Differentiation of Yeast-Inoculated and Uninoculated Tomatoes Using Fluorescence Spectroscopy Combined with Machine Learning
Abstract
1. Introduction
- Various ML methods are used for the analysis of the spectroscopic data.
- Computer aided systems are used to distinguish uninoculated and yeast-inoculated tomato samples.
- Different ML methods distinguish between uninoculated and yeast-inoculated tomato varieties with a high accuracy.
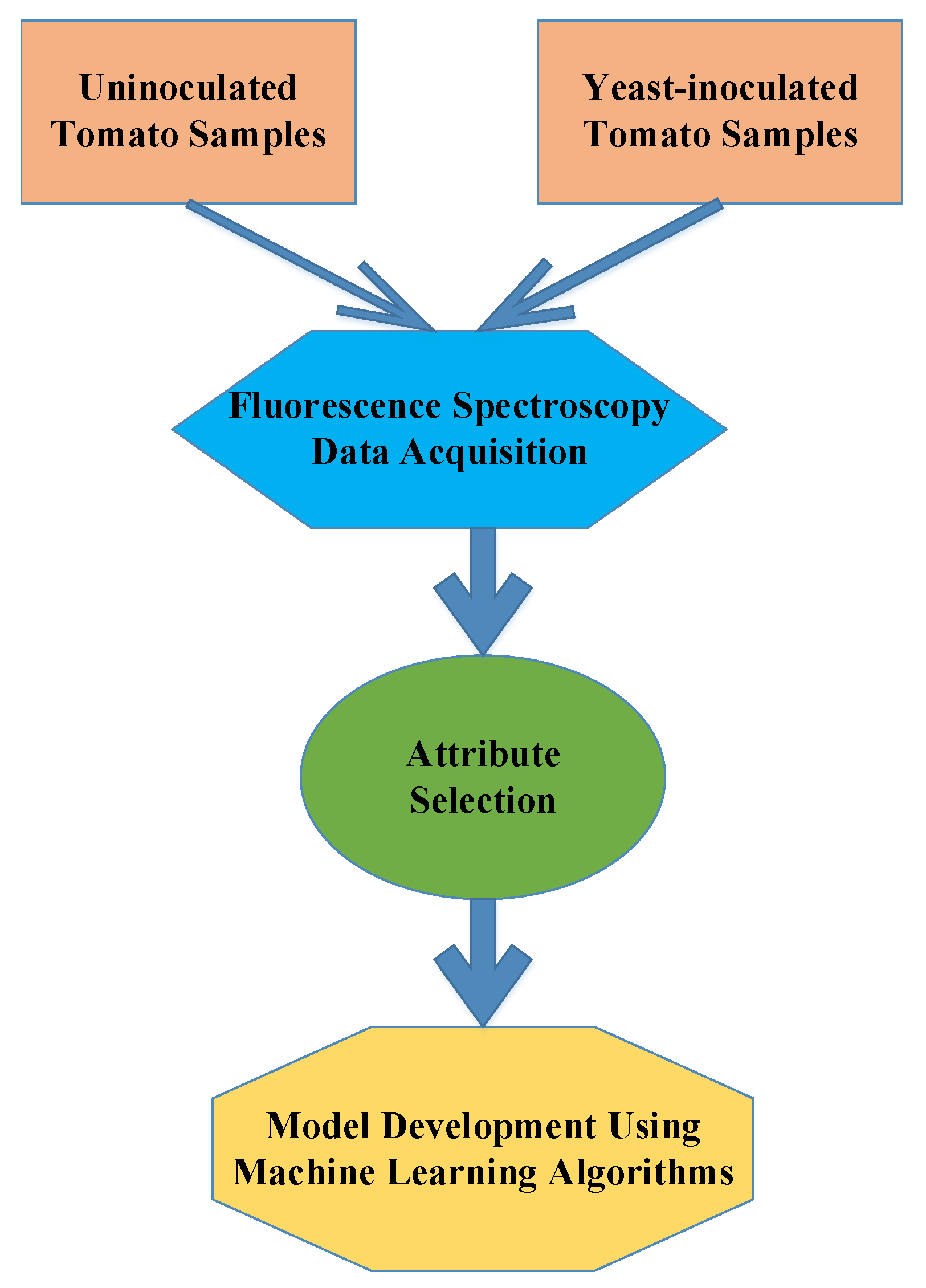
2. Materials and Methods
2.1. Material
2.2. Isolation and Molecular Identification of Yeast
2.3. Colonization on the Development of Tomato in Field Experiments
2.4. Fluorescence Spectroscopy
2.5. Discriminant Analysis
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Osae, R.; Apaliya, M.T.; Alolga, R.N.; Kwaw, E.; Otu, P.N.Y.; Akaba, S. Influence of shea butter, bee wax and cassava starch coatings on enzyme inactivation, antioxidant properties, phenolic compounds and quality retention of tomato (Solanum lycopersicum) fruits. Appl. Food Res. 2022, 2, 100041. [Google Scholar] [CrossRef]
- Saini, R.K.; Moon, S.H.; Keum, Y.-S. An updated review on use of tomato pomace and crustacean processing waste to recover commercially vital carotenoids. Food Res. Int. 2018, 108, 516–529. [Google Scholar] [CrossRef]
- Vallecilla-Yepez, L.; Ciftci, O.N. Increasing cis-lycopene content of the oleoresin from tomato processing byproducts using supercritical carbon dioxide. LWT 2018, 95, 354–360. [Google Scholar] [CrossRef]
- Vigneshwaran, G.; More, P.R.; Arya, S.S. Non-thermal hydrodynamic cavitation processing of tomato juice for physicochemical, bioactive, and enzyme stability: Effect of process conditions, kinetics, and shelf-life extension. Curr. Res. Food Sci. 2022, 5, 313–324. [Google Scholar] [CrossRef]
- Akbudak, M.A.; Filiz, E.; Çetin, D. Genome-wide identification and characterization of high-affinity nitrate transporter 2 (NRT2) gene family in tomato (Solanum lycopersicum) and their transcriptional responses to drought and salinity stresses. J. Plant Physiol. 2022, 272, 153684. [Google Scholar] [CrossRef] [PubMed]
- El-Sitiny, M.F.; Habeba, M.; El-Shehawi, A.M.; Elseehy, M.M.; El-Tahan, A.M.; El-Saadony, M.T.; Selem, G.S. Biochemical and molecular diagnosis of different tomato cultivars susceptible and resistant to Tuta absoluta (meyrick) infestation. Saudi J. Biol. Sci. 2022, 29, 2904–2910. [Google Scholar] [CrossRef]
- Guo, J.-E. Histone deacetylase gene SlHDT1 regulates tomato fruit ripening by affecting carotenoid accumulation and ethylene biosynthesis. Plant Sci. 2022, 318, 111235. [Google Scholar] [CrossRef]
- Nassiri, S.M.; Tahavoor, A.; Jafari, A. Fuzzy logic classification of mature tomatoes based on physical properties fusion. Inf. Process. Agric. 2021, 9, 547–555. [Google Scholar] [CrossRef]
- Morales-Rabanales, Q.N.; Coyotl-Pérez, W.A.; Rubio-Rosas, E.; Cortes-Ramírez, G.S.; Ramírez, J.F.S.; Villa-Ruano, N. Antifungal properties of hybrid films containing the essential oil of Schinus molle: Protective effect against postharvest rot of tomato. Food Control 2022, 134, 108766. [Google Scholar] [CrossRef]
- Li, Q.; Xie, F.; Zhao, Y.; Cao, J. Inhibitory effect of postharvest yeast mannan treatment on Alternaria rot of tomato fruit involving the enhancement of hemicellulose polysaccharides and antioxidant metabolism. Sci. Hortic. 2021, 277, 109798. [Google Scholar] [CrossRef]
- Li, H.; Sun, Q.; Liu, S.; Liu, L.; Shi, Y. A Novel Tomato Volume Measurement Method based on Machine Vision. Teh. Vjesn. 2021, 28, 1674–1680. [Google Scholar] [CrossRef]
- Xie, F.; Yuan, S.; Pan, H.; Wang, R.; Cao, J.; Jiang, W. Effect of yeast mannan treatments on ripening progress and modification of cell wall polysaccharides in tomato fruit. Food Chem. 2017, 218, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Guo, Y.; Wang, Q.; Luo, H.; He, C.; An, B. Expression of flagellin at yeast surface increases biocontrol efficiency of yeast cells against postharvest disease of tomato caused by Botrytis cinerea. Postharvest Biol. Technol. 2020, 162, 111112. [Google Scholar] [CrossRef]
- Guo, J.; Sun, K.; Zhang, Y.; Hu, K.; Zhao, X.; Liu, H.; Wu, S.; Hu, Y.; Zhang, Y.; Wang, Y. SlMAPK3, a key mitogen-activated protein kinase, regulates the resistance of cherry tomato fruit to Botrytis cinerea induced by yeast cell wall and β-glucan. Postharvest Biol. Technol. 2021, 171, 111350. [Google Scholar] [CrossRef]
