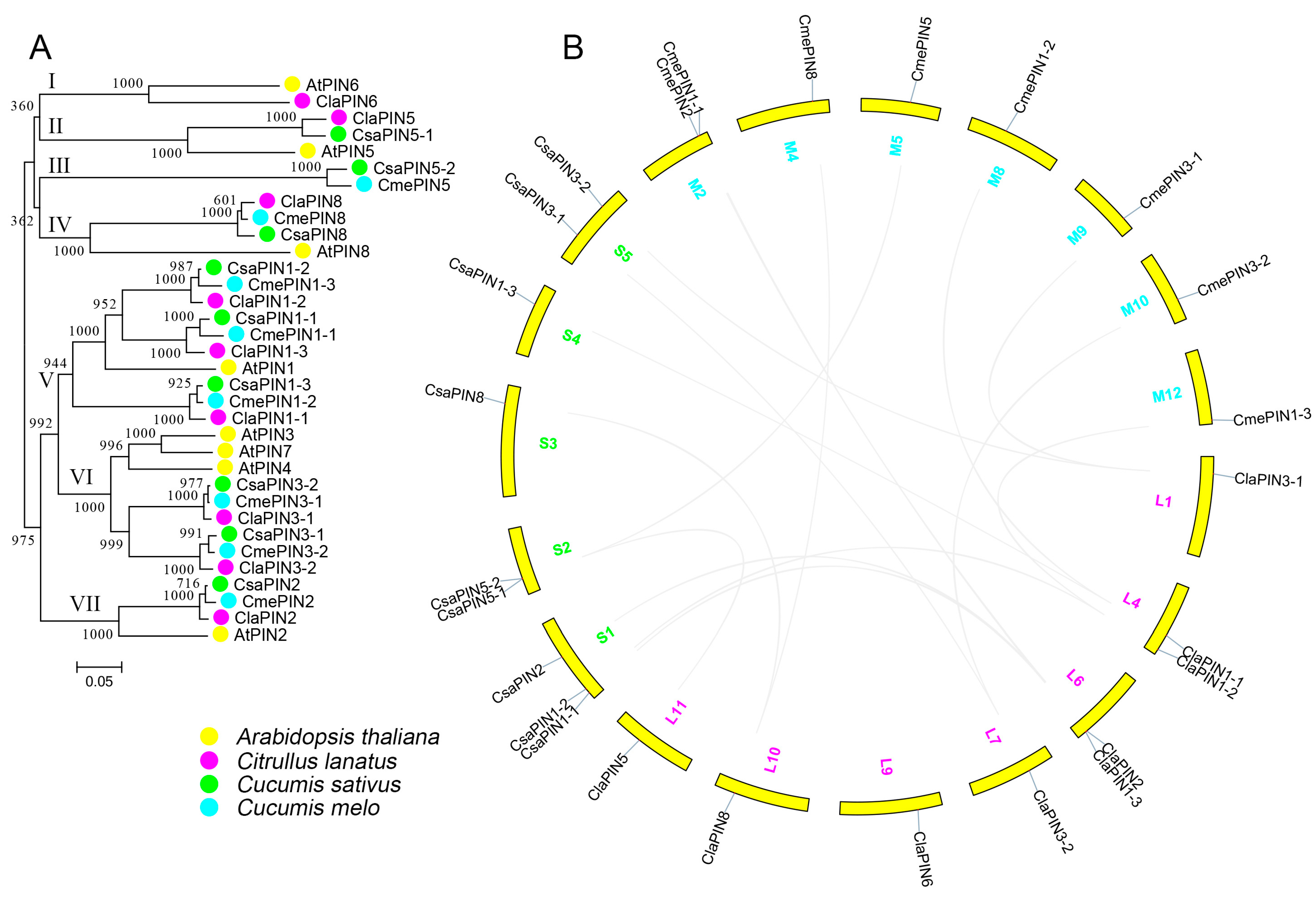
The PIN proteins of Arabidopsis and the three Cucurbitaceae species were aligned for phylogenetic analysis. These proteins were grouped into seven clusters (Figure 1A). One PIN gene from watermelon clustered together with AtPIN6 from Arabidopsis in cluster I. Clusters IV, V, VI, and VII contained an equal number of PIN genes to the Cucurbitaceae species. Arabidopsis PINs were also clustered in these groups. We further examined the conserved domains of all the PIN proteins (Figure S2). Most sequences contained two conserved domains at both the N and C-termini. The middle parts of these sequences were predicted to be located inside the transmembrane.
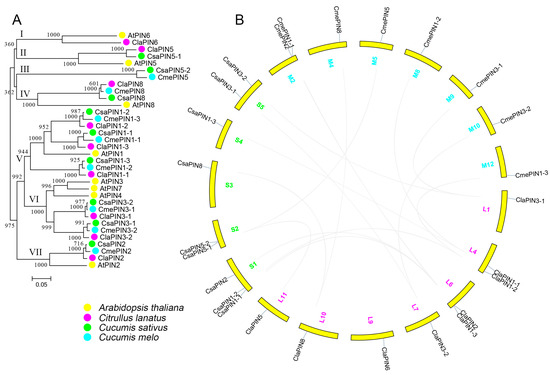
Figure 1.
Phylogenetic (A) and chromosomal localization (B) of the Cucurbitaceae PIN family. The proteins from Arabidopsis thaliana, cucumber, melon and watermelon are shown in yellow, blue, green, and pink, respectively (A). The chromosome numbers are listed beside chromosomes (yellow) for watermelon (pink), cucumber (blue), and melon (green), respectively. The homologous gene pairs are connected with grey solid lines.
3.2. Cis-Element and Gene Structure Analysis of Watermelon PINs
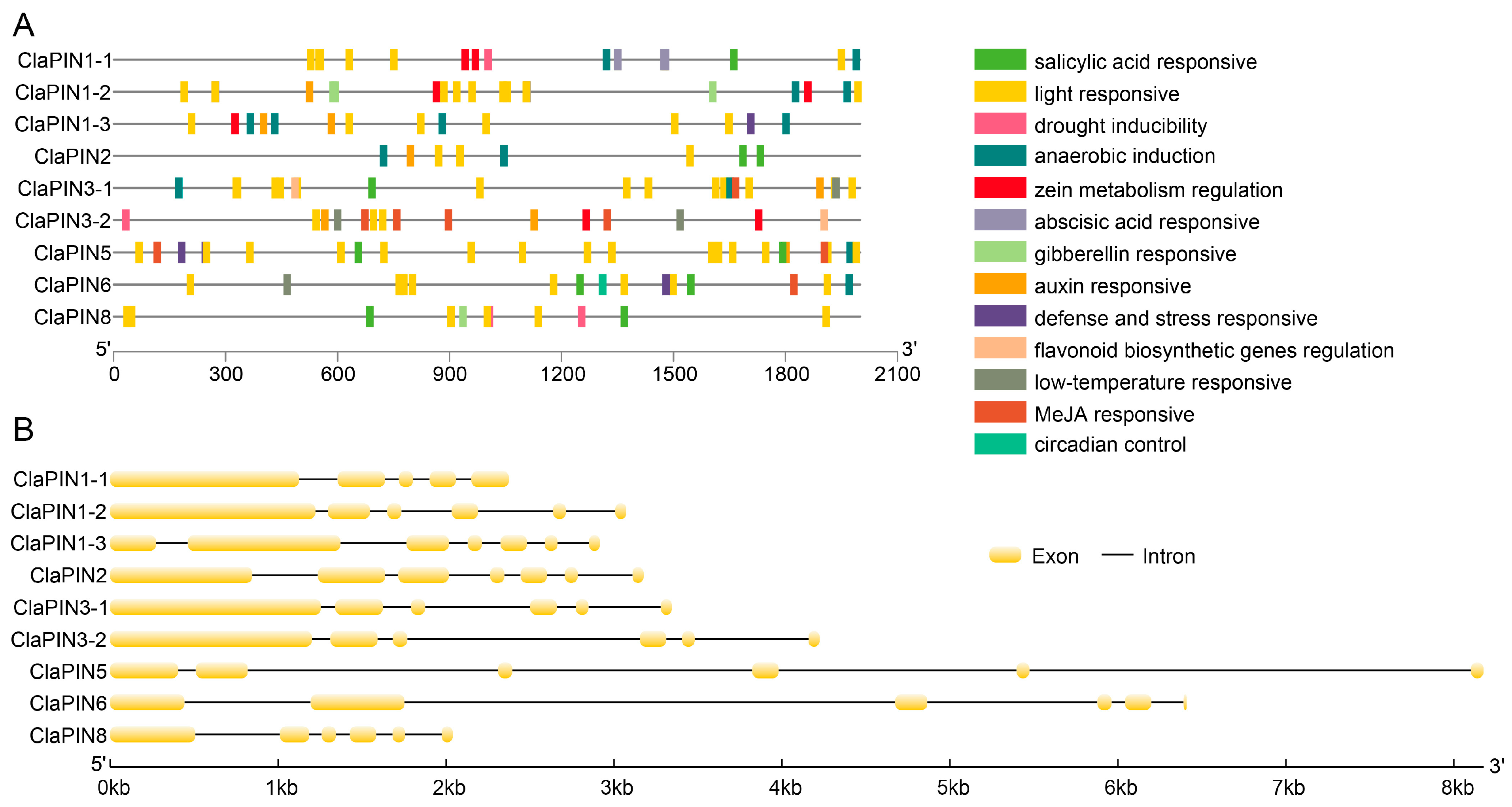
The upstream sequences (2000 bp) of watermelon PINs were selected for scanning cis-regulatory elements (Table S1). We annotated 13 kinds of cis-elements that are related to salicylic acid, gibberellin, auxin, MeJA, light, low-temperature, defense/stress responses, drought, and anaerobic induction, regulation of zein metabolism, flavonoid biosynthesis, and circadian control (Figure 2A). Among these, the number of light responsive cis-element (78) was the most abundant (Table S1). The numbers of MeJA responsive, anaerobic induction responsive, and salicylic acid responsive elements were 16, 14, and 11, respectively. We further analyzed the introns of ClaPINs (Figure 2B). Most sequences contained 5 introns. There were 4 introns in ClaPIN1-1 and 6 introns in both ClaPIN1-3 and ClaPIN2.
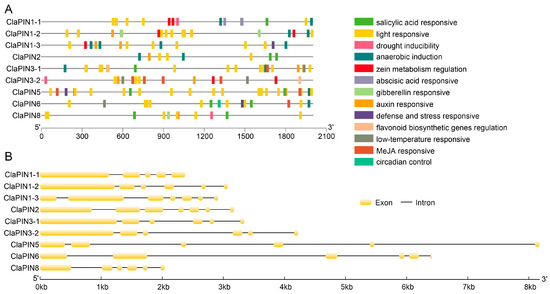
Figure 2.
Cis-elements (A) and intron (B) structures of watermelon PIN family. The cis-element prediction was conducted with 2000 bp upstream sequences by the PlantCARE software [41]. The intron structure analysis was conducted with genomic and coding sequences of watermelon PIN family.
3.3. In Silico Expression of PINs in Watermelon Tissues
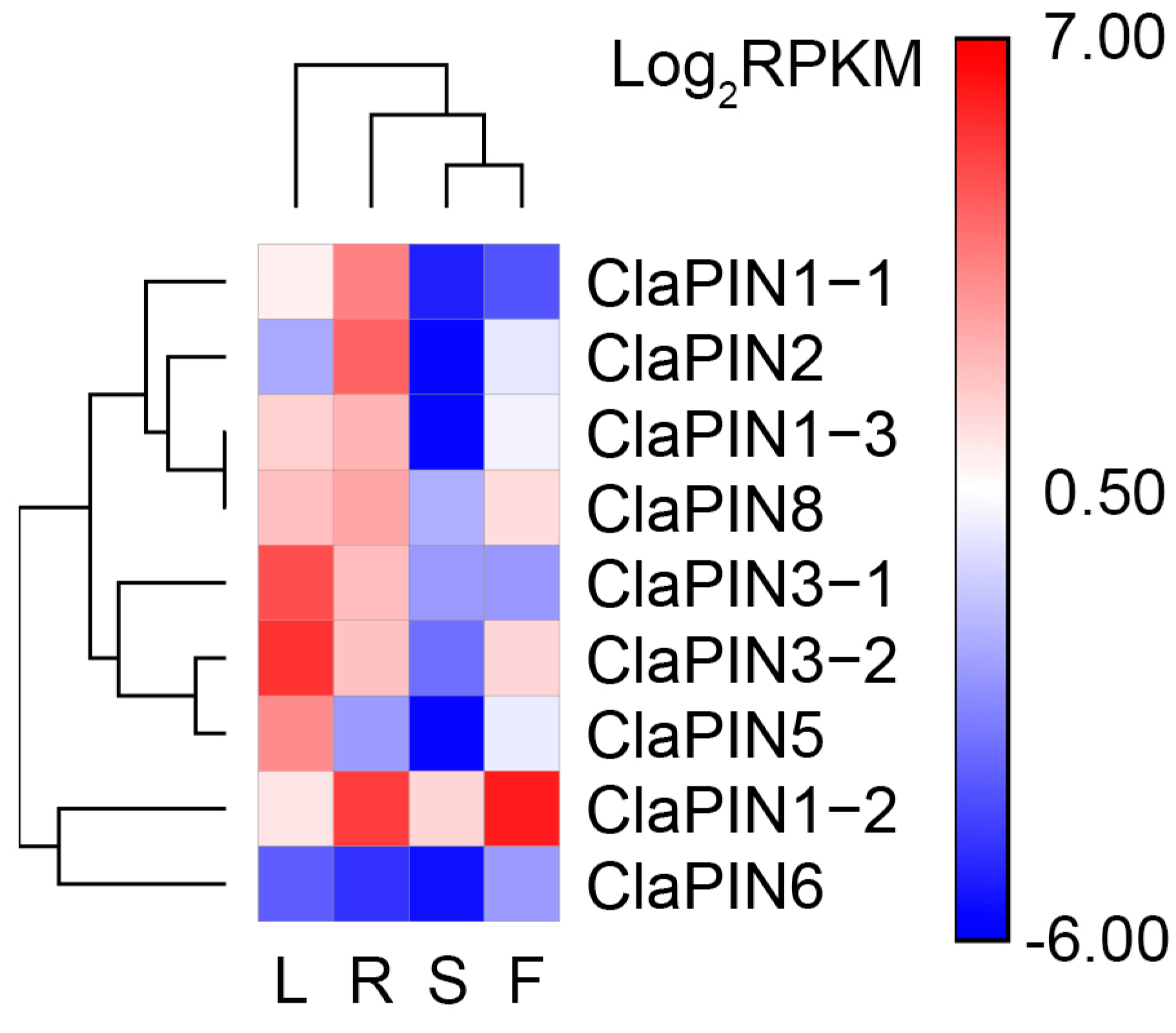
To estimate the expression patterns of ClaPINs, in silico analysis was carried out in watermelon tissues (Figure 3). Most ClaPINs were highly expressed in roots and leaves compared with flowers and seeds. CaPIN1–2 showed high expression in both roots and flowers. ClaPIN5 was highly expressed in seeds and leaves. ClaPIN6, meanwhile, showed relatively low expression in all tissues.
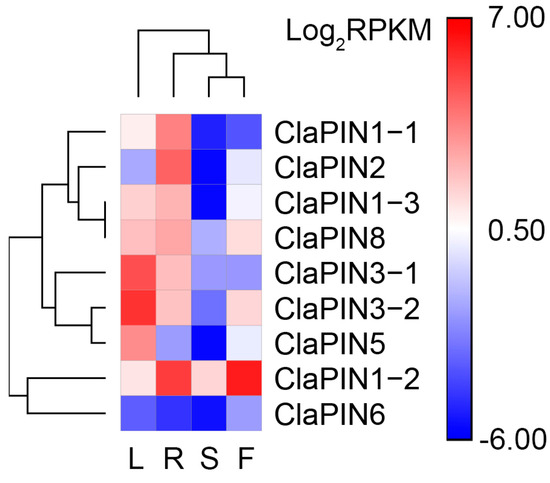
Figure 3.
In silico expression of ClaPINs in watermelon tissues. The single capital letters represent root (R), leaf (L), flower (F), and seed (S).
3.4. Expression Profiles of Watermelon PINs during Shoot Regeneration
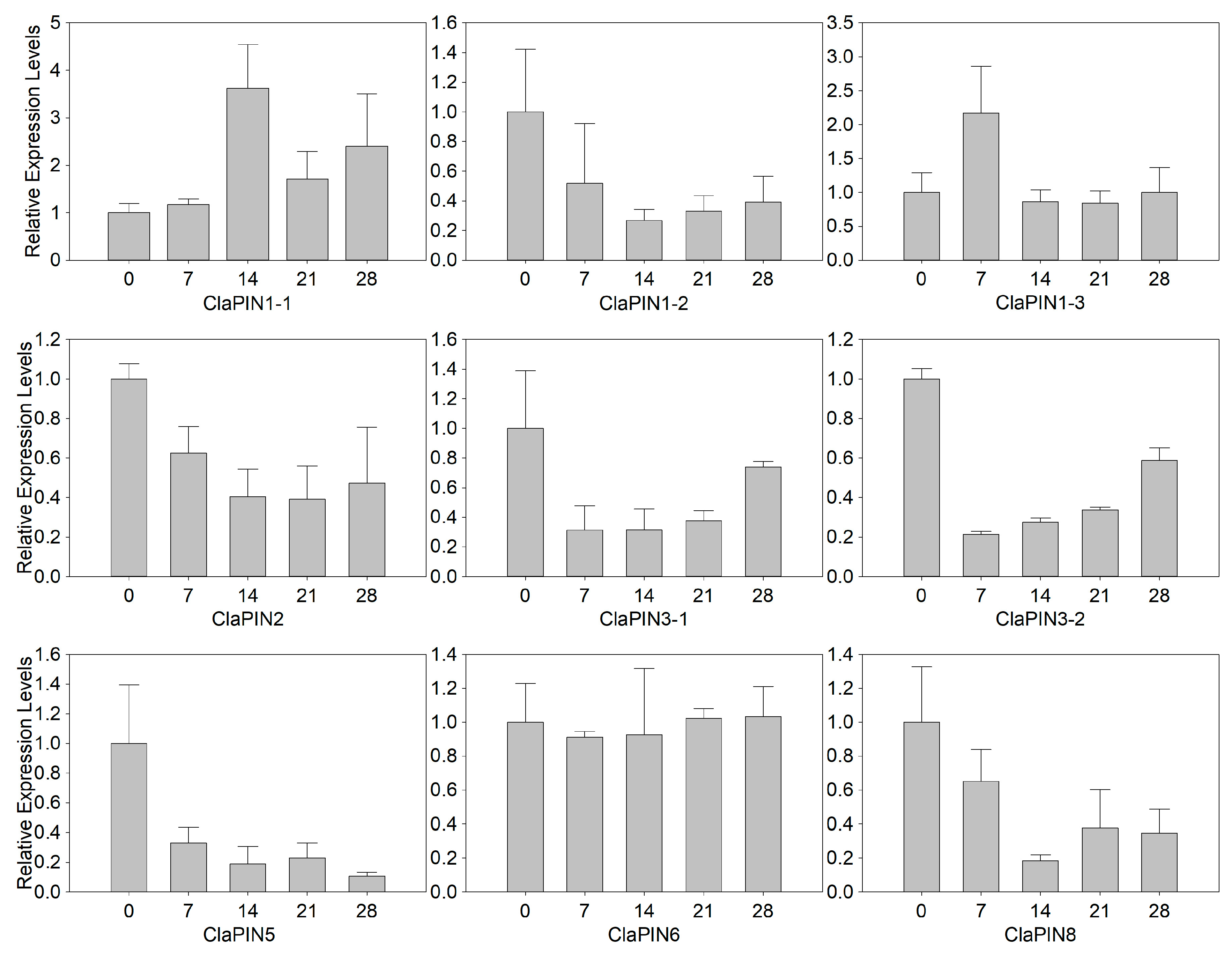
To further examine the expression patterns of ClaPINs, the expression profiles of ClaPINs were examined during shoot regeneration by qRT-PCR. The results indicated that nine ClaPINs followed four different types of expression patterns (Figure 4, Table S2). ClaPIN1-1 and ClaPIN1-3 were up-regulated at early stages of shoot regeneration and then down-regulated during the process. Five ClaPINs that include ClaPIN1-2, ClaPIN2, ClaPIN3-1, ClaPIN3-2 and ClaPIN8 showed the highest expression at the beginning of shoot generation, but had lower levels of expression during successive stages. While there was no significant change in expression levels of ClaPIN6, the expression of ClaPIN5 was gradually down regulated during shoot regeneration.
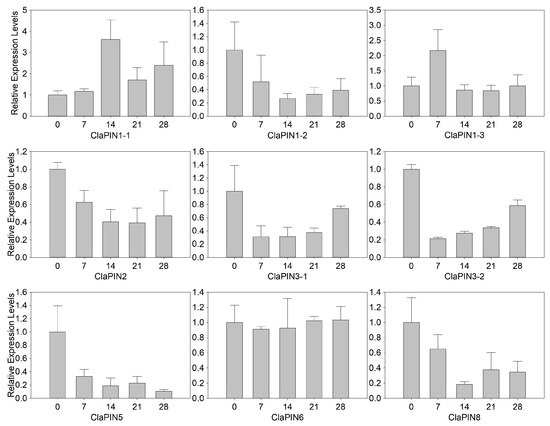
Figure 4.
Expression pattern of ClaPINs during shoot regeneration by qRT-PCR. The numbers of the X-axis represent that the samples were collected at 0, 7, 14, 21, and 28 days after incubation.
4. Discussion
4.1. Identification of PINs in Cucurbitaceae Species
In the present study, PIN genes of watermelon (9 genes), cucumber (9 genes), and melon (8 genes) were partially characterized (Table 1). There might be evolutionary differences between watermelon and cucumber/melon genes. Previous studies revealed the absence of recent whole-genome duplications in cucumber and melon after the ancient eudicot triplication [43,44]. The evolution of PINs tends to be conserved between cucumber and melon. The phylogenetic results indicated that cucumber and melon shared the same number of PINs in each sub-cluster (Figure 1A). In this study, we carried out a new genome-wide characterization of the PIN gene family in watermelon using version 2 of the watermelon 97103 genome, which has been generated by using PacBio long reads, combined with BioNano optical and Hi-C chromatin interaction maps [23]. The results presented here are also different from the previous study [31]. Two of the PINs (Cla011708 and Cla011709) in genome version 1 were reassembled into a single gene as ClaPIN3-2 in genome version 2 (Table 1). Moreover, despite the existence of it genomic sequence, Cla015026 (ClaPIN6) was not annotated in genome version 2. Interestingly, only watermelon contained PIN6 homologue, which also indicated the different evolutionary history of watermelon compared with cucumber or melon.
4.2. Putative Regulators in ClaPINs
The in silico expression analysis revealed a series of ClaPINs that are mainly expressed in roots (ClaPIN1-1 and ClaPIN2) and leaves (ClaPIN3-1, ClaPIN3-2 and ClaPIN5) (Figure 3). ClaPIN1-2 was mainly and highly expressed in both roots and flowers. These results indicated the tissue specific expression of PINs in watermelon, which has also been reported in Arabidopsis [3,31]. There were four types of expression patterns for ClaPINs during the process of shoot regeneration (Figure 4). Among these, the up-regulation of ClaPIN1-1 and ClaPIN1-3 at a very early stage of shoot regeneration indicated their important functions in regulating auxin transport to generate auxin maxima, which is necessary for fast developing tissues like calli [29]. In contrast, several ClaPINs such as ClaPIN1-2, ClaPIN2, ClaPIN3-1, ClaPIN3-2, ClaPIN5 and ClaPIN8 were down-regulated during the callus stage but up-regulated during the shoot stage. They seemed to have functions in mature tissues after cell differentiation, which might be related to their tissue specific expression patterns. On the other hand, the expression of ClaPIN6 was considerably low at all stages of shoot regeneration and did not show any significant differences among different stages suggesting that it may not play an essential role in shoot regeneration. The loss of PIN6 in cucumber and melon genomes could partly explain the non-essential role of ClaPIN6, which also indicated a species-specific evolution of the PIN family in the Cucurbitaceae species. Besides, auxin responsive cis-elements were identified in six ClaPINs, including ClaPIN1-2/3, ClaPIN2, ClaPIN3-1/2, and ClaPIN5 (Figure 2), which might contribute to their functions during shoot regeneration. There were other types of phytohormone responsive cis-elements identified in ClaPIN1-1, ClaPIN6, and ClaPIN8. This observation suggests that their potential roles might be regulated by auxin crosstalk with other phytohormones [45]. We could draw the preliminary conclusion that ClaPIN1-1 and ClaPIN1-3 are possible candidates for regulators of shoot regeneration. However, more work is still needed to reveal the functions of other ClaPINs.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/agriculture11050447/s1: Figure S1—The results of transmembrane topology analysis for watermelon PIN proteins. The blue and purple lines represent inside and outside the transmembrane, respectively; Figure S2—Conserved domains of watermelon PIN proteins; Table S1—Cis-element analysis in upstream sequences of watermelon PIN genes; Table S2—The raw data of qRT-PCR analysis.
Author Contributions
Conceptualization, H.S., L.Y., and X.H.; formal analysis, H.S., N.Z., and X.H.; investigation, H.S. and N.Z.; resources, N.Z. and L.Y.; writing—original draft preparation, H.S. and X.H.; writing—review and editing, Z.X., S.D., and L.Y.; supervision, L.Y. and X.H.; funding acquisition, N.Z. and L.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Hubei Natural Science Foundation (2018CFB686), Hubei Key R&D Program (2020BBA037), and the Earmarked Fund for China Agriculture Research System (CARS-25).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are contained within the article and supplementary materials.
Acknowledgments
We would like to thank Yaning Bao from Guizhou University (Guiyang 550025, China) for her help with the bioinformatics software.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, Y.; Jiao, Y. Auxin and above-ground meristems. J. Exp. Bot. 2018, 69, 147–154. [Google Scholar] [CrossRef]
- Adamowski, M.; Friml, J. PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef]
- Krecek, P.; Skupa, P.; Libus, J.; Naramoto, S.; Tejos, R.; Friml, J.; Zazímalová, E. The PIN-FORMED (PIN) protein family of auxin transporters. Genome Biol. 2009, 10, 249. [Google Scholar] [CrossRef]
- Zhou, J.J.; Luo, J. The PIN-FORMED Auxin Efflux Carriers in Plants. Int. J. Mol. Sci. 2018, 19, 2759. [Google Scholar] [CrossRef]
- Ding, Z.; Wang, B.; Moreno, I.; Duplakova, N.; Simon, S.; Carraro, N.; Reemmer, J.; Pencik, A.; Chen, X.; Tejos, R.; et al. ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 2012, 3, 941. [Google Scholar] [CrossRef]
- Mravec, J.; Skupa, P.; Bailly, A.; Hoyerova, K.; Krecek, P.; Bielach, A.; Petrasek, J.; Zhang, J.; Gaykova, V.; Stierhof, Y.D. Subcellular homeostasis of phytohormone auxin is mediated by the ER localized PIN5 transporter. Nature 2009, 459, 1136–1140. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.; Skupa, P.; Viaene, T.; Zwiewka, M.; Tejos, R.; Klima, P.; Carna, M.; Rolcik, J.; De Rycke, R.; Moreno, I.; et al. PIN6 auxin transporter at endoplasmic reticulum and plasma membrane mediates auxin homeostasis and organogenesis in Arabidopsis. New Phytol. 2016, 211, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Rakusová, H.; Abbas, M.; Han, H.; Song, S.; Robert, H.S.; Friml, J. Termination of shoot gravitropic responses by auxin feedback on PIN3 polarity. Curr. Biol. 2016, 26, 3026–3032. [Google Scholar] [CrossRef] [PubMed]
- Rigo, G.; Ayaydin, F.; Tietz, O.; Zsigmond, L.; Kovacs, H.; Pay, A.; Salchert, K.; Darula, Z.; Medzihradszky, K.F.; Szabados, L.; et al. Inactivation of plasma membrane-localized CDPK-RELATED KINASE5 decelerates PIN2 exocytosis and root gravitropic response in Arabidopsis. Plant Cell 2013, 25, 1592–1608. [Google Scholar] [CrossRef]
- Rosquete, M.R.; Waidmann, S.; Kleine, V.J. PIN7 auxin carrier has a preferential role in terminating radial root expansion in Arabidopsis thaliana. Int. J. Mol. Sci. 2018, 19, 1238. [Google Scholar] [CrossRef]
- Xi, W.; Gong, X.; Yang, Q.; Yu, H.; Liou, Y.C. Pin1At regulates PIN1 polar localization and root gravitropism. Nat. Commun. 2016, 7, 10430. [Google Scholar] [CrossRef]
- Haga, K.; Sakai, T. PIN auxin efflux carriers are necessary for pulse-induced but not continuous light-induced phototropism in Arabidopsis. Plant Physiol. 2012, 160, 763–776. [Google Scholar] [CrossRef]
- Zadnikova, P.; Petrasek, J.; Marhavy, P.; Raz, V.; Vandenbussche, F.; Ding, Z.; Schwarzerova, K.; Morita, M.T.; Tasaka, M.; Hejatko, J.; et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 2010, 137, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Okada, K.; Ueda, J.; Komaki, M.K.; Bell, C.J.; Shimura, Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell 1991, 3, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Dal Bosco, C.; Dovzhenko, A.; Liu, X.; Woerner, N.; Rensch, T.; Eismann, M.; Eimer, S.; Hegermann, J.; Paponov, I.A.; Ruperti, B.; et al. The endoplasmic reticulum localized PIN8 is a pollen-specific auxin carrier involved in intracellular auxin homeostasis. Plant J. 2012, 71, 860–870. [Google Scholar] [CrossRef]
- Willige, B.C.; Chory, J. A current perspective on the role of AGCVIII kinases in PIN-mediated apical hook development. Front. Plant Sci. 2015, 6, 767. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Takahashi, M.; Shibasaki, K.; Wu, S.; Inaba, T.; Tsurumi, S.; Baskin, T.I. Gravitropism of Arabidopsis thaliana roots requires the polarization of PIN2 toward the root tip in meristematic cortical cells. Plant Cell 2010, 22, 1762–1776. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Liu, Y.; Maere, S.; Lee, E.; Van, I.G.; Xie, Z.; Xuan, W.; Lucas, J.; Vassileva, V.; Kitakura, S.; et al. A coherent transcriptional feed-forward motif model for mediating auxin-sensitive PIN3 expression during lateral root development. Nat. Commun. 2015, 6, 8821. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Benková, E.; Blilou, I.; Wisniewska, J.; Hamann, T.; Ljung, K.; Woody, S.; Sandberg, G.; Scheres, B.; Jürgens, G.; et al. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 2002, 108, 661–673. [Google Scholar] [CrossRef]
- Vieten, A.; Vanneste, S.; Wisniewska, J.; Benková, E.; Benjamins, R.; Beeckman, T.; Luschnig, C.; Friml, J. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 2005, 132, 4521–4531. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations. Available online: Faostat.fao.org (accessed on 6 January 2021).
- Ellis, A.C.; Dudenbostel, T.; Crowe-White, K. Watermelon Juice: A Novel Functional Food to Increase Circulating Lycopene in Older Adult Women. Plant Foods Hum. Nutr. 2019, 74, 200–203. [Google Scholar] [CrossRef]
- Guo, S.; Zhao, S.; Sun, H.; Wang, X.; Wu, S.; Lin, T.; Ren, Y.; Gao, L.; Deng, Y.; Zhang, J.; et al. Resequencing of 414 cultivated and wild watermelon accessions identifies selection for fruit quality traits. Nat. Genet. 2019, 51, 1616–1623. [Google Scholar] [CrossRef]
- Choi, P.S.; Soh, W.Y.; Kim, Y.S.; Yoo, O.J.; Liu, J.R. Genetic transformation and plant regeneration of watermelon using Agrobacterium tumefaciens. Plant Cell Rep. 1994, 13, 344–348. [Google Scholar] [CrossRef]
- Tian, S.; Jiang, L.; Cui, X.; Zhang, J.; Guo, S.; Li, M.; Zhang, H.; Ren, Y.; Gong, G.; Zong, M.; et al. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 2018, 37, 1353–1356. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Huang, X.; Bao, Y.; Wang, B.; Liu, L.; Dai, L.; Chen, J.; An, X.; Sun, Y.; Peng, D. Genome-wide identification and expression profiling of WUSCHEL-related homeobox (WOX) genes during adventitious shoot regeneration of watermelon (Citrullus lanatus). Acta Physiol. Plant. 2015, 37, 224. [Google Scholar] [CrossRef]
- Zhang, N.; Huang, X.; Bao, Y.; Wang, B.; Zeng, H.; Cheng, W.; Tang, M.; Li, Y.; Ren, J.; Sun, Y. Genome-wide identification of SAUR genes in watermelon (Citrullus lanatus). Physiol. Mol. Biol. Plants 2017, 23, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Radhakrishnan, D.; Kareem, A.; Durgaprasad, K.; Sreeraj, E.; Sugimoto, K.; Prasad, K. Shoot regeneration: A journey from acquisition of competence to completion. Curr. Opin. Plant Biol. 2018, 41, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Duclercq, J.; Sangwan-Norreel, B.; Catterou, M.; Sangwan, R.S. De novo shoot organogenesis: From art to science. Trends Plant Sci. 2011, 16, 597–606. [Google Scholar] [CrossRef]
- Krogan, N.T.; Marcos, D.; Weiner, A.I.; Berleth, T. The auxin response factor MONOPTEROS controls meristem function and organogenesis in both the shoot and root through the direct regulation of PIN genes. New Phytol. 2016, 212, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Dong, W.; Zhan, Y.; Huang, Z.A.; Li, Z.; Kim, I.S.; Zhang, C. Genome-wide identification and expression analysis of ClLAX, ClPIN and ClABCB genes families in Citrullus lanatus under various abiotic stresses and grafting. BMC Genet. 2017, 18, 33. [Google Scholar] [CrossRef]
- Cucurbit Genomics Database. Available online: Cucurbitgenomics.org (accessed on 6 January 2021).
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- ProtParam tool. Available online: Web.expasy.org/protparam/ (accessed on 6 January 2021).
- Yu, C.S.; Chen, Y.C.; Lu, C.H.; Hwang, J.K. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- DNAMAN-Bioinformatics Solutions. Available online: www.lynnon.com (accessed on 6 January 2021).
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools—An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Peer, Y.V.D.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef]
- Garcia-Mas, J.; Benjak, A.; Sanseverino, W.; Bourgeois, M.; Mir, G.; González, V.M.; Hénaff, E.; Câmara, F.; Cozzuto, L.; Lowy, E.; et al. The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. USA 2012, 109, 11872–11877. [Google Scholar] [CrossRef]
- Altmann, M.; Altmann, S.; Rodriguez, P.A.; Weller, B.; Elorduy Vergara, L.; Palme, J.; Marín-de la Rosa, N.; Sauer, M.; Wenig, M.; Villaécija-Aguilar, J.A.; et al. Extensive signal integration by the phytohormone protein network. Nature 2020, 583, 271–276. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).