Eco-Friendly Biocontrol Strategies of Alternaria Phytopathogen Fungus: A Focus on Gene-Editing Techniques
Abstract
:1. Introduction
2. Alternaria spp.
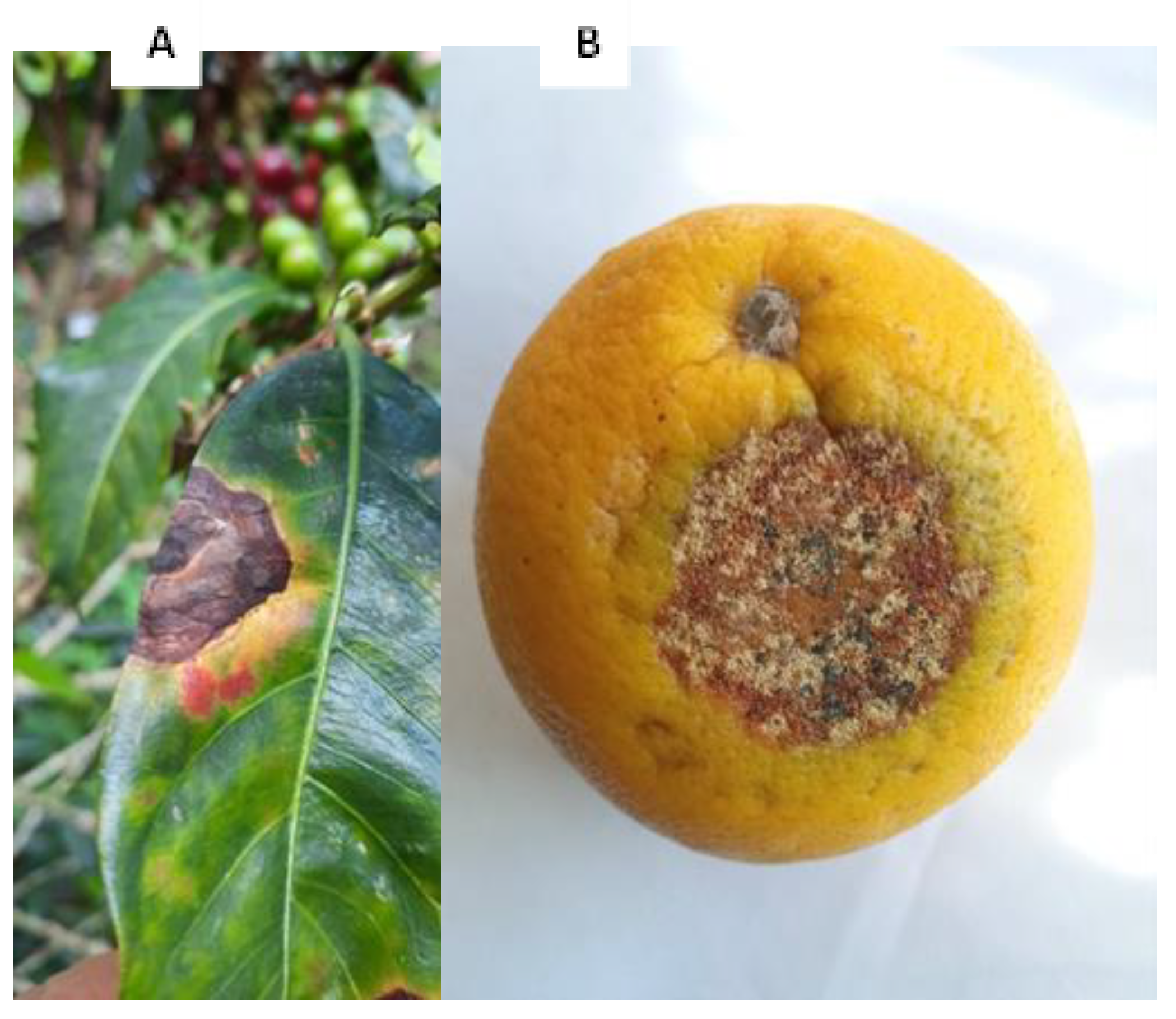
2.1. Alternaria Infection
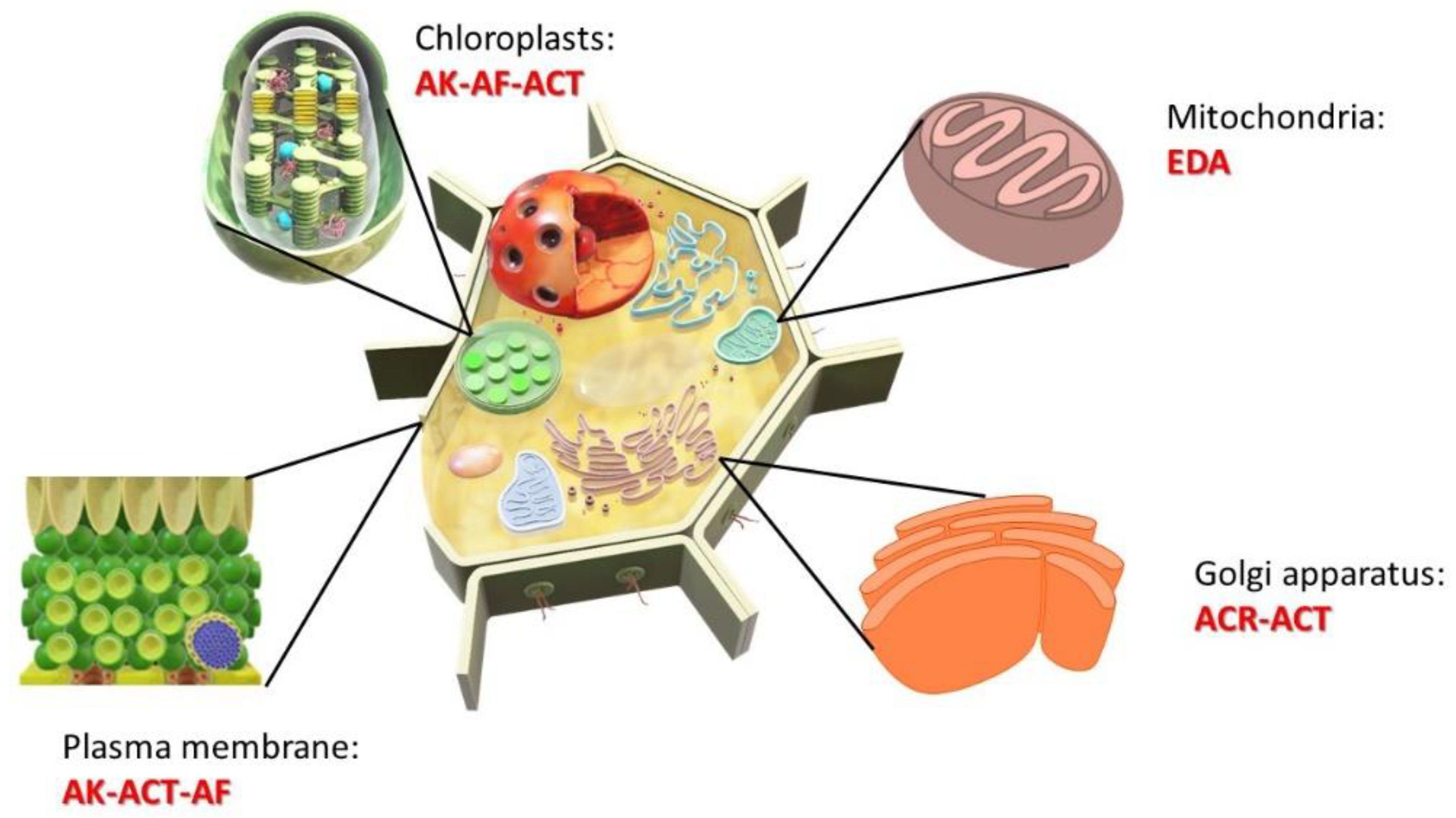
2.2. Phytopathogenic Genes in Alternaria spp.
3. Alternaria Control Strategies
3.1. Chemical Control Strategies
3.2. Green Control Strategies
3.2.1. Essential Oils and Biopolymers Used to Control Alternaria Fungi
3.2.2. Antagonistic Microorganisms to Control Alternaria
3.3. Molecular-Based Methods for Controlling Alternaria
3.3.1. Control of Alternaria Pathogenicity Based on CRISPR/Cas9
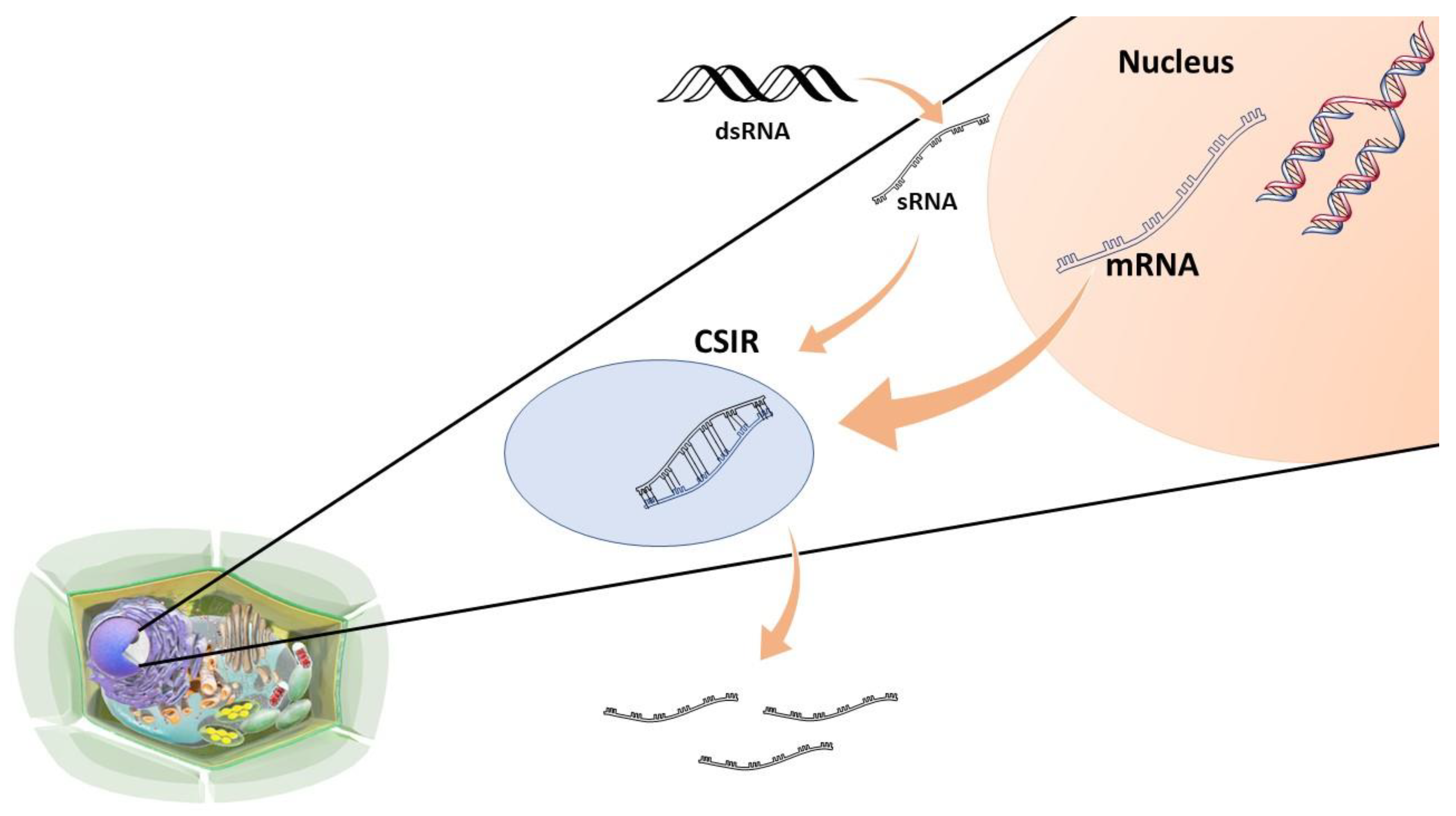
3.3.2. RNAi in the Control of Alternaria
3.3.3. Control of Alternaria Phytopathogenicity through Regulation of Transcriptional Factors
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Teng, P.S.; Shane, W.W.; MacKenzie, D.R. Crop Losses Due to Plant Pathogens. CRC Crit. Rev. Plant Sci. 1984, 2, 21–47. [Google Scholar] [CrossRef]
- Adikaram, N.K.B.; Yakandawala, D.M.D. A Checklist of Plant Pathogenic Fungi and Oomycota in Sri Lanka. Ceylon J. Sci. 2020, 49, 93. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The Global Burden of Pathogens and Pests on Major Food Crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef] [PubMed]
- Blagojević, J.D.; Vukojević, J.B.; Ivanović, Ž.S. Occurrence and Characterization of Alternaria Species Associated with Leaf Spot Disease in Rapeseed in Serbia. Plant Pathol. 2020, 69, 883–900. [Google Scholar] [CrossRef]
- Claeys, L.; Romano, C.; De Ruyck, K.; Wilson, H.; Fervers, B.; Korenjak, M.; Zavadil, J.; Gunter, M.J.; De Saeger, S.; De Boevre, M.; et al. Mycotoxin Exposure and Human Cancer Risk: A Systematic Review of Epidemiological Studies. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1449–1464. [Google Scholar] [CrossRef]
- Ma, G.; Bao, S.; Zhao, J.; Sui, Y.; Wu, X. Morphological and Molecular Characterization of Alternaria Species Causing Leaf Blight on Watermelon in China. Plant Dis. 2020, 105, 60–70. [Google Scholar] [CrossRef]
- Witte, T.E.; Villeneuve, N.; Boddy, C.N.; Overy, D.P. Accessory Chromosome-Acquired Secondary Metabolism in Plant Pathogenic Fungi: The Evolution of Biotrophs Into Host-Specific Pathogens. Front. Microbiol. 2021, 12, 664276. [Google Scholar] [CrossRef]
- González-Estrada, R.R.; Carvajal-Millán, E.; Ragazzo-Sánchez, J.A.; Bautista-Rosales, P.U.; Calderón-Santoyo, M. Control of Blue Mold Decay on Persian Lime: Application of Covalently Cross-Linked Arabinoxylans Bioactive Coatings with Antagonistic Yeast Entrapped. LWT Food Sci. Technol. 2017, 85, 187–196. [Google Scholar] [CrossRef]
- Mehrabi, R.; Bahkali, A.H.; Abd-Elsalam, K.A.; Moslem, M.; Ben M’Barek, S.; Gohari, A.M.; Jashni, M.K.; Stergiopoulos, I.; Kema, G.H.J.; de Wit, P.J.G.M. Horizontal Gene and Chromosome Transfer in Plant Pathogenic Fungi Affecting Host Range. FEMS Microbiol. Rev. 2011, 35, 542–554. [Google Scholar] [CrossRef] [Green Version]
- Muñoz, I.V.; Sarrocco, S.; Malfatti, L.; Baroncelli, R.; Vannacci, G. CRISPR-Cas for Fungal Genome Editing: A New Tool for the Management of Plant Diseases. Front. Plant Sci. 2019, 10, 135. [Google Scholar] [CrossRef]
- Lu, K.; Zhang, M.; Yang, R.; Zhang, M.; Guo, Q.; Baek, K.H.; Xu, H. The MAP Kinase Kinase Gene Abste7 Regulates Multiple Aspects of Alternaria Brassicicola Pathogenesis. Plant Pathol. J. 2019, 35, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.-R.; Wu, P.-C.; Chen, Y.-K.; Yago, J.I. The Siderophore Repressor SreA Maintains Growth, Hydrogen Peroxide Resistance, and Cell Wall Integrity in the Phytopathogenic Fungus Alternaria Alternata. Fungal Genet. Biol. 2020, 139, 103384. [Google Scholar] [CrossRef] [PubMed]
- Rokas, A.; Mead, M.E.; Steenwyk, J.L.; Raja, H.A.; Oberlies, N.H. Biosynthetic Gene Clusters and the Evolution of Fungal Chemodiversity. Nat. Prod. Rep. 2020, 37, 868–878. [Google Scholar] [CrossRef]
- Arazoe, T. Development of Genome-Editing Technologies for Plant Pathogenic Fungi. J. Gen. Plant Pathol. 2020, 86, 523–525. [Google Scholar] [CrossRef]
- Goulin, E.H.; Galdeano, D.M.; Granato, L.M.; Matsumura, E.E.; Dalio, R.J.D.; Machado, M.A. RNA Interference and CRISPR: Promising Approaches to Better Understand and Control Citrus Pathogens. Microbiol. Res. 2019, 226, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kwasiborski, A.; Bastide, F.; Hamon, B.; Poupard, P.; Simoneau, P.; Guillemette, T. In Silico Analysis of RNA Interference Components and MiRNAs-like RNAs in the Seed-Borne Necrotrophic Fungus Alternaria Brassicicola. Fungal Biol. 2022, 126, 224–234. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira Filho, J.G.; Silva, G.d.C.; Cipriano, L.; Gomes, M.; Egea, M.B. Control of Postharvest Fungal Diseases in Fruits Using External Application of RNAi. J. Food Sci. 2021, 86, 3341–3348. [Google Scholar] [CrossRef]
- Das, P.R.; Sherif, S.M. Application of Exogenous DsRNAs-Induced RNAi in Agriculture: Challenges and Triumphs. Front. Plant Sci. 2020, 11, 946. [Google Scholar] [CrossRef]
- Aslam, M.F.; Irshad, G.; Naz, F. Evaluation of the Antifungal Activity of Essential Oils against Alternaria Alternata Causing Fruit Rot of Eriobotrya Japonica Eriobotrya Japonica. Turk. J. Biochem. 2022, 47. [Google Scholar] [CrossRef]
- Chrapačienė, S.; Rasiukevičiūtė, N.; Valiuškaitė, A. Biocontrol of Carrot Disease-Causing Pathogens Using Essential Oils. Plants 2021, 10, 2231. [Google Scholar] [CrossRef]
- Lei, X.; Deng, B.; Ruan, C.; Deng, L.; Zeng, K. Phenylethanol as a Quorum Sensing Molecule to Promote Biofilm Formation of the Antagonistic Yeast Debaryomyces Nepalensis for the Control of Black Spot Rot on Jujube. Postharvest Biol. Technol. 2022, 185, 111788. [Google Scholar] [CrossRef]
- Tsyganenko, K.S.; Savchuk, Y.I.; Nakonechna, L.T.; Kurchenko, I.M. The Biological Activity of Alternaria Species. Mikrobiol. Zh. 2018, 80, 78–87. [Google Scholar] [CrossRef] [Green Version]
- Meng, J.-W.; He, D.-C.; Zhu, W.; Yang, L.-N.; Wu, E.-J.; Xie, J.-H.; Shang, L.-P.; Zhan, J. Human-Mediated Gene Flow Contributes to Metapopulation Genetic Structure of the Pathogenic Fungus Alternaria Alternata from Potato. Front. Plant Sci. 2018, 9, 198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puvača, N.; Bursić, V.; Vuković, G.; Budakov, D.; Petrović, A. Ascomycete Fungi (Alternaria spp.) Characterization as Major Feed Grains Pathogens. J. Agron. Technol. Eng. Manag. 2020, 3, 499–505. [Google Scholar]
- Somma, S.; Amatulli, M.T.; Masiello, M.; Moretti, A.; Logrieco, A.F. Alternaria Species Associated to Wheat Black Point Identified through a Multilocus Sequence Approach. Int. J. Food Microbiol. 2019, 293, 34–43. [Google Scholar] [CrossRef]
- Ahmadpour, A.; Ghosta, Y.; Poursafar, A. Novel Species of Alternaria Section Nimbya from Iran as Revealed by Morphological and Molecular Data. Mycologia 2021, 113, 1073–1088. [Google Scholar] [CrossRef]
- Abbo, A.S.; Idris, M.O.; Elballa, M.A.; Hammad, A.M.; Rahman El Siddig, M.A.; Karlovsky, P. Genetic Variability and Host Specialization in Alternaria Alternata Colonizing Solanaceous Crops in Sudan. J. Plant Prot. Res. 2018, 58, 246–256. [Google Scholar] [CrossRef]
- Luo, Y.; Hou, L.; Förster, H.; Pryor, B.; Adaskaveg, J.E. Identification of Alternaria Species Causing Heart Rot of Pomegranates in California. Plant Dis. 2016, 101, 421–427. [Google Scholar] [CrossRef] [Green Version]
- Guillemette, T.; Iacomi-Vasilescu, B.; Simoneau, P. Conventional and Real-Time PCR-Based Assay for Detecting Pathogenic Alternaria Brassicae in Cruciferous Seed. Plant Dis. 2004, 88, 490–496. [Google Scholar] [CrossRef] [Green Version]
- Adama, S.; Abdoulaye, S.; Kadidia, K.; Elisabeth, Z.; Abel, T.N.; Drissa, S.; Théodore, N.; Mahamadou, S. Characterization of Alternaria Brassicicola Isolated from Tomato in Burkina Faso, and Use of Two Essential Oils for Its Control in Vitro. Afr. J. Agric. Res. 2021, 17, 1371–1379. [Google Scholar] [CrossRef]
- Koike, S.T.; Smith, R.F.; Cahn, M.D.; Pryor, B.M. Association of the Carrot Pathogen Alternaria Dauci With New Diseases, Alternaria Leaf Speck, of Lettuce and Celery in California. Plant Health Prog. 2017, 18, 136–143. [Google Scholar] [CrossRef]
- Pavón, M.Á.; González, I.; Pegels, N.; Martín, R.; García, T. PCR Detection and Identification of Alternaria Species-Groups in Processed Foods Based on the Genetic Marker Alt a 1. Food Control 2010, 21, 1745–1756. [Google Scholar] [CrossRef]
- Khan, M.; Wang, R.; Li, B.; Liu, P.; Weng, Q.; Chen, Q. Comparative Evaluation of the LAMP Assay and PCR-Based Assays for the Rapid Detection of Alternaria Solani. Front. Microbiol. 2018, 9, 2089. [Google Scholar] [CrossRef] [Green Version]
- DAR, A.A.; SHARMA, S.; MAHAJAN, R.; MUSHTAQ, M.; SALATHIA, A.; AHAMAD, S.; SHARMA, J.P. Overview of Purple Blotch Disease and Understanding Its Management through Chemical, Biological and Genetic Approaches. J. Integr. Agric. 2020, 19, 3013–3024. [Google Scholar] [CrossRef]
- Li, L.; Pan, H.; Deng, L.; Wang, Z.P.; Li, D.W.; Zhang, Q.; Chen, M.Y.; Zhong, C.H. First Report of Alternaria Tenuissima Causing Brown Spot Disease of Kiwifruit Foliage in China. Plant Dis. 2018, 103, 582. [Google Scholar] [CrossRef]
- Zwickel, T.; Kahl, S.M.; Rychlik, M.; Müller, M.E.H. Chemotaxonomy of Mycotoxigenic Small-Spored Alternaria Fungi—Do Multitoxin Mixtures Act as an Indicator for Species Differentiation? Front. Microbiol. 2018, 9, 1368. [Google Scholar] [CrossRef] [Green Version]
- Stinson, E.E.; Bills, D.D.; Osman, S.F.; Siciliano, J.; Ceponis, M.J.; Heisler, E.G. Mycotoxin Production by Alternaria Species Grown on Apples, Tomatoes, and Blueberries. J. Agric. Food Chem. 1980, 28, 960–963. [Google Scholar] [CrossRef]
- Nizamani, S.; Khaskheli, A.J.; Khaskheli, A.A.; Jiskani, A.M. The Intensity of Tomato Post-Harvest Rot in the Surroundings of Tandojam. Turk. J. Agric. —Food Sci. Technol. 2021, 9, 288–295. [Google Scholar] [CrossRef]
- da Cruz Cabral, L.; Rodríguez, A.; Delgado, J.; Patriarca, A. Understanding the Effect of Postharvest Tomato Temperatures on Two Toxigenic Alternaria Spp. Strains: Growth, Mycotoxins and Cell-Wall Integrity-Related Gene Expression. J. Sci. Food Agric. 2019, 99, 6689–6695. [Google Scholar] [CrossRef]
- Iwebor, M.; Frolov, S.; Frolova, I.; Shabaldas, O.; Chernikova, M. The Role of Insects in the Spreading of Pathogens and Development of Diseases on Sunflower in the Krasnodar Region of the Russian Federation. E3S Web Conf. 2020, 222, 02025. [Google Scholar] [CrossRef]
- Taj, G.; Meena, P.D.; Giri, P.; Pandey, D.; Kumar, A.; Kumar, A. Pathogenesis Mechanisms Employed by Alternaria Species. J. Oilseed Brassica 2015, 6, 213–240. [Google Scholar]
- Bose, S.K.; Howlader, P.; Wang, W.; Yin, H. Oligosaccharide Is a Promising Natural Preservative for Improving Postharvest Preservation of Fruit: A Review. Food Chem. 2021, 341, 128178. [Google Scholar] [CrossRef] [PubMed]
- Ngolong Ngea, G.L.; Qian, X.; Yang, Q.; Dhanasekaran, S.; Ianiri, G.; Ballester, A.R.; Zhang, X.; Castoria, R.; Zhang, H. Securing Fruit Production: Opportunities from the Elucidation of the Molecular Mechanisms of Postharvest Fungal Infections. Compr. Rev. Food Sci. Food Saf. 2021, 20, 2508–2533. [Google Scholar] [CrossRef]
- Fontaine, K.; Fourrier-Jeandel, C.; Armitage, A.D.; Boutigny, A.-L.; Crépet, M.; Caffier, V.; Gnide, D.C.; Shiller, J.; Le Cam, B.; Giraud, M.; et al. Identification and Pathogenicity of Alternaria Species Associated with Leaf Blotch Disease and Premature Defoliation in French Apple Orchards. PeerJ 2021, 9, e12496. [Google Scholar] [CrossRef]
- Hu, J.; Chen, C.; Peever, T.; Dang, H.; Lawrence, C.; Mitchell, T. Genomic Characterization of the Conditionally Dispensable Chromosome in Alternaria Arborescens Provides Evidence for Horizontal Gene Transfer. BMC Genom. 2012, 13, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, H.; Cai, G.; Luo, J.; Bhattacharya, D.; Zhang, N. Extensive Horizontal Gene Transfers between Plant Pathogenic Fungi. BMC Biol. 2016, 14, 41. [Google Scholar] [CrossRef] [Green Version]
- Meena, M.; Gupta, S.K.; Swapnil, P.; Zehra, A.; Dubey, M.K.; Upadhyay, R.S. Alternaria Toxins: Potential Virulence Factors and Genes Related to Pathogenesis. Front. Microbiol. 2017, 8, 1451. [Google Scholar] [CrossRef] [Green Version]
- Meena, M.; Samal, S. Alternaria Host-Specific (HSTs) Toxins: An Overview of Chemical Characterization, Target Sites, Regulation and Their Toxic Effects. Toxicol. Reports 2019, 6, 745–758. [Google Scholar] [CrossRef]
- Praveen, B.; Nagaraja, A.; Prasanna Kumar, M.K.; Pramesh, D.; Palanna, K.B.; Buella, P.P. First Report of Alternaria Alternata Causing Leaf Blight on Little Millet (Panicum Sumatrense) in India. Plant Dis. 2020, 105, 1202. [Google Scholar] [CrossRef]
- Panel, E.; Chain, F. Scientific Opinion on the Risks for Animal and Public Health Related to the Presence of Alternaria Toxins in Feed and Food. EFSA J. 2011, 9, 2407. [Google Scholar] [CrossRef]
- Meena, M.; Swapnil, P.; Upadhyay, R.S. Isolation, Characterization and Toxicological Potential of Alternaria-Mycotoxins (TeA, AOH and AME) in Different Alternaria Species from Various Regions of India. Sci. Rep. 2017, 7, 8777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.; Liu, D.; Yang, X.; Zhang, L.; Yang, M. Detection of Seven Alternaria Toxins in Edible and Medicinal Herbs Using Ultra-High Performance Liquid Chromatography-Tandem Mass Spectrometry. Food Chem. X 2022, 13, 100186. [Google Scholar] [CrossRef] [PubMed]
- Saleem, A.; El-Shahir, A.A. Morphological and Molecular Characterization of Some Alternaria Species Isolated from Tomato Fruits Concerning Mycotoxin Production and Polyketide Synthase Genes. Plants 2022, 11, 1168. [Google Scholar] [CrossRef]
- Garganese, F.; Schena, L.; Siciliano, I.; Prigigallo, M.I.; Spadaro, D.; De Grassi, A.; Ippolito, A.; Sanzani, S.M. Characterization of Citrus-Associated Alternaria Species in Mediterranean Areas. PLoS ONE 2016, 11, e0163255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dettman, J.R.; Eggertson, Q. Phylogenomic Analyses of Alternaria Section Alternaria: A High-Resolution, Genome-Wide Study of Lineage Sorting and Gene Tree Discordance. Mycologia 2021, 113, 1218–1232. [Google Scholar] [CrossRef]
- Gai, Y.; Ma, H.; Chen, Y.; Li, L.; Cao, Y.; Wang, M.; Sun, X.; Jiao, C.; Riely, B.K.; Li, H. Chromosome-Scale Genome Sequence of Alternaria Alternata Causing Alternaria Brown Spot of Citrus. Mol. Plant-Microbe Interact. 2021, 34, 726–732. [Google Scholar] [CrossRef]
- Félix, C.; Meneses, R.; Gonçalves, M.F.M.; Tilleman, L.; Duarte, A.S.; Jorrín-Novo, J.V.; Van de Peer, Y.; Deforce, D.; Van Nieuwerburgh, F.; Esteves, A.C.; et al. A Multi-Omics Analysis of the Grapevine Pathogen Lasiodiplodia Theobromae Reveals That Temperature Affects the Expression of Virulence- and Pathogenicity-Related Genes. Sci. Rep. 2019, 9, 13144. [Google Scholar] [CrossRef] [Green Version]
- Wolters, P.J.; Faino, L.; van den Bosch, T.B.M.; Evenhuis, B.; Visser, R.G.F.; Seidl, M.F.; Vleeshouwers, V.G.A.A. Gapless Genome Assembly of the Potato and Tomato Early Blight Pathogen Alternaria Solani. Mol. Plant-Microbe Interact. 2018, 31, 692–694. [Google Scholar] [CrossRef] [Green Version]
- Dang, H.X.; Pryor, B.; Peever, T.; Lawrence, C.B. The Alternaria Genomes Database: A Comprehensive Resource for a Fungal Genus Comprised of Saprophytes, Plant Pathogens, and Allergenic Species. BMC Genomics 2015, 16, 239. [Google Scholar] [CrossRef] [Green Version]
- Tsuzuki, R.; Pintado, R.M.C.; Thorndike, J.A.B.; Reynoso, D.L.G.; Guerra, C.A.A.; Abad, J.C.G.; Caballero, L.M.A.; Zaquinaula, M.H.H.; Sierra, C.U.; Cruz, O.I.A.; et al. Mutations Found in the Asc1 Gene That Confer Susceptibility to the Aal-Toxin in Ancestral Tomatoes from Peru and Mexico. Plants 2021, 10, 47. [Google Scholar] [CrossRef]
- Spassieva, S.D.; Markham, J.E.; Hille, J. The Plant Disease Resistance Gene Asc-1 Prevents Disruption of Sphingolipid Metabolism during AAL-Toxin-Induced Programmed Cell Death. Plant J. 2002, 32, 561–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Zhang, M.; Gao, L.; Yang, Q.; Kalaji, H.M.; Qiang, S.; Strasser, R.J.; Chen, S. Tenuazonic Acid-Triggered Cell Death Is the Essential Prerequisite for Alternaria Alternata (Fr.) Keissler to Infect Successfully Host Ageratina Adenophora. Cells 2021, 10, 1010. [Google Scholar] [CrossRef] [PubMed]
- Pontes, J.G.d.M.; Fernandes, L.S.; dos Santos, R.V.; Tasic, L.; Fill, T.P. Virulence Factors in the Phytopathogen–Host Interactions: An Overview. J. Agric. Food Chem. 2020, 68, 7555–7570. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.W.; Perez, F.G. Assessing Possible Mechanisms of Resistance to Early Blight Caused by Alternaria Solani. Potato Res. 2019, 62, 423–434. [Google Scholar] [CrossRef]
- Løbner-Olesen, A.; Skovgaard, O.; Marinus, M.G. Dam Methylation: Coordinating Cellular Processes. Curr. Opin. Microbiol. 2005, 8, 154–160. [Google Scholar] [CrossRef]
- Minami, A.; Tajima, N.; Higuchi, Y.; Toyomasu, T.; Sassa, T.; Kato, N.; Dairi, T. Identification and Functional Analysis of Brassicicene C Biosynthetic Gene Cluster in Alternaria Brassicicola. Bioorg. Med. Chem. Lett. 2009, 19, 870–874. [Google Scholar] [CrossRef]
- Bonthala, B.; Small, C.S.; Lutz, M.A.; Graf, A.; Krebs, S.; Sepúlveda, G.; Stam, R. ONT-Based Draft Genome Assembly and Annotation of Alternaria Atra. Mol. Plant-Microbe Interact. 2021, 34, 870–873. [Google Scholar] [CrossRef]
- Vaquera, S.; Patriarca, A.; Cabrera, G.; Fernández Pinto, V. Temperature and Water Activity Influence on Simultaneous Production of AAL Toxins by Alternaria Arborescens on Tomato Medium. Eur. J. Plant Pathol. 2017, 148, 1003–1009. [Google Scholar] [CrossRef]
- Wenderoth, M.; Garganese, F.; Schmidt-Heydt, M.; Soukup, S.T.; Ippolito, A.; Sanzani, S.M.; Fischer, R. Alternariol as Virulence and Colonization Factor of Alternaria Alternata during Plant Infection. Mol. Microbiol. 2019, 112, 131–146. [Google Scholar] [CrossRef]
- Liu, M.; Xu, L.; Liu, W.; Yu, J.; Jing, G.; Liu, H. Cinnamaldehyde Regulates the Synthesis of Alternaria Alternata Non-Host Selective Toxins by Influencing PKS Gene Expression and Oxidoreductase Activity. Ind. Crops Prod. 2020, 145, 112074. [Google Scholar] [CrossRef]
- Carvalho, F.P. Pesticides, Environment, and Food Safety. Food Energy Secur. 2017, 6, 48–60. [Google Scholar] [CrossRef]
- Pereira, P.C.G.; Parente, C.E.T.; Carvalho, G.O.; Torres, J.P.M.; Meire, R.O.; Dorneles, P.R.; Malm, O. A Review on Pesticides in Flower Production: A Push to Reduce Human Exposure and Environmental Contamination. Environ. Pollut. 2021, 289, 117817. [Google Scholar] [CrossRef]
- Zhao, W.; Sun, C.; Wei, L.; Chen, W.; Wang, B.; Li, F.; Wei, M.; Lou, T.; Zhang, P.; Zheng, H.; et al. Detection and Fitness of Dicarboximide-Resistant Isolates of Alternaria Alternata from Dendrobium Officinale, a Chinese Indigenous Medicinal Herb. Plant Dis. 2020, 105, 2222–2230. [Google Scholar] [CrossRef] [PubMed]
- Lichtemberg, P.; Puckett, R.; Felts, D.; Luo, Y.; Doster, L.; Rodriguez, D.; Michailides, T. Survey of the Pathogen of Alternaria Late Blight Reveals Different Levels of Carboxamide Fungicide Resistance in the Main Pistachio Producing Regions of California. Calif. Agric. 2018, 72, 170–178. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.C.; Zhang, C.Q. Multi-Resistance to Thiophanate-Methyl, Diethofencarb, and Procymidone among Alternaria Alternata Populations from Tobacco Plants, and the Management of Tobacco Brown Spot with Azoxystrobin. Phytoparasitica 2018, 46, 677–687. [Google Scholar] [CrossRef]
- Ishii, H.; Bryson, P.K.; Kayamori, M.; Miyamoto, T.; Yamaoka, Y.; Schnabel, G. Cross-Resistance to the New Fungicide Mefentrifluconazole in DMI-Resistant Fungal Pathogens. Pestic. Biochem. Physiol. 2021, 171, 104737. [Google Scholar] [CrossRef]
- Yang, L.-N.; He, M.-H.; Ouyang, H.-B.; Zhu, W.; Pan, Z.-C.; Sui, Q.-J.; Shang, L.-P.; Zhan, J. Cross-Resistance of the Pathogenic Fungus Alternaria Alternata to Fungicides with Different Modes of Action. BMC Microbiol. 2019, 19, 205. [Google Scholar] [CrossRef] [Green Version]
- Chrapačienė, S.; Rasiukevičiūtė, N.; Valiuškaitė, A. Control of Seed-Borne Fungi by Selected Essential Oils. Horticulturae 2022, 8, 220. [Google Scholar] [CrossRef]
- Sazvar, E.; Jahani, M.; Aminifard, M.H.; Hosseini, S.A. In Vitro and In Vivo Control of Alternaria Alternata in Barberry (Berberis Vulgaris) by Some Essential Oils. Erwerbs-Obstbau 2022, 64, 413–423. [Google Scholar] [CrossRef]
- Feng, W.; Zheng, X. Essential Oils to Control Alternaria Alternata in Vitro and in Vivo. Food Control 2007, 18, 1126–1130. [Google Scholar] [CrossRef]
- Zaker, M.; Mosallanejad, H. Antifungal Activity of Some Plant Extracts on Alternaria Alternata, the Causal Agent of Alternaria Leaf Spot of Potato. Pakistan J. Biol. Sci. PJBS 2010, 13, 1023–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Chen, S.; Zhang, Y.; Wang, Y.; Zhang, X.; Bi, Z.; Yuan, H. Chemical Composition and Antifungal Activity of Essential Oils from Three Artemisia Species Against Alternaria Solani. J. Essent. Oil Bear. Plants 2019, 22, 1581–1592. [Google Scholar] [CrossRef]
- Dethoup, T.; Songkumarn, P.; Rueangrit, S.; Suesa-ard, S.; Kaewkrajay, C. Fungicidal Activity of Thai Medicinal Plant Extracts against Alternaria Brassicicola Causing Black Spot of Chinese Kale. Eur. J. Plant Pathol. 2018, 152, 157–167. [Google Scholar] [CrossRef]
- Izadi, M.; Moosawi Jorf, S.A.; Nikkhah, M.; Moradi, S. Antifungal Activity of Hydrocolloid Nano Encapsulated Carum Copticum Essential Oil and Peganum Harmala Extract on the Pathogenic Fungi Alternaria Alternata. Physiol. Mol. Plant Pathol. 2021, 116, 101714. [Google Scholar] [CrossRef]
- Nguyen, M.-H.; Tran, T.-N.-M.; Vu, N.-B.-D. Antifungal Activity of Essential Oil-Encapsulated Lipid Nanoemulsions Formulations against Leaf Spot Disease on Tomato Caused by Alternaria Alternata. Arch. Phytopathol. Plant Prot. 2022, 55, 235–257. [Google Scholar] [CrossRef]
- Perveen, K.; Bokhari, N.A.; Al-Rashid, S.A.I.; Al-Humaid, L.A. Chemical Composition of Essential Oil of Ocimum Basilicum L. and Its Potential in Managing the Alternaria Rot of Tomato. J. Essent. Oil Bear. Plants 2020, 23, 1428–1437. [Google Scholar] [CrossRef]
- Guo, Q.; Du, G.; Jia, H.; Fan, Q.; Wang, Z.; Gao, Z.; Yue, T.; Yuan, Y. Essential Oils Encapsulated by Biopolymers as Antimicrobials in Fruits and Vegetables: A Review. Food Biosci. 2021, 44, 101367. [Google Scholar] [CrossRef]
- Dhumal, C.V.; Sarkar, P. Composite Edible Films and Coatings from Food-Grade Biopolymers. J. Food Sci. Technol. 2018, 55, 4369–4383. [Google Scholar] [CrossRef]
- Piekarska, K. The Use of Carbohydrate Biopolymers in Plant Protection against Pathogenic Fungi. Polymers 2022, 14, 2854. [Google Scholar]
- Chakraborty, M.; Hasanuzzaman, M.; Rahman, M.; Khan, M.A.; Bhowmik, P.; Mahmud, N.U.; Tanveer, M.; Islam, T. Mechanism of Plant Growth Promotion and Disease Suppression by Chitosan Biopolymer. Agriculture 2020, 10, 624. [Google Scholar] [CrossRef]
- Abdolahi, A.L.I.; Hassani, A.; Ghosta, Y.; Javadi, T.; Meshkatalsadat, M.H. Essential oils as control agents of postaharvest alternaria and penicillium rots on tomato fruits. J. Food Saf. 2010, 30, 341–352. [Google Scholar] [CrossRef]
- Moghaddam, M.; Taheri, P.; Pirbalouti, A.G.; Mehdizadeh, L. Chemical Composition and Antifungal Activity of Essential Oil from the Seed of Echinophora Platyloba DC. against Phytopathogens Fungi by Two Different Screening Methods. LWT Food Sci. Technol. 2015, 61, 536–542. [Google Scholar] [CrossRef]
- Puškárová, A.; Bučková, M.; Kraková, L.; Pangallo, D.; Kozics, K. The Antibacterial and Antifungal Activity of Six Essential Oils and Their Cyto/Genotoxicity to Human HEL 12469 Cells. Sci. Rep. 2017, 7, 8211. [Google Scholar] [CrossRef] [PubMed]
- Sapper, M.; Wilcaso, P.; Santamarina, M.P.; Roselló, J.; Chiralt, A. Antifungal and Functional Properties of Starch-Gellan Films Containing Thyme (Thymus Zygis) Essential Oil. Food Control 2018, 92, 505–515. [Google Scholar] [CrossRef]
- Xu, S.; Yan, F.; Ni, Z.; Chen, Q.; Zhang, H.; Zheng, X. In Vitro and in Vivo Control of Alternaria Alternata in Cherry Tomato by Essential Oil from Laurus Nobilis of Chinese Origin. J. Sci. Food Agric. 2014, 94, 1403–1408. [Google Scholar] [CrossRef]
- Chen, Q.; Xu, S.; Wu, T.; Guo, J.; Sha, S.; Zheng, X.; Yu, T. Effect of Citronella Essential Oil on the Inhibition of Postharvest Alternaria Alternata in Cherry Tomato. J. Sci. Food Agric. 2014, 94, 2441–2447. [Google Scholar] [CrossRef]
- Soylu, E.M.; Kose, F. Antifungal Activities of Essential Oils Against Citrus Black Rot Disease Agent Alternaria Alternata. J. Essent. Oil Bear. Plants 2015, 18, 894–903. [Google Scholar] [CrossRef]
- Slathia, S.; Sharma, Y.P.; Hakla, H.R.; Urfan, M.; Yadav, N.S.; Pal, S. Post-Harvest Management of Alternaria Induced Rot in Tomato Fruits With Essential Oil of Zanthoxylum Armatum DC. Front. Sustain. Food Syst. 2021, 5, 679830. [Google Scholar] [CrossRef]
- Fraternale, D.; Flamini, G.; Ricci, D. Essential Oil Composition of Angelica archangelica L. (Apiaceae) Roots and Its Antifungal Activity against Plant Pathogenic Fungi. Plant Biosyst. 2016, 150, 558–563. [Google Scholar] [CrossRef]
- Amri, I.; Gargouri, S.; Hamrouni, L.; Hanana, M.; Fezzani, T.; Jamoussi, B. Chemical Composition, Phytotoxic and Antifungal Activities of Pinus Pinea Essential Oil. J. Pest Sci. 2012, 85, 199–207. [Google Scholar] [CrossRef]
- Znini, M.; Cristofari, G.; Majidi, L.; Paolini, J.; Desjobert, J.M.; Costa, J. Essential Oil Composition and Antifungal Activity of Pulicaria Mauritanica Coss., against Postharvest Phytopathogenic Fungi in Apples. LWT Food Sci. Technol. 2013, 54, 564–569. [Google Scholar] [CrossRef]
- Znini, M.; Cristofari, G.; Majidi, L.; El Harrak, A.; Paolini, J.; Costa, J. In Vitro Antifungal Activity and Chemical Composition of Warionia Saharae Essential Oil against 3 Apple Phytopathogenic Fungi. Food Sci. Biotechnol. 2013, 22, 113–119. [Google Scholar] [CrossRef]
- McDaniel, A.; Tonyali, B.; Yucel, U.; Trinetta, V. Formulation and Development of Lipid Nanoparticle Antifungal Packaging Films to Control Postharvest Disease. J. Agric. Food Res. 2019, 1, 100013. [Google Scholar] [CrossRef]
- Dan, Y.; Liu, H.-Y.; Gao, W.-W.; Chen, S.-L. Activities of Essential Oils from Asarum Heterotropoides Var. Mandshuricum against Five Phytopathogens. Crop Prot. 2010, 29, 295–299. [Google Scholar] [CrossRef]
- Marco, A.; Santos, S.; Caetano, J.; Pintado, M.; Vieira, E.; Moreira, P.R. Basil Essential Oil as an Alternative to Commercial Biocides against Fungi Associated with Black Stains in Mural Painting. Build. Environ. 2020, 167, 106459. [Google Scholar] [CrossRef]
- Ho, W.-C.; Wu, T.-Y.; Su, H.-J.; Ko, W.-H. Effect of Oriental Medicinal Plant Extracts on Spore Germination of Alternaria Brassicicola and Nature of Inhibitory Substances from Speedweed. Plant Dis. 2007, 91, 1621–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawar, V.C.; Thaker, V.S. Evaluation of the Anti-Fusarium Oxysporum f. Sp Cicer and Anti-Alternaria Porri Effects of Some Essential Oils. World J. Microbiol. Biotechnol. 2007, 23, 1099–1106. [Google Scholar] [CrossRef]
- Plotto, A.; Roberts, D.D.; Roberts, R.G. Evaluation of plant essential oils as natural postharvest disease control of tomato (Lycopersicon esculentum). In Proceedings of the Acta Horticulturae, International Society for Horticultural Science (ISHS), Leuven, Belgium, December 2003; pp. 737–745. [Google Scholar]
- Saharan, V.; Mehrotra, A.; Khatik, R.; Rawal, P.; Sharma, S.S.; Pal, A. Synthesis of Chitosan Based Nanoparticles and Their in Vitro Evaluation against Phytopathogenic Fungi. Int. J. Biol. Macromol. 2013, 62, 677–683. [Google Scholar] [CrossRef]
- González-Saucedo, A.; Barrera-Necha, L.L.; Ventura-Aguilar, R.I.; Correa-Pacheco, Z.N.; Bautista-Baños, S.; Hernández-López, M. Extension of the Postharvest Quality of Bell Pepper by Applying Nanostructured Coatings of Chitosan with Byrsonima Crassifolia Extract (L.) Kunth. Postharvest Biol. Technol. 2019, 149, 74–82. [Google Scholar] [CrossRef]
- Feliziani, E.; Santini, M.; Landi, L.; Romanazzi, G. Pre- and Postharvest Treatment with Alternatives to Synthetic Fungicides to Control Postharvest Decay of Sweet Cherry. Postharvest Biol. Technol. 2013, 78, 133–138. [Google Scholar] [CrossRef]
- Zhang, C.; Gong, H.; Liu, Y. Effects of Postharvest Coating Using Chitosan Combined with Natamycin on Physicochemical and Microbial Properties of Sweet Cherry during Cold Storage. Int. J. Biol. Macromol. 2022, 214, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hernández, A.; Ruiz-Moyano, S.; Galván, A.I.; Merchán, A.V.; Pérez Nevado, F.; Aranda, E.; Serradilla, M.J.; Córdoba, M.d.G.; Martín, A. Anti-Fungal Activity of Phenolic Sweet Orange Peel Extract for Controlling Fungi Responsible for Post-Harvest Fruit Decay. Fungal Biol. 2021, 125, 143–152. [Google Scholar] [CrossRef]
- Abbas, E.; Osman, A.; Sitohy, M. Biochemical Control of Alternaria Tenuissima Infecting Post-Harvest Fig Fruit by Chickpea Vicilin. J. Sci. Food Agric. 2020, 100, 2889–2897. [Google Scholar] [CrossRef] [PubMed]
- Panth, M.; Hassler, S.C.; Baysal-Gurel, F. Methods for Management of Soilborne Diseases in Crop Production. Agriculture 2020, 10, 16. [Google Scholar] [CrossRef] [Green Version]
- Kong, G.A.; Kochman, J.K.; Brown, J.F. Phylloplane Bacteria Antagonistic to the Sunflower Pathogen Alternaria Helianthi. Australas. Plant Pathol. 1997, 26, 85–97. [Google Scholar] [CrossRef]
- Kurniawan, O.; Wilson, K.; Mohamed, R.; Avis, T.J. Bacillus and Pseudomonas Spp. Provide Antifungal Activity against Gray Mold and Alternaria Rot on Blueberry Fruit. Biol. Control 2018, 126, 136–141. [Google Scholar] [CrossRef]
- Komhorm, A.; Thongmee, S.; Thammakun, T.; Oiuphisittraiwat, T.; Jantasorn, A. In Vivo Testing of Antagonistic Fungi against Alternaria Brassicicola Causing Chinese Kale Black Spot Disease. J. Plant Dis. Prot. 2021, 128, 183–189. [Google Scholar] [CrossRef]
- Attia, M.S.; El-Sayyad, G.S.; Abd Elkodous, M.; El-Batal, A.I. The Effective Antagonistic Potential of Plant Growth-Promoting Rhizobacteria against Alternaria Solani-Causing Early Blight Disease in Tomato Plant. Sci. Hortic. 2020, 266, 109289. [Google Scholar] [CrossRef]
- Al-Maawali, S.S.; Al-Sadi, A.M.; Ali Khalifa Alsheriqi, S.; Nasser Al-Sabahi, J.; Velazhahan, R. The Potential of Antagonistic Yeasts and Bacteria from Tomato Phyllosphere and Fructoplane in the Control of Alternaria Fruit Rot of Tomato. All Life 2021, 14, 34–48. [Google Scholar] [CrossRef]
- Sharma, R.; Sindhu, S.; Sindhu, S.S. Suppression of Alternaria Blight Disease and Plant Growth Promotion of Mustard (Brassica Juncea L.) by Antagonistic Rhizosphere Bacteria. Appl. Soil Ecol. 2018, 129, 145–150. [Google Scholar] [CrossRef]
- Li, Z.; Guo, B.K.; Wan, K.; Cong, M.; Huang, H.; Ge, Y.Y. Effects of Bacteria-Free Filtrate from Bacillus Megaterium Strain L2 on the Mycelium Growth and Spore Germination of Alternaria Alternata. Biotechnol. Biotechnol. Equip. 2015, 29, 1062–1068. [Google Scholar] [CrossRef] [Green Version]
- Trivedi, P.; Pandey, A.; Palni, L.M.S. In Vitro Evaluation of Antagonistic Properties of Pseudomonas Corrugata. Microbiol. Res. 2008, 163, 329–336. [Google Scholar] [CrossRef] [PubMed]
- Atay, M.; Kara, M.; Uysal, A.; Soylu, S.; Kurt, S.; Soylu, E.M. In Vitro Antifungal Activities of Endophytic Bacterial Isolates against Postharvest Heart Rot Disease Agent Alternaria Alternata in Pomegranate Fruits. Acta Hortic. 2020, 1289, 309–313. [Google Scholar] [CrossRef]
- Saeed, I.; Khan, S.H.; Rasheed, A.; Jahangir, M.M.; Jabbar, A.; Shaheen, H.M.F.; Din, W.U.; Mazhar, K. Assessment of Antagonistic Potential of Bacteria As Biocontrol Agent Against Alternaria Leaf Spot of Turnip. Pak. J. Phytopathol. 2021, 33, 401–409. [Google Scholar] [CrossRef]
- Leng, J.; Dai, Y.; Qiu, D.; Zou, Y.; Wu, X. Utilization of the Antagonistic Yeast, Wickerhamomyces Anomalus, Combined with UV-C to Manage Postharvest Rot of Potato Tubers Caused by Alternaria Tenuissima. Int. J. Food Microbiol. 2022, 377, 109782. [Google Scholar] [CrossRef]
- Li, W.; Long, Y.; Mo, F.; Shu, R.; Yin, X.; Wu, X.; Zhang, R.; Zhang, Z.; He, L.; Chen, T.; et al. Antifungal Activity and Biocontrol Mechanism of Fusicolla Violacea J-1 against Soft Rot in Kiwifruit Caused by Alternaria Alternata. J. Fungi 2021, 7, 937. [Google Scholar] [CrossRef]
- Müller, T.; Ruppel, S.; Behrendt, U.; Lentzsch, P.; Müller, M.E.H. Antagonistic Potential of Fluorescent Pseudomonads Colonizing Wheat Heads against Mycotoxin Producing Alternaria and Fusaria. Front. Microbiol. 2018, 9, 2124. [Google Scholar] [CrossRef]
- Yin, W.; Keller, N.P. Transcriptional Regulatory Elements in Fungal Secondary Metabolism. J. Microbiol. 2011, 49, 329–339. [Google Scholar] [CrossRef] [Green Version]
- Tsuge, T.; Harimoto, Y.; Hanada, K.; Akagi, Y.; Kodama, M.; Akimitsu, K.; Yamamoto, M. Evolution of Pathogenicity Controlled by Small, Dispensable Chromosomes in Alternaria Alternata Pathogens. Physiol. Mol. Plant Pathol. 2016, 95, 27–31. [Google Scholar] [CrossRef]
- Liu, R.; Chen, L.; Jiang, Y.; Zhou, Z.; Zou, G. Efficient Genome Editing in Filamentous Fungus Trichoderma Reesei Using the CRISPR/Cas9 System. Cell Discov. 2015, 1, 15007. [Google Scholar] [CrossRef] [Green Version]
- Nødvig, C.S.; Nielsen, J.B.; Kogle, M.E.; Mortensen, U.H. A CRISPR-Cas9 System for Genetic Engineering of Filamentous Fungi. PLoS ONE 2015, 10, e0133085. [Google Scholar] [CrossRef] [Green Version]
- Wenderoth, M.; Pinecker, C.; Voß, B.; Fischer, R. Establishment of CRISPR/Cas9 in Alternaria Alternata. Fungal Genet. Biol. 2017, 101, 55–60. [Google Scholar] [CrossRef]
- Redman, M.; King, A.; Watson, C.; King, D. What Is CRISPR/Cas9? Arch. Dis. Child. Educ. Pract. Ed. 2016, 101, 213–215. [Google Scholar] [CrossRef]
- Roux, I.; Woodcraft, C.; Hu, J.; Wolters, R.; Gilchrist, C.L.M.; Chooi, Y.-H. CRISPR-Mediated Activation of Biosynthetic Gene Clusters for Bioactive Molecule Discovery in Filamentous Fungi. ACS Synth. Biol. 2020, 9, 1843–1854. [Google Scholar] [CrossRef] [PubMed]
- Zou, G.; Xiao, M.; Chai, S.; Zhu, Z.; Wang, Y.; Zhou, Z. Efficient Genome Editing in Filamentous Fungi via an Improved CRISPR-Cas9 Ribonucleoprotein Method Facilitated by Chemical Reagents. Microb. Biotechnol. 2021, 14, 2343–2355. [Google Scholar] [CrossRef]
- Rozhkova, A.M.; Kislitsin, V.Y. CRISPR/Cas Genome Editing in Filamentous Fungi. Biochem. 2021, 86, S120–S139. [Google Scholar] [CrossRef]
- Wang, Q.; Coleman, J.J. Progress and Challenges: Development and Implementation of CRISPR/Cas9 Technology in Filamentous Fungi. Comput. Struct. Biotechnol. J. 2019, 17, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.-Z.; Zhu, Y.-L.; Huang, P.-W.; Yang, Q.; Dai, C.-C. Strategies for Gene Disruption and Expression in Filamentous Fungi. Appl. Microbiol. Biotechnol. 2019, 103, 6041–6059. [Google Scholar] [CrossRef] [PubMed]
- Eid, A.; Alshareef, S.; Mahfouz, M.M. CRISPR Base Editors: Genome Editing without Double-Stranded Breaks. Biochem. J. 2018, 475, 1955–1964. [Google Scholar] [CrossRef]
- Terns, M.P. CRISPR-Based Technologies: Impact of RNA-Targeting Systems. Mol. Cell 2018, 72, 404–412. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimaraes, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C.; et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159, 647–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brakhage, A.A.; Schroeckh, V. Fungal Secondary Metabolites—Strategies to Activate Silent Gene Clusters. Fungal Genet. Biol. 2011, 48, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Coleman, J.J. CRISPR/Cas9-Mediated Endogenous Gene Tagging in Fusarium Oxysporum. Fungal Genet. Biol. 2019, 126, 17–24. [Google Scholar] [CrossRef]
- Yamato, T.; Handa, A.; Arazoe, T.; Kuroki, M.; Nozaka, A.; Kamakura, T.; Ohsato, S.; Arie, T.; Kuwata, S. Single Crossover-Mediated Targeted Nucleotide Substitution and Knock-in Strategies with CRISPR/Cas9 System in the Rice Blast Fungus. Sci. Rep. 2019, 9, 7427. [Google Scholar] [CrossRef] [Green Version]
- Dort, E.N.; Tanguay, P.; Hamelin, R.C. CRISPR/Cas9 Gene Editing: An Unexplored Frontier for Forest Pathology. Front. Plant Sci. 2020, 11, 1126. [Google Scholar] [CrossRef]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.M.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and Classification of the CRISPR-Cas Systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Xia, X.; Cheng, X.; Li, R.; Yao, J.; Li, Z.; Cheng, Y. Advances in Application of Genome Editing in Tomato and Recent Development of Genome Editing Technology. Theor. Appl. Genet. 2021, 134, 2727–2747. [Google Scholar] [CrossRef]
- Jiang, C.; Lv, G.; Tu, Y.; Cheng, X.; Duan, Y.; Zeng, B.; He, B. Applications of CRISPR/Cas9 in the Synthesis of Secondary Metabolites in Filamentous Fungi. Front. Microbiol. 2021, 12, 638096. [Google Scholar] [CrossRef]
- Nowara, D.; Gay, A.; Lacomme, C.; Shaw, J.; Ridout, C.; Douchkov, D.; Hensel, G.; Kumlehn, J.; Schweizer, P. HIGS: Host-Induced Gene Silencing in the Obligate Biotrophic Fungal Pathogen Blumeria Graminis. Plant Cell 2010, 22, 3130–3141. [Google Scholar] [CrossRef] [Green Version]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A.; et al. An RNAi-Based Control of Fusarium Graminearum Infections Through Spraying of Long DsRNAs Involves a Plant Passage and Is Controlled by the Fungal Silencing Machinery. PLoS Pathog. 2016, 12, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Sang, H.; Kim, J.-I. Advanced Strategies to Control Plant Pathogenic Fungi by Host-Induced Gene Silencing (HIGS) and Spray-Induced Gene Silencing (SIGS). Plant Biotechnol. Rep. 2020, 14, 1–8. [Google Scholar] [CrossRef]
- NUNES, C.C.; DEAN, R.A. Host-Induced Gene Silencing: A Tool for Understanding Fungal Host Interaction and for Developing Novel Disease Control Strategies. Mol. Plant Pathol. 2012, 13, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; Kumar, N.; Weber, L.; Keller, H.; Imani, J.; Kogel, K.-H. Host-Induced Gene Silencing of Cytochrome P450 Lanosterol C14α-Demethylase–Encoding Genes Confers Strong Resistance to Fusarium Species. Proc. Natl. Acad. Sci. USA 2013, 110, 19324–19329. [Google Scholar] [CrossRef] [Green Version]
- Andrade, C.M.; Tinoco, M.L.P.; Rieth, A.F.; Maia, F.C.O.; Aragão, F.J.L. Host-Induced Gene Silencing in the Necrotrophic Fungal Pathogen Sclerotinia Sclerotiorum. Plant Pathol. 2016, 65, 626–632. [Google Scholar] [CrossRef] [Green Version]
- Elbashir, S.M.; Lendeckel, W.; Tuschl, T. RNA Interference Is Mediated by 21- and 22-Nucleotide RNAs. Genes Dev. 2001, 15, 188–200. [Google Scholar] [CrossRef] [Green Version]
- Tijsterman, M.; Plasterk, R.H.A. Dicers at RISC: The Mechanism of RNAi. Cell 2004, 117, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Robinson, R. RNAi Therapeutics: How Likely, How Soon? PLoS Biol. 2004, 2, 18–21. [Google Scholar] [CrossRef] [Green Version]
- Thakur, O.; Prasad, R. Engineering Resistance to Alternaria Cyamopsidis by RNAi Mediated Gene Silencing of Chitin Synthase Export Chaperone CHS7 in Guar. Physiol. Mol. Plant Pathol. 2020, 112, 101541. [Google Scholar] [CrossRef]
- Zhai, X.; Kong, Q.; An, P.; Ren, X. The Function and Mechanism of Pathogenesis-Related 5 Protein Resistance in Cherry Tomato in Response to Alternaria Alternata. Food Biotechnol. 2018, 32, 178–190. [Google Scholar] [CrossRef]
- Zhang, Q.; Xu, C.; Wei, H.; Fan, W.; Li, T. Two Pathogenesis-Related Proteins Interact with Leucine-Rich Repeat Proteins to Promote Alternaria Leaf Spot Resistance in Apple. Hortic. Res. 2021, 8, 219. [Google Scholar] [CrossRef] [PubMed]
- Meng, D.; Li, C.; Park, H.-J.; González, J.; Wang, J.; Dandekar, A.M.; Turgeon, B.G.; Cheng, L. Sorbitol Modulates Resistance to Alternaria Alternata by Regulating the Expression of an NLR Resistance Gene in Apple. Plant Cell 2018, 30, 1562–1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghag, S.B. Chapter 24—RNAi Strategy for Management of Phytopathogenic Fungi. In Nanobiotechnology for Plant Protection; Abd-Elsalam, K.A., Lim, K.-T.B.T.-C., Rna, .S., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 535–550. ISBN 978-0-12-821910-2. [Google Scholar]
- Song, N.; Ma, L.; Wang, W.; Sun, H.; Wang, L.; Baldwin, I.T.; Wu, J. An ERF2-like Transcription Factor Regulates Production of the Defense Sesquiterpene Capsidiol upon Alternaria Alternata Infection. J. Exp. Bot. 2019, 70, 5895–5908. [Google Scholar] [CrossRef] [PubMed]
- John, E.; Singh, K.B.; Oliver, R.P.; Tan, K.-C. Transcription Factor Control of Virulence in Phytopathogenic Fungi. Mol. Plant Pathol. 2021, 22, 858–881. [Google Scholar] [CrossRef]
- Park, S.-Y.; Choi, J.; Lim, S.-E.; Lee, G.-W.; Park, J.; Kim, Y.; Kong, S.; Kim, S.R.; Rho, H.-S.; Jeon, J.; et al. Global Expression Profiling of Transcription Factor Genes Provides New Insights into Pathogenicity and Stress Responses in the Rice Blast Fungus. PLoS Pathog. 2013, 9, e1003350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Estrada, C.; Domínguez-Santos, R.; Kosalková, K.; Martín, J.-F. Transcription Factors Controlling Primary and Secondary Metabolism in Filamentous Fungi: The β-Lactam Paradigm. Fermentation 2018, 4, 47. [Google Scholar] [CrossRef] [Green Version]
- Shelest, E. Transcription Factors in Fungi: TFome Dynamics, Three Major Families, and Dual-Specificity TFs. Front. Genet. 2017, 8, 53. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Cao, Y.; Gai, Y.; Ma, H.; Zhu, Z.; Chung, K.-R.; Li, H. Genome-Wide Identification and Functional Characterization of GATA Transcription Factor Gene Family in Alternaria Alternata. J. Fungi 2021, 7, 1013. [Google Scholar] [CrossRef]
- Gai, Y.; Li, L.; Liu, B.; Ma, H.; Chen, Y.; Zheng, F.; Sun, X.; Wang, M.; Jiao, C.; Li, H. Distinct and Essential Roles of BZIP Transcription Factors in the Stress Response and Pathogenesis in Alternaria Alternata. Microbiol. Res. 2022, 256, 126915. [Google Scholar] [CrossRef]
- Yang, Y.; Xie, P.; Yuan, J.; Liu, Y.; Zhang, M.; Li, Y.; Bi, Y.; Prusky, D.B. The Calcineurin-Responsive Transcription Factor Crz1 Is Required for Regulation of Infection Structure Differentiation, Calcium Homeostasis and Cell Wall Integrity in Alternaria Alternata. Postharvest Biol. Technol. 2022, 194, 112064. [Google Scholar] [CrossRef]
- John, E.; Singh, K.B.; Oliver, R.P.; Tan, K.-C. Transcription Factor Lineages in Plant-Pathogenic Fungi, Connecting Diversity with Fungal Virulence. Fungal Genet. Biol. 2022, 161, 103712. [Google Scholar] [CrossRef] [PubMed]



| Species | Hosts | Disease | Ref. |
|---|---|---|---|
| A. alternata (Fr) Keissl | Potato (Solanum tuberosum) | Leaf spots | [27,28] |
| Tomato (Lycopersicum esculentum) | |||
| Pepper (Capsicum annuum) | |||
| Eggplant (Solanum melongena) | |||
| Pomegranate (Punica granatum) | |||
| A. brassicae (Berk) Sacc. | Oilseeds | Black spots | [29] |
| Vegetables | |||
| Canola (Brassica napus) | |||
| Mustard (Brassica juncea) | |||
| A. brassicicola (Schwein) Wiltshire | Cabbage varieties | Black spots | [30] |
| Mustard (B. juncea) | |||
| Potato (Solanum tuberosum) | |||
| Carrot (Daucus carota L.) | |||
| Tomato (Lycopersicum esculentum) | |||
| A. dauci (Kuhn) J. W. Groves y Akolko | Carrot (Daucus carota L.) | Round or oval white spots with brown edges 2–4mm in diameter (disease known as Alternaria leaf spot) | [31] |
| Lettuce (Lactuca sativa) | |||
| Celery (Apium graveolens) | |||
| Spinach (Spinacea oleracea) | |||
| A. infectoria | Rice (Oryza sativa) | Necrotic leaf lesions | [32] |
| Cereal varieties | |||
| A. solani (Ellis y G. Martin) L. R. Jones | Potato (Solanum tuberosum) | Necrotic leaf spots | [33] |
| A. porri (Ellis) Ciffer | Onion (Allium cepa) | Purple and black lesions | [34] |
| A. tenuissima (Nees y T. Nees: Fr) Wiltshire | Kiwifruit (Actinidia spp.) | Circular brown necrotic lesions up to 8 mm | [35,36,37] |
| Wheat (Triticum) | |||
| Apples (Malus domestica) | |||
| Tomatoes (Lycopersicum esculentum) | |||
| Blueberries (Vaccinium myrtillus) | |||
| Pomegranate (Punica granatum) |
| Phytopathogenic Fungus | Disease Caused | Host/Type of Study | Source Of Essential Oil | Polymer Matrix * | Ref. |
|---|---|---|---|---|---|
| A. alternata | Leaf spot—Alternaria rot | Berberis vulgaris; Lycopersicum esculentum; in vivo and in vitro | Carum carvi L., Thimus vulgaris L., Pimpinella anisum, Chamaemelum nobile, Origanum majorana | - | [79] |
| In vivo and in vitro | Trachyspermum copticum, Foeniculum vulgare L., and Carum carvi | - | [91] | ||
| In vitro | Echinophora platyloba | - | [92] | ||
| - | Origanum vulgare L., Thimus vulgaris L. and Syzygium aromaticum | - | [93] | ||
| In vitro | Thymus zygiis | - | [94] | ||
| - | |||||
| Cherry tomato/in vitro and in vivo | Laurus nobilis L. | - | [95] | ||
| Cherry tomato/in vitro and in vivo | Cymbopogon nardus | - | [96] | ||
| Origanum onites L., Thymbra spicata L., Lavandula stoechas L., subsp. Stoechas L., F. vulgare Mill, Laurus nobilis L. | - | [97] | |||
| Zanthoxylum armatum DC | - | [98] | |||
| A. solani | Early blight | In vivo and in vitro | Angelica archangelica | - | [99] |
| Alternaria spp. | Alternariose—black leaf spot | Malus domestica in vivo and in vitro | Pinus pinea | [100] | |
| Pulicaria mauritanica | [101] | ||||
| Warionia saharae | [102] | ||||
| Cocos nucifera and hydrogenated Trachycarpus fortunei oil | [103] | ||||
| A. humicola | Alternariose | In vivo and in vitro | Asarum heterotropoides | - | [104] |
| A. tenuissima | Early blight | In vivo and in vitro | F. vulgari—Ocimum basilicum—Citrus lemmon—Rosmarinus officinalis—Salvia officinalis | - | [105] |
| A. brassicicola | Black leaf spot | In vivo and in vitro | Polygonum perfoliatum | - | [106] |
| A. porri | Early blight | In vivo and in vitro | Cymbopocon citratus—Syzygium aromaticum—Cinnamomum aromaticum—Cinnamomum verum—Ocimum basilicum—Foeniculum vulgare, and Oenothera biennis | - | [107] |
| A. arborescens | Alternaria rot | In vivo and in vitro | O. vulgare—Thymus vulgaris—Cymbopogon citratus—Coriandrum sativum | - | [108] |
| Essential oils with Biopolymers | |||||
| A. alternata | Leaf spot | In vitro | CS **, CS-saponin NPs and CS-Cu NPs ** | [109] | |
| In vivo and in vitro | Byrsonima crassifolia extract | CS NPs | [110] | ||
| A. alternata | Alternaria rot | Prunus avium | Nettle extract—spruce extract | CS—oligosaccharides | [111] |
| Alternaria species | Alternaria rot | In vivo and in vitro | - | CS combined with natamycin | [112] |
| A. alternata | Alternaria rot—leaf spot—early blight | In vivo and in vitro | Polyphenolic extracts from orange peel | - | [113] |
| A. tenuissima | Leaf spot | In vivo and in vitro | Cicer arietinum | Chickpea vicilin | [114] |
| Pathogen | Disease | Host | Antagonist | Inhibition | Ref. |
|---|---|---|---|---|---|
| Bacteria | |||||
| A. alternata | Leaf spot | Nicotiana tabacum | Bacillus megaterium | 76% inhibition in mycelial growth after seven days | [122] |
| Pseudomonas corrugata | 58% inhibition of mycelial growth after five days | [123] | |||
| Alternaria Species | Leaf spot | Punica granatum L. | Bacillus subtilis—Bacillus pumilus—Bacillus amyloliquefaciens—Bacillus mojavensis—Bacillus vallismortis—Solibacillus silvestris—B. megaterium—Corynebacterium glutamicum—Erwinia herbicola—Pantoea dispersa—Bacillus cereus—Bacillus endophyticus | Bacillus mojavensis presented the highest efficiency in mycelial growth inhibition with 80%. Bacillus myloliquefaciens 78.9%, Bacillus vallismortis 76.7%, and B. subtilis 75.6% | [124] |
| A. brassicicola | Leaf spot | Brassica rapa | B. subtilis—Pseudomonas fluorescens—Streptomyces hydrogenans | B. subtilis generated the highest inhibition of mycelial growth (24%) after seven days, followed by Pseudomonas fluorescens with 21.7% inhibition after 15 days and Streptomyces hydrogenans with 22.2% after 12 days | [125] |
| Fungi | |||||
| A. tenuissima | Early blight | Solanum tuberosum | Wickerhamomyces anomalus | W. anomalus inhibits infection by A. tenuissima. Combined with UV, it stimulates and improves biocontrol activity against the disease. Treatment with W. anomalus–UV generates a defense response in potatoes | [126] |
| A. alternata | Stem rot | Actinidia cinensis | Fusicolla violacea (J-1) | J-1 exhibited 66.1% inhibition under in vitro conditions | [127] |
| A. alternata (CABI strain 353822), A. tenuissima (CABI strain 352931), and A. infectoria (CBS strain 120149) | Leaf spot | Oriza sativa | Pseudomonmas libanensis—Pseudomonas rhodesiae | After inoculation of Alternaria species with a total of 110 Pseudomonas isolates, a slight reduction in fungal growth was recorded, with a significant decrease in mycotoxin production | [128] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrascal-Hernández, D.C.; Flórez-López, E.; Peralta-Ruiz, Y.; Chaves-López, C.; Grande-Tovar, C.D. Eco-Friendly Biocontrol Strategies of Alternaria Phytopathogen Fungus: A Focus on Gene-Editing Techniques. Agriculture 2022, 12, 1722. https://doi.org/10.3390/agriculture12101722
Carrascal-Hernández DC, Flórez-López E, Peralta-Ruiz Y, Chaves-López C, Grande-Tovar CD. Eco-Friendly Biocontrol Strategies of Alternaria Phytopathogen Fungus: A Focus on Gene-Editing Techniques. Agriculture. 2022; 12(10):1722. https://doi.org/10.3390/agriculture12101722
Chicago/Turabian StyleCarrascal-Hernández, Domingo Cesar, Edwin Flórez-López, Yeimmy Peralta-Ruiz, Clemencia Chaves-López, and Carlos David Grande-Tovar. 2022. "Eco-Friendly Biocontrol Strategies of Alternaria Phytopathogen Fungus: A Focus on Gene-Editing Techniques" Agriculture 12, no. 10: 1722. https://doi.org/10.3390/agriculture12101722
APA StyleCarrascal-Hernández, D. C., Flórez-López, E., Peralta-Ruiz, Y., Chaves-López, C., & Grande-Tovar, C. D. (2022). Eco-Friendly Biocontrol Strategies of Alternaria Phytopathogen Fungus: A Focus on Gene-Editing Techniques. Agriculture, 12(10), 1722. https://doi.org/10.3390/agriculture12101722









