Functional Interpretation of Cross-Talking Pathways with Emphasis on Amino Acid Metabolism in Rhizosphere Microbiome of the Wild Plant Moringa oleifera
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. Whole-Genome Shotgun Sequencing and Bioinformatics Analysis
3. Results
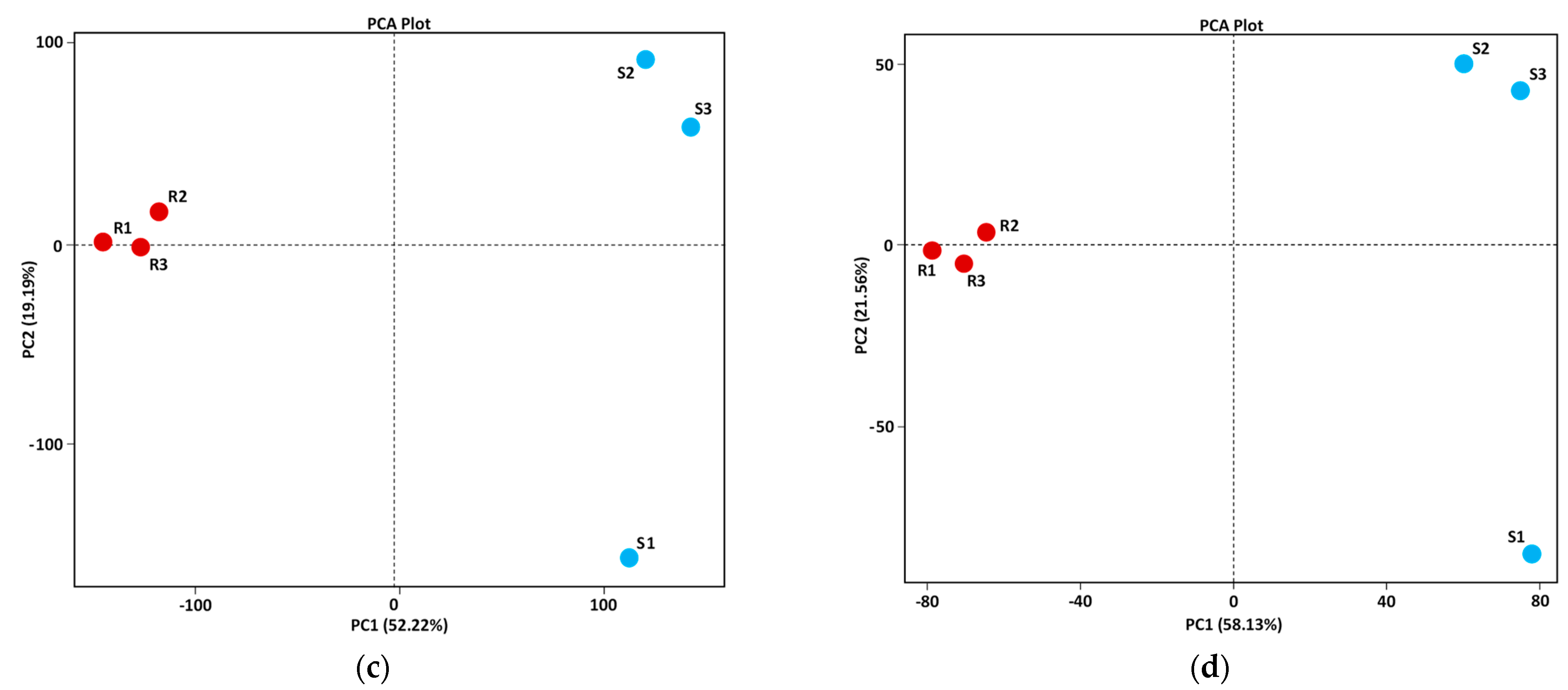
3.1. Validation and Fidelity Testing of KEGG Datasets
3.2. Description of Assembled Raw Sequence Data
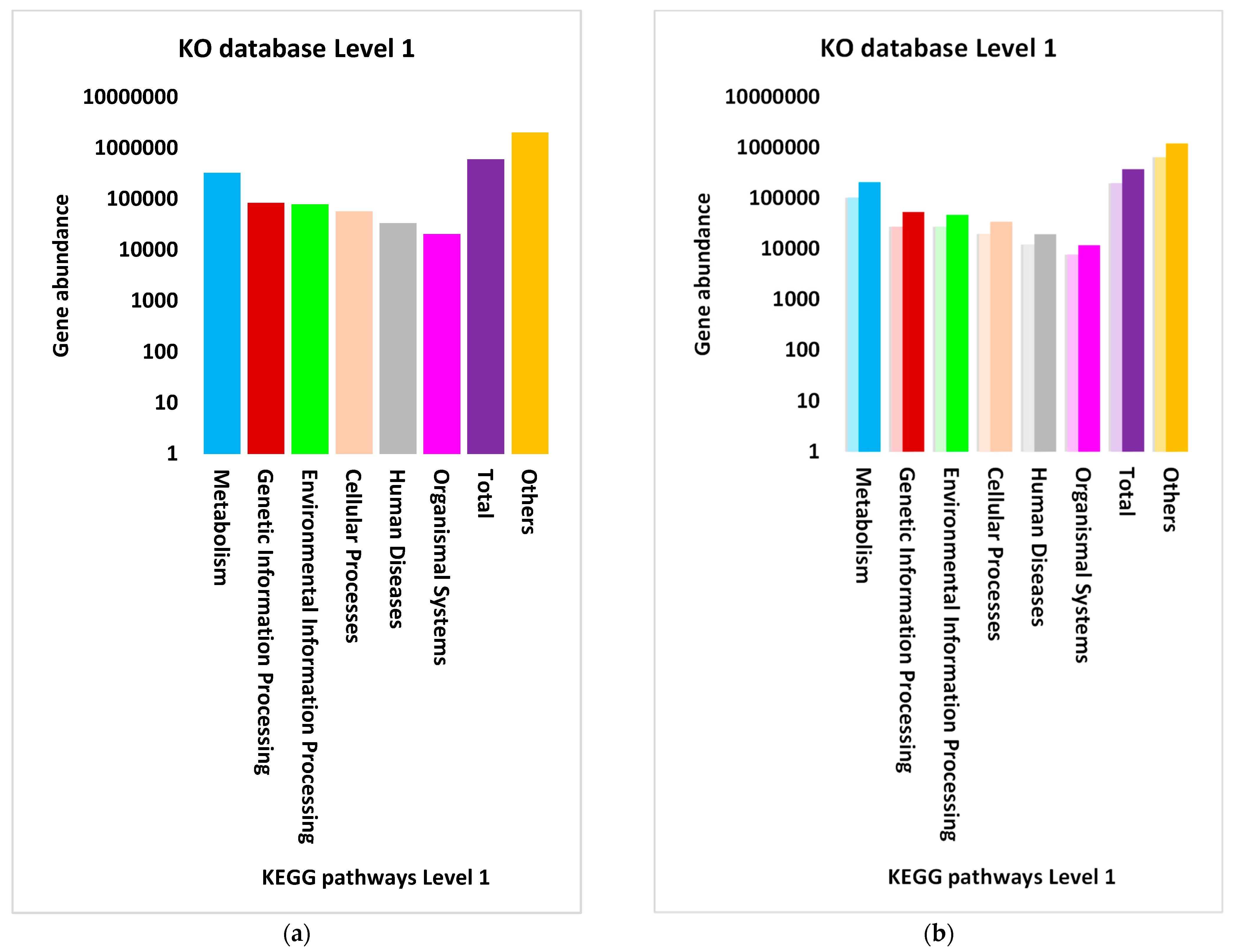
3.3. Differential Abundance of Genes Encoding KEGG Enzymes
4. Discussion
4.1. Major Contributors to Plant Growth Parameters and Defense against Phytopathogens in the Rhizosphere Microbiome of M. oleifera
4.2. Chemotaxis in the Rhizosphere Microbiome of M. oleifera
4.3. Mineral Solubilization in the Rhizosphere Microbiome of M. oleifera
4.4. Mobility and Restoration of Nutrients in the Rhizosphere Microbiome of M. oleifera
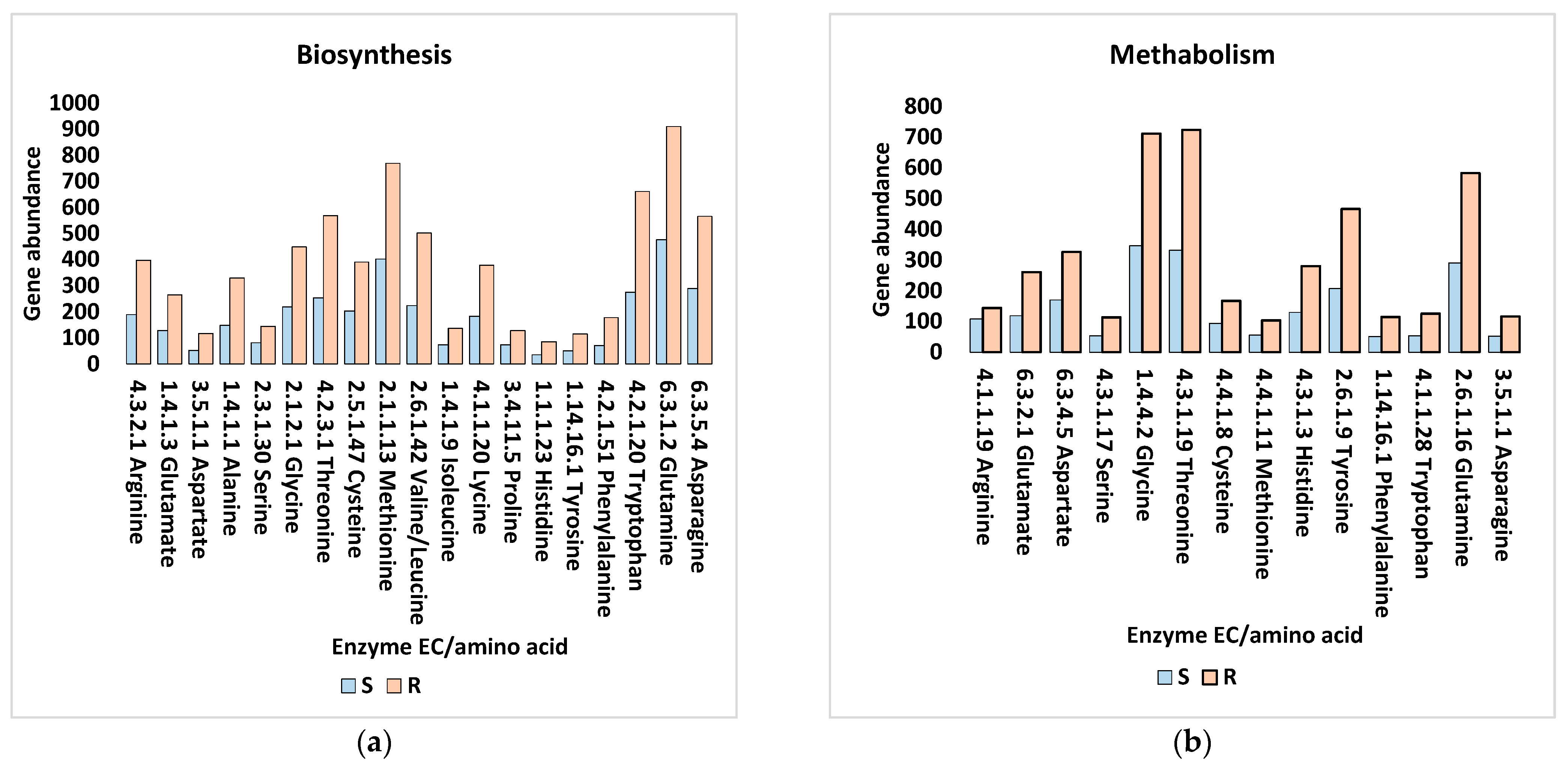
4.5. Amino Acid Transport in the Rhizosphere Microbiome of M. oleifera
4.6. Amino Acids and Biofilm Formation in the Rhizosphere Microbiome of M. oleifera
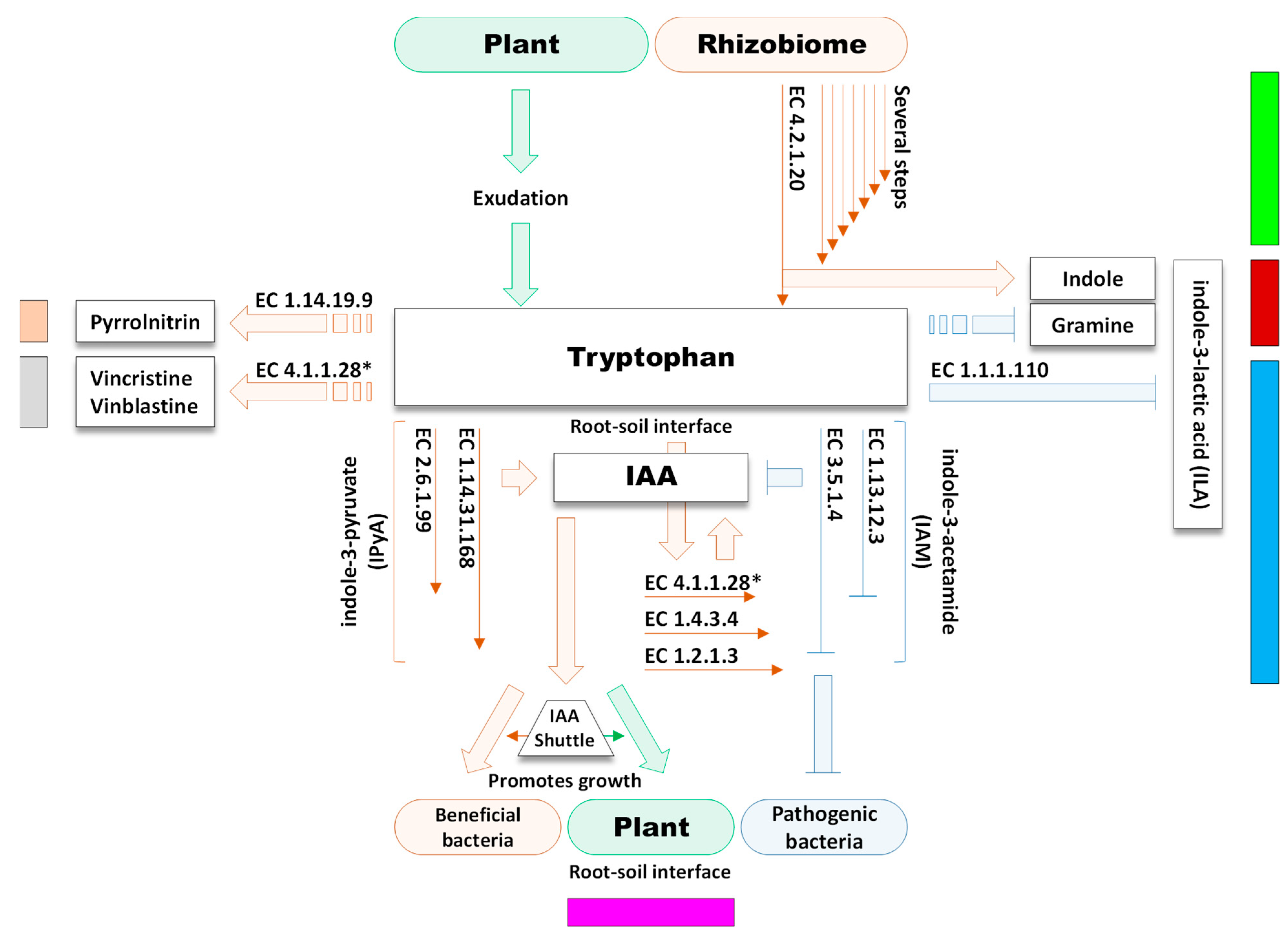
4.7. Production of Indole Acetic Acid in the Rhizosphere Microbiome of M. oleifera
4.8. Tryptophan as a Core Substrate in a Number of Reactions in the Rhizosphere Microbiome of M. oleifera
4.9. Role of Amino Acyl tRNA in the Rhizosphere Microbiome of M. oleifera
4.10. Glucose as the Ultimate Target of Carbohydrate Metabolism in M. oleifera
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Olson, M.; Flora of North America Committee. eFlora summary: Moringaceae: Drumstick family. Flora N. Am. North Mex. 2010, 7, 167–169. [Google Scholar]
- Fozia, F.; Meenu, R.; Avinash, T.; Abdul, A.K.; Shaila, F. Medicinal properties of Moringa oleifera: An overview of promising healer. J. Med. Plants Res. 2012, 6, 4368–4374. [Google Scholar]
- Al-Eisawi, D.M.; Al-Ruzayza, S. The flora of holy Mecca district, Saudi Arabia. Int. J. Biodivers. Conserv. 2015, 7, 173–189. [Google Scholar]
- Gopalakrishnan, L.; Doriya, K.; Kumar, D.S. Moringa oleifera: A review on nutritive importance and its medicinal application. Food Sci. Hum. Wellness 2016, 5, 49–56. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlton, J.M.; Angiuoli, S.V.; Suh, B.B.; Kooij, T.W.; Pertea, M.; Silva, J.C.; Ermolaeva, M.D.; Allen, J.E.; Selengut, J.D.; Koo, H.L.; et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 2002, 419, 512–519. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Zhou, Y.; Fu, H.; Xiong, X.; Fang, S.; Jiang, H.; Wu, J.; Yang, H.; Gao, J.; Huang, L. Expanded catalog of microbial genes and metagenome-assembled genomes from the pig gut microbiome. Nat. Commun. 2021, 12, 1106. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Wang, Q.; O’Sullivan, O.; Greene-Diniz, R.; Cole, J.R.; Ross, R.P.; O’Toole, P.W. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010, 38, e200. [Google Scholar] [CrossRef]
- Kennedy, N.A.; Walker, A.W.; Berry, S.H.; Duncan, S.H.; Farquarson, F.M.; Louis, P.; Thomson, J.M.; Satsangi, J.; Flint, H.J. The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS ONE 2014, 9, e88982. [Google Scholar] [CrossRef]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef] [Green Version]
- Tringe, S.G.; von Mering, C.; Kobayashi, A.; Salamov, A.A.; Chen, K.; Chang, H.W.; Podar, M.; Short, J.M.; Mathur, E.J.; Detter, J.C.; et al. Comparative metagenomics of microbial communities. Science 2005, 308, 554–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raes, J.; Foerstner, K.U.; Bork, P. Get the most out of your metagenome: Computational analysis of environmental sequence data. Curr. Opin. Microbiol. 2007, 10, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, L.G.; Ettinger, C.L.; Jospin, G.; Eisen, J.A. Metagenome-assembled genomes provide new insight into the microbial diversity of two thermal pools in Kamchatka, Russia. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.D.; Auffret, M.D.; Warr, A.; Wiser, A.H.; Press, M.O.; Langford, K.W.; Liachko, I.; Snelling, T.J.; Dewhurst, R.J.; Walker, A.W.; et al. Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat. Commun. 2018, 9, 870. [Google Scholar] [CrossRef] [Green Version]
- Dilthey, A.T.; Jain, C.; Koren, S.; Phillippy, A.M. Strain-level metagenomic assignment and compositional estimation for long reads with MetaMaps. Nat. Commun. 2019, 10, 3066. [Google Scholar] [CrossRef] [Green Version]
- Scholz, M.; Ward, D.V.; Pasolli, E.; Tolio, T.; Zolfo, M.; Asnicar, F.; Truong, D.T.; Tett, A.; Morrow, A.L.; Segata, N. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat. Methods 2016, 13, 435–438. [Google Scholar] [CrossRef]
- Sugiyama, A.; Ueda, Y.; Zushi, T.; Takase, H.; Yazaki, K. Changes in the bacterial community of soybean rhizospheres during growth in the field. PLoS ONE 2014, 9, e100709. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, A. The soybean rhizosphere: Metabolites, microbes, and beyond—A review. J. Adv. Res. 2019, 19, 67–73. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Saleem, M.; Law, A.D.; Sahib, M.R.; Pervaiz, Z.H.; Zhang, Q. Impact of root system architecture on rhizosphere and root microbiome. Rhizosphere 2018, 6, 47–51. [Google Scholar] [CrossRef]
- Huang, X.-F.; Chaparro, J.M.; Reardon, K.F.; Zhang, R.; Shen, Q.; Vivanco, J.M. Rhizosphere interactions: Root exudates, microbes, and microbial communities. Botany 2014, 92, 267–275. [Google Scholar] [CrossRef]
- Holden, N. You are what you can find to eat: Bacterial metabolism in the rhizosphere. Curr. Issues Mol. Biol. 2019, 30, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kolton, M.; Green, S.J.; Harel, Y.M.; Sela, N.; Elad, Y.; Cytryn, E. Draft genome sequence of Flavobacterium sp. strain F52, isolated from the rhizosphere of bell pepper (Capsicum annuum L. cv. Maccabi). J. Bacteriol. 2012, 194, 5462–5463. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Hulbert, S.H.; Schroeder, K.L.; Mavrodi, O.; Mavrodi, D.; Dhingra, A.; Schillinger, W.F.; Paulitz, T.C. Role of bacterial communities in the natural suppression of Rhizoctonia solani bare patch disease of wheat (Triticum aestivum L.). Appl. Environ. Microbiol. 2013, 79, 7428–7438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartman, K.; van der Heijden, M.G.; Roussely-Provent, V.; Walser, J.C.; Schlaeppi, K. Deciphering composition and function of the root microbiome of a legume plant. Microbiome 2017, 5, 2. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Jaramillo, J.E.; Carrión, V.J.; Bosse, M.; Ferrão, L.F.; De Hollander, M.; Garcia, A.A.; Ramírez, C.A.; Mendes, R.; Raaijmakers, J.M. Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J. 2017, 11, 2244–2257. [Google Scholar] [CrossRef] [Green Version]
- Schlaeppi, K.; Dombrowski, N.; Oter, R.G.; Ver Loren van Themaat, E.; Schulze-Lefert, P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc. Natl. Acad. Sci. USA 2014, 111, 585–592. [Google Scholar] [CrossRef] [Green Version]
- Zachow, C.; Müller, H.; Tilcher, R.; Berg, G. Differences between the rhizosphere microbiome of Beta vulgaris ssp. maritima—Ancestor of all beet crops—And modern sugar beets. Front. Microbiol. 2014, 5, 415. [Google Scholar] [CrossRef] [Green Version]
- Bulgarelli, D.; Garrido-Oter, R.; Munch, P.C.; Weiman, A.; Droge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef] [Green Version]
- Pett-Ridge, J.; Shi, S.; Estera-Molina, K.; Nuccio, E.; Yuan, M.; Rijkers, R.; Swenson, T.; Zhalnina, K.; Northen, T.; Zhou, J. Rhizosphere carbon turnover from cradle to grave: The role of microbe–plant interactions. In Rhizosphere Biology: Interactions between Microbes and Plants; Springer: Berlin/Heidelberg, Germany, 2021; pp. 51–73. [Google Scholar]
- Pett-Ridge, J.; Firestone, M.K. Using stable isotopes to explore root-microbe-mineral interactions in soil. Rhizosphere 2017, 3, 244–253. [Google Scholar] [CrossRef]
- Hurt, R.A.; Qiu, X.; Wu, L.; Roh, Y.; Palumbo, A.V.; Tiedje, J.M.; Zhou, J. Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 2001, 67, 4495–4503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mende, D.R.; Waller, A.S.; Sunagawa, S.; Järvelin, A.I.; Chan, M.M.; Arumugam, M.; Raes, J.; Bork, P. Assessment of metagenomic assembly using simulated next generation sequencing data. PLoS ONE 2012, 7, e31386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlsson, F.H.; Fåk, F.; Nookaew, I.; Tremaroli, V.; Fagerberg, B.; Petranovic, D.; Bäckhed, F.; Nielsen, J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 2012, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Byrd, A.L.; Deming, C.; Conlan, S.; Program, N.C.S.; Kong, H.H.; Segre, J.A. Biogeography and individuality shape function in the human skin metagenome. Nature 2014, 514, 59–64. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, H.B.; Almeida, M.; Juncker, A.S.; Rasmussen, S.; Li, J.; Sunagawa, S.; Plichta, D.R.; Gautier, L.; Pedersen, A.G.; Le Chatelier, E. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat. Biotechnol. 2014, 32, 822–828. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef]
- Huson, D.H.; Beier, S.; Flade, I.; Górska, A.; El-Hadidi, M.; Mitra, S.; Ruscheweyh, H.-J.; Tappu, R. MEGAN community edition-interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 2016, 12, e1004957. [Google Scholar] [CrossRef] [Green Version]
- Huson, D.H.; Mitra, S.; Ruscheweyh, H.-J.; Weber, N.; Schuster, S.C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011, 21, 1552–1560. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, D354–D357. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 2014, 42, D199–D205. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlsson, F.H.; Tremaroli, V.; Nookaew, I.; Bergstrom, G.; Behre, C.J.; Fagerberg, B.; Nielsen, J.; Backhed, F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013, 498, 99–103. [Google Scholar] [CrossRef]
- Bahieldin, A.; Atef, A.; Sabir, J.S.; Gadalla, N.O.; Edris, S.; Alzohairy, A.M.; Radhwan, N.A.; Baeshen, M.N.; Ramadan, A.M.; Eissa, H.F.; et al. RNA-Seq analysis of the wild barley (H. spontaneum) leaf transcriptome under salt stress. Comptes Rendus Biol. 2015, 338, 285–297. [Google Scholar] [CrossRef]
- Naik, K.; Mishra, S.; Srichandan, H.; Singh, P.K.; Sarangi, P.K. Plant growth promoting microbes: Potential link to sustainable agriculture and environment. Biocatal. Agric. Biotechnol. 2019, 21, 101326. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [Green Version]
- Devi, R.; Kaur, T.; Kour, D.; Yadav, A.; Yadav, A.N.; Suman, A.; Ahluwalia, A.S.; Saxena, A.K. Minerals solubilizing and mobilizing microbiomes: A sustainable approaches for managing minerals deficiency in agricultural soil. J. Appl. Microbiol. 2022, 133, 1245–1272. [Google Scholar] [CrossRef]
- Moe, L.A. Amino acids in the rhizosphere: From plants to microbes. Am. J. Bot. 2013, 100, 1692–1705. [Google Scholar] [CrossRef]
- Harris, S.J.; Shih, Y.L.; Bentley, S.D.; Salmond, G.P. The hexA gene of Erwinia carotovora encodes a LysR homologue and regulates motility and the expression of multiple virulence determinants. Mol. Microbiol. 1998, 28, 705–717. [Google Scholar] [CrossRef] [Green Version]
- Silby, M.W.; Cerdeno-Tarraga, A.M.; Vernikos, G.S.; Giddens, S.R.; Jackson, R.W.; Preston, G.M.; Zhang, X.X.; Moon, C.D.; Gehrig, S.M.; Godfrey, S.A.; et al. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol. 2009, 10, R51. [Google Scholar] [CrossRef] [Green Version]
- Badri, D.V.; Quintana, N.; El Kassis, E.G.; Kim, H.K.; Choi, Y.H.; Sugiyama, A.; Verpoorte, R.; Martinoia, E.; Manter, D.K.; Vivanco, J.M. An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol. 2009, 151, 2006–2017. [Google Scholar] [CrossRef] [Green Version]
- Hartmann, N.; Erbs, M.; Forrer, H.R.; Vogelgsang, S.; Wettstein, F.E.; Schwarzenbach, R.P.; Bucheli, T.D. Occurrence of zearalenone on Fusarium graminearum infected wheat and maize fields in crop organs, soil, and drainage water. Environ. Sci. Technol. 2008, 42, 5455–5460. [Google Scholar] [CrossRef]
- Jones, D.; Kemmitt, S.; Wright, D.; Cuttle, S.; Bol, R.; Edwards, A. Rapid intrinsic rates of amino acid biodegradation in soils are unaffected by agricultural management strategy. Soil Biol. Biochem. 2005, 37, 1267–1275. [Google Scholar] [CrossRef]
- Rudrappa, T.; Bais, H.P. Curcumin, a known phenolic from Curcuma longa, attenuates the virulence of Pseudomonas aeruginosa PAO1 in whole plant and animal pathogenicity models. J. Agric. Food Chem. 2008, 56, 1955–1962. [Google Scholar] [CrossRef]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2, a012427. [Google Scholar] [CrossRef]
- Torres-Cerna, C.E.; Morales, J.A.; Hernandez-Vargas, E.A. Modeling Quorum Sensing Dynamics and Interference on Escherichia coli. Front Microbiol. 2019, 10, 1835. [Google Scholar] [CrossRef] [Green Version]
- Ng, W.L.; Bassler, B.L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009, 43, 197–222. [Google Scholar] [CrossRef] [Green Version]
- Williams, P.; Camara, M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: A tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 2009, 12, 182–191. [Google Scholar] [CrossRef]
- Parsek, M.R.; Val, D.L.; Hanzelka, B.L.; Cronan, J.E., Jr.; Greenberg, E.P. Acyl homoserine-lactone quorum-sensing signal generation. Proc. Natl. Acad. Sci. USA 1999, 96, 4360–4365. [Google Scholar] [CrossRef] [Green Version]
- Perego, M.; Glaser, P.; Hoch, J.A. Aspartyl-phosphate phosphatases deactivate the response regulator components of the sporulation signal transduction system in Bacillus subtilis. Mol. Microbiol. 1996, 19, 1151–1157. [Google Scholar] [CrossRef]
- Kalamara, M.; Spacapan, M.; Mandic-Mulec, I.; Stanley-Wall, N.R. Social behaviours by Bacillus subtilis: Quorum sensing, kin discrimination and beyond. Mol. Microbiol. 2018, 110, 863–878. [Google Scholar] [CrossRef] [Green Version]
- Gallegos-Monterrosa, R.; Christensen, M.N.; Barchewitz, T.; Koppenhofer, S.; Priyadarshini, B.; Balint, B.; Maroti, G.; Kempen, P.J.; Dragos, A.; Kovacs, A.T. Impact of Rap-Phr system abundance on adaptation of Bacillus subtilis. Commun. Biol. 2021, 4, 468. [Google Scholar] [CrossRef]
- De Keersmaecker, S.C.; Sonck, K.; Vanderleyden, J. Let LuxS speak up in AI-2 signaling. Trends Microbiol. 2006, 14, 114–119. [Google Scholar] [CrossRef]
- Miller, M.B.; Bassler, B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [Green Version]
- Hayat, R.; Ali, S.; Amara, U.; Khalid, R.; Ahmed, I. Soil beneficial bacteria and their role in plant growth promotion: A review. Ann. Microbiol. 2010, 60, 579–598. [Google Scholar] [CrossRef]
- James, E. Nitrogen fixation in endophytic and associative symbiosis. Field Crops Res. 2000, 65, 197–209. [Google Scholar] [CrossRef]
- de Bruijn, F.J. The quest for biological nitrogen fixation in cereals: A perspective and prospective. Biol. Nitrogen Fixat. 2015, 1087–1101. [Google Scholar]
- Duan, B.; Li, L.; Chen, G.; Su-Zhou, C.; Li, Y.; Merkeryan, H.; Liu, W.; Liu, X. 1-Aminocyclopropane-1-Carboxylate Deaminase-Producing Plant Growth-Promoting Rhizobacteria Improve Drought Stress Tolerance in Grapevine (Vitis vinifera L.). Front. Plant Sci. 2021, 12, 706990. [Google Scholar]
- Shen, X.; Hu, H.; Peng, H.; Wang, W.; Zhang, X. Comparative genomic analysis of four representative plant growth-promoting rhizobacteria in Pseudomonas. BMC Genom. 2013, 14, 271. [Google Scholar] [CrossRef] [Green Version]
- Blake, C.; Christensen, M.N.; Kovacs, A.T. Molecular Aspects of Plant Growth Promotion and Protection by Bacillus subtilis. Mol. Plant Microbe Interact. 2021, 34, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Sun, B.; Shi, H.; Gao, T.; He, Y.; Li, Y.; Liu, Y.; Li, X.; Zhang, L.; Li, S.; et al. Sucrose triggers a novel signaling cascade promoting Bacillus subtilis rhizosphere colonization. ISME J. 2021, 15, 2723–2737. [Google Scholar] [CrossRef]
- Mishra, S.; Upadhyay, R.S.; Nautiyal, C.S. Unravelling the beneficial role of microbial contributors in reducing the allelopathic effects of weeds. Appl. Microbiol. Biotechnol. 2013, 97, 5659–5668. [Google Scholar] [CrossRef]
- Andreote, F.D.; Gumiere, T.; Durrer, A. Exploring interactions of plant microbiomes. Sci. Agrícola 2014, 71, 528–539. [Google Scholar] [CrossRef] [Green Version]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Doornbos, R.F.; van Loon, L.C.; Bakker, P.A. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron. Sustain. Dev. 2012, 32, 227–243. [Google Scholar] [CrossRef] [Green Version]
- Oku, S.; Komatsu, A.; Tajima, T.; Nakashimada, Y.; Kato, J. Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0-1 and their involvement in chemotaxis to tomato root exudate and root colonization. Microbes Environ. 2012, 27, ME12005. [Google Scholar] [CrossRef] [Green Version]
- de Weert, S.; Vermeiren, H.; Mulders, I.H.; Kuiper, I.; Hendrickx, N.; Bloemberg, G.V.; Vanderleyden, J.; De Mot, R.; Lugtenberg, B.J. Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol. Plant-Microbe Interact. 2002, 15, 1173–1180. [Google Scholar] [CrossRef] [Green Version]
- Alabouvette, C.; Couteaudier, Y. Biological control of Fusarium wilts with nonpathogenic Fusaria. Biol. Control. Plant Dis. 1992, 415–426. [Google Scholar]
- Falke, J.J.; Bass, R.B.; Butler, S.L.; Chervitz, S.A.; Danielson, M.A. The two-component signaling pathway of bacterial chemotaxis: A molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu. Rev. Cell Dev. Biol. 1997, 13, 457–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bren, A.; Eisenbach, M. How signals are heard during bacterial chemotaxis: Protein-protein interactions in sensory signal propagation. J. Bacteriol. 2000, 182, 6865–6873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourret, R.B.; Hess, J.F.; Borkovich, K.A.; Pakula, A.A.; Simon, M.I. Protein phosphorylation in chemotaxis and two-component regulatory systems of bacteria. J. Biol. Chem. 1989, 264, 7085–7088. [Google Scholar] [CrossRef]
- Bibikov, S.I.; Biran, R.; Rudd, K.E.; Parkinson, J.S. A signal transducer for aerotaxis in Escherichia coli. J. Bacteriol. 1997, 179, 4075–4079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Divjot, K.; Rana, K.L.; Tanvir, K.; Yadav, N.; Yadav, A.N.; Kumar, M.; Kumar, V.; Dhaliwal, H.S.; Saxena, A.K. Biodiversity, current developments and potential biotechnological applications of phosphorus-solubilizing and-mobilizing microbes: A review. Pedosphere 2021, 31, 43–75. [Google Scholar]
- Costacurta, A.; Vanderleyden, J. Synthesis of phytohormones by plant-associated bacteria. Crit. Rev. Microbiol. 1995, 21, 1–18. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef] [Green Version]
- Francis, I.; Holsters, M.; Vereecke, D. The Gram-positive side of plant–microbe interactions. Environ. Microbiol. 2010, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Römheld, V. Root-induced changes in the availability of nutrients in the rhizosphere. Plant Roots Hidden Half 2002, 617–649. [Google Scholar]
- Vranova, V.; Rejsek, K.; Skene, K.R.; Formanek, P. Non-protein amino acids: Plant, soil and ecosystem interactions. Plant Soil 2011, 342, 31–48. [Google Scholar] [CrossRef]
- Saier, M.H., Jr. Families of transmembrane transporters selective for amino acids and their derivatives the information presented in this review was initially prepared for presentation at the FASEB meeting on amino acid transport held in Copper Mountain, Colorado, June 26–July 1, 1999 and was updated in January 2000 following the meeting of the Transport Nomenclature Panel of the International Union of Biochemistry and Molecular Biology (IUBMB) in Geneva, November 28–30, 1999. The system of classification described in this review reflects the recommendations of that panel. Microbiology 2000, 146, 1775–1795. [Google Scholar] [PubMed] [Green Version]
- Tegeder, M. Transporters for amino acids in plant cells: Some functions and many unknowns. Curr. Opin. Plant Biol. 2012, 15, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Bao, Y.; Li, P.; Hu, H.; Ding, C.; Wang, S.; Li, T.; Qi, J.; Wang, X.; Yu, S. The putative amino acid ABC transporter substrate-binding protein AapJ2 is necessary for Brucella virulence at the early stage of infection in a mouse model. Vet. Res. 2018, 49, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Kolodkin-Gal, I.; Romero, D.; Cao, S.; Clardy, J.; Kolter, R.; Losick, R. D-amino acids trigger biofilm disassembly. Science 2010, 328, 627–629. [Google Scholar] [CrossRef] [Green Version]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef] [Green Version]
- Valle, J.; Da Re, S.; Schmid, S.; Skurnik, D.; d’Ari, R.; Ghigo, J.-M. The amino acid valine is secreted in continuous-flow bacterial biofilms. J. Bacteriol. 2008, 190, 264–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Toole, G.A.; Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: A genetic analysis. Mol. Microbiol. 1998, 28, 449–461. [Google Scholar] [CrossRef]
- Staswick, P.E. The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol. 2009, 150, 1310–1321. [Google Scholar] [CrossRef] [Green Version]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef] [Green Version]
- Teale, W.D.; Paponov, I.A.; Palme, K. Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 2006, 7, 847–859. [Google Scholar] [CrossRef] [PubMed]
- Brandl, M.T.; Lindow, S.E. Environmental signals modulate the expression of an indole-3-acetic acid biosynthetic gene in Erwinia herbicola. Mol. Plant-Microbe Interact. 1997, 10, 499–505. [Google Scholar] [CrossRef] [Green Version]
- Patten, C.L.; Glick, B.R. Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 2002, 68, 3795–3801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubeikovsky, A.; Mordukhova, E.; Kochetkov, V.t.; Polikarpova, F.; Boronin, A. Growth promotion of blackcurrant softwood cuttings by recombinant strain Pseudomonas fluorescens BSP53a synthesizing an increased amount of indole-3-acetic acid. Soil Biol. Biochem. 1993, 25, 1277–1281. [Google Scholar] [CrossRef]
- Persello-Cartieaux, F.; Nussaume, L.; Robaglia, C. Tales from the underground: Molecular plant–rhizobacteria interactions. Plant Cell Environ. 2003, 26, 189–199. [Google Scholar] [CrossRef]
- Kwan, G.; Pisithkul, T.; Amador-Noguez, D.; Barak, J. De novo amino acid biosynthesis contributes to salmonella enterica growth in Alfalfa seedling exudates. Appl. Environ. Microbiol. 2015, 81, 861–873. [Google Scholar] [CrossRef] [Green Version]
- Dobbelaere, S.; Croonenborghs, A.; Thys, A.; Vande Broek, A.; Vanderleyden, J. Phytostimulatory effect of Azospirillum brasilense wild type and mutant strains altered in IAA production on wheat. Plant Soil 1999, 212, 153–162. [Google Scholar] [CrossRef]
- Prinsen, E.; Chauvaux, N.; Schmidt, J.; John, M.; Wieneke, U.; De Greef, J.; Schell, J.; Van Onckelen, H. Stimulation of indole-3-acetic acid production in Rhizobium by flavonoids. FEBS Lett. 1991, 282, 53–55. [Google Scholar] [CrossRef]
- BAR, T.; OKON, Y. Induction of indole-3-acetic acid synthesis and possible toxicity of tryptophan in Azospirillum brasilense Sp7. Symbiosis 1992, 13, 191–198. [Google Scholar]
- Steenhoudt, O.; Vanderleyden, J. Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: Genetic, biochemical and ecological aspects. FEMS Microbiol. Rev. 2000, 24, 487–506. [Google Scholar] [CrossRef]
- Sprunck, S.; Jacobsen, H.-J.; Reinard, T. Indole-3-lactic acid is a weak auxin analogue but not an anti-auxin. J. Plant Growth Regul. 1995, 14, 191–197. [Google Scholar] [CrossRef]
- Glass, N.L.; Kosuge, T. Cloning of the gene for indoleacetic acid-lysine synthetase from Pseudomonas syringae subsp. savastanoi. J. Bacteriol. 1986, 166, 598–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, J.-C.; Huang, Y.-B. Induction and characterization of an indole-3-acetyl-L-alanine hydrolase from Arthrobacter ilicis. J. Plant Growth Regul. 2005, 24, 11–18. [Google Scholar] [CrossRef]
- Lambrecht, M.; Okon, Y.; Broek, A.V.; Vanderleyden, J. Indole-3-acetic acid: A reciprocal signalling molecule in bacteria–plant interactions. Trends Microbiol. 2000, 8, 298–300. [Google Scholar] [CrossRef]
- Kim, J.; Hong, H.; Heo, A.; Park, W. Indole toxicity involves the inhibition of adenosine triphosphate production and protein folding in Pseudomonas putida. FEMS Microbiol. Lett. 2013, 343, 89–99. [Google Scholar] [CrossRef] [Green Version]
- Ma, Q.; Zhang, X.; Qu, Y. Biodegradation and Biotransformation of Indole: Advances and Perspectives. Front Microbiol. 2018, 9, 2625. [Google Scholar] [CrossRef] [Green Version]
- Maver, M.; Escudero-Martinez, C.; Abbott, J.; Morris, J.; Hedley, P.E.; Mimmo, T.; Bulgarelli, D. Applications of the indole-alkaloid gramine modulate the assembly of individual members of the barley rhizosphere microbiota. PeerJ 2021, 9, e12498. [Google Scholar] [CrossRef]
- Burd, V.N.; Van Pee, K.H. Halogenating enzymes in the biosynthesis of antibiotics. Biochemistry 2003, 68, 1132–1135. [Google Scholar] [CrossRef]
- Alam, M.M.; Naeem, M.; Khan, M.; Uddin, M. Vincristine and vinblastine anticancer catharanthus alkaloids: Pharmacological applications and strategies for yield improvement. In Catharanthus Roseus; Springer: Berlin/Heidelberg, Germany, 2017; pp. 277–307. [Google Scholar]
- Wada, A. Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells 1998, 3, 203–208. [Google Scholar] [CrossRef]
- Crozier, L.; Hedley, P.E.; Morris, J.; Wagstaff, C.; Andrews, S.C.; Toth, I.; Jackson, R.W.; Holden, N.J. Whole-transcriptome analysis of verocytotoxigenic Escherichia coli O157: H7 (Sakai) suggests plant-species-specific metabolic responses on exposure to spinach and lettuce extracts. Front. Microbiol. 2016, 7, 1088. [Google Scholar]
- Maughan, R. Carbohydrate metabolism. Surgery 2009, 27, 6–10. [Google Scholar]
- Nakrani, M.N.; Wineland, R.H.; Anjum, F. Physiology, Glucose Metabolism; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Chaudhuri, B.; Hörmann, F.; Lalonde, S.; Brady, S.M.; Orlando, D.A.; Benfey, P.; Frommer, W.B. Protonophore-and pH-insensitive glucose and sucrose accumulation detected by FRET nanosensors in Arabidopsis root tips. Plant J. 2008, 56, 948–962. [Google Scholar] [CrossRef] [Green Version]
- Lemoine, R.; La Camera, S.; Atanassova, R.; Dedaldechamp, F.; Allario, T.; Pourtau, N.; Bonnemain, J.L.; Laloi, M.; Coutos-Thevenot, P.; Maurousset, L.; et al. Source-to-sink transport of sugar and regulation by environmental factors. Front Plant Sci. 2013, 4, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badri, D.V.; Chaparro, J.M.; Zhang, R.; Shen, Q.; Vivanco, J.M. Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic-related compounds predominantly modulate the soil microbiome. J. Biol. Chem. 2013, 288, 4502–4512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, J.M.; Kühn, C.; Tegeder, M.; Frommer, W.B. Sucrose transport in higher plants. Int. Rev. Cytol. 1997, 178, 41–71. [Google Scholar]
- Bais, H.P.; Fall, R.; Vivanco, J.M. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 2004, 134, 307–319. [Google Scholar] [CrossRef] [Green Version]
- Veillet, F.; Gaillard, C.; Coutos-Thevenot, P.; La Camera, S. Targeting the AtCWIN1 Gene to Explore the Role of Invertases in Sucrose Transport in Roots and during Botrytis cinerea Infection. Front Plant Sci. 2016, 7, 1899. [Google Scholar] [CrossRef]


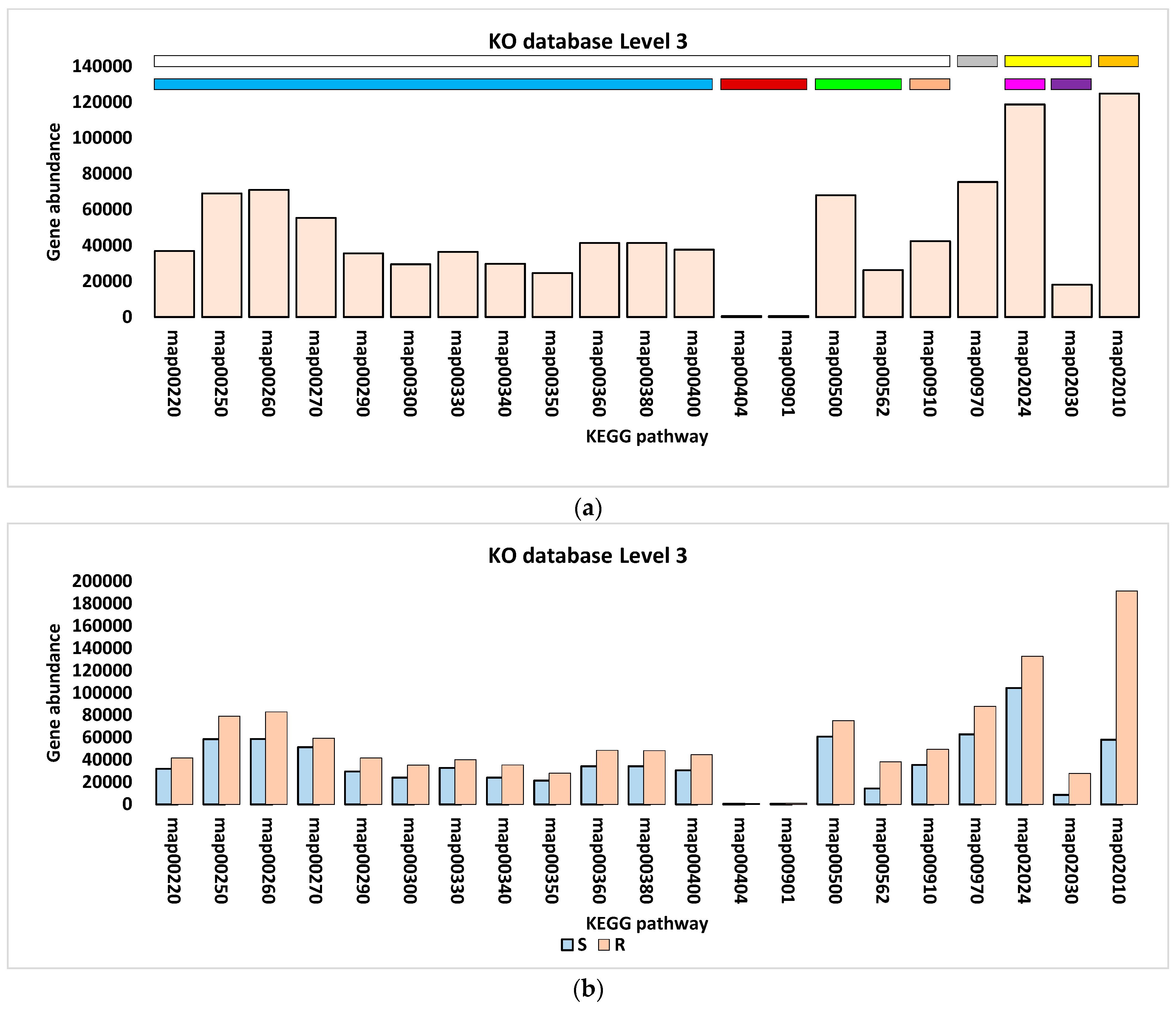
| KEGG Level 1 (Category) | KEGG Level 2 (Sub-Category) | KEGG Level 3 (Pathway) * | Pathway ID |
|---|---|---|---|
| Metabolism | Amino acid metabolism | Arginine biosynthesis | map00220 |
| Alanine, aspartate, and glutamate metabolism | map00250 | ||
| Glycine, serine, and threonine metabolism | map00260 | ||
| Cysteine and methionine metabolism | map00270 | ||
| Valine, leucine, and isoleucine biosynthesis | map00290 | ||
| Lysine biosynthesis | map00300 | ||
| Arginine and proline metabolism | map00330 | ||
| Histidine metabolism | map00340 | ||
| Tyrosine metabolism | map00350 | ||
| Phenylalanine metabolism | map00360 | ||
| Tryptophan metabolism | map00380 | ||
| Phenylalanine, tyrosine, and tryptophan biosynthesis | map00400 | ||
| Biosynthesis of other secondary metabolites | Staurosporine biosynthesis | Map00404 | |
| Indole alkaloid biosynthesis | map00901 | ||
| Biosynthesis of various plant secondary metabolites | Map00999 ** | ||
| Carbohydrate metabolism | Starch and sucrose metabolism | map00500 | |
| Inositol phosphate metabolism | map00562 | ||
| Energy metabolism | Nitrogen metabolism | map00910 | |
| Genetic Information Processing | Translation | Aminoacyl-tRNA biosynthesis | map00970 |
| Cellular Processes | Cellular community—prokaryotes | Quorum sensing | Map02024 |
| Cell motility | Bacterial chemotaxis | Map02030 | |
| Environmental Information Processing | Membrane transport | ABC transporters | map02010 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tashkandi, M.A.; Jalal, R.S.; Baz, L.; Refai, M.Y.; Shami, A.; Ashy, R.A.; Abuauf, H.W.; Alshehrei, F.M.; Alshubaily, F.A.; Barqawi, A.A.; et al. Functional Interpretation of Cross-Talking Pathways with Emphasis on Amino Acid Metabolism in Rhizosphere Microbiome of the Wild Plant Moringa oleifera. Agriculture 2022, 12, 1814. https://doi.org/10.3390/agriculture12111814
Tashkandi MA, Jalal RS, Baz L, Refai MY, Shami A, Ashy RA, Abuauf HW, Alshehrei FM, Alshubaily FA, Barqawi AA, et al. Functional Interpretation of Cross-Talking Pathways with Emphasis on Amino Acid Metabolism in Rhizosphere Microbiome of the Wild Plant Moringa oleifera. Agriculture. 2022; 12(11):1814. https://doi.org/10.3390/agriculture12111814
Chicago/Turabian StyleTashkandi, Manal A., Rewaa S. Jalal, Lina Baz, Mohammed Y. Refai, Ashwag Shami, Ruba Abdulrahman Ashy, Haneen W. Abuauf, Fatimah M. Alshehrei, Fawzia A. Alshubaily, Aminah A. Barqawi, and et al. 2022. "Functional Interpretation of Cross-Talking Pathways with Emphasis on Amino Acid Metabolism in Rhizosphere Microbiome of the Wild Plant Moringa oleifera" Agriculture 12, no. 11: 1814. https://doi.org/10.3390/agriculture12111814