- Castanheira, M.; Deshpande, L.M.; Davis, A.P.; Rhomberg, P.R.; Pfaller, M.A. Monitoring Antifungal Resistance in a Global Collection of Invasive Yeasts and Molds: Application of CLSI Epidemiological Cutoff Values and Whole-Genome Sequencing Analysis for Detection of Azole Resistance in Candida Albicans. Antimicrob. Agents Chemother. 2017, 61, e00906-17. [Google Scholar] [CrossRef]
- Freimoser, F.M.; Rueda-Mejia, M.P.; Tilocca, B.; Migheli, Q. Biocontrol yeasts: Mechanisms and applications. World J. Microbiol. Biotechnol. 2019, 35, 154. [Google Scholar] [CrossRef]
- Ling, L.; Tu, Y.; Ma, W.; Feng, S.; Yang, C.; Zhao, Y.; Wang, N.; Li, Z.; Lu, L.; Zhang, J. A potentially important resource: Endophytic yeasts. World J. Microbiol. Biotechnol. 2020, 36, 110. [Google Scholar] [CrossRef]
- Hassan, S.E.-D. Plant growth-promoting activities for bacterial and fungal endophytes isolated from medicinal plant of Teucrium polium L. J. Adv. Res. 2017, 8, 687–695. [Google Scholar] [CrossRef]
- Buzzini, P.; Branda, E.; Goretti, M.; Turchetti, B. Psychrophilic yeasts from worldwide glacial habitats: Diversity, adaptation strategies and biotechnological potential. FEMS Microbiol. Ecol. 2012, 82, 217–241. [Google Scholar] [CrossRef]
- Sundh, I.; Melin, P. Safety and regulation of yeasts used for biocontrol or biopreservation in the food or feed chain. Antonie Van Leeuwenhoek 2010, 99, 113–119. [Google Scholar] [CrossRef]
- Van Lenteren, J.C.; Bolckmans, K.; Köhl, J.; Ravensberg, W.J.; Urbaneja, A. Biological control using invertebrates and microorganisms: Plenty of new opportunities. BioControl 2018, 63, 39–59. [Google Scholar] [CrossRef]
- Spadaro, D.; Droby, S. Development of biocontrol products for postharvest diseases of fruit: The importance of elucidating the mechanisms of action of yeast antagonists. Trends Food Sci. Technol. 2016, 47, 39–49. [Google Scholar] [CrossRef]
- Droby, S.; Wisniewski, M.; Teixidó, N.; Spadaro, D.; Jijakli, M.H. The science, development, and commercialization of postharvest biocontrol products. Postharvest Biol. Technol. 2016, 122, 22–29. [Google Scholar] [CrossRef]
- Petkova, M.; Petrova, S.; Spasova-Apostolova, V.; Naydenov, M. Tobacco Plant Growth-Promoting and Antifungal Activities of Three Endophytic Yeast Strains. Plants 2022, 11, 751. [Google Scholar] [CrossRef] [PubMed]
- Valeur, B.; Berberan-Santos, M.N. Molecular Fluorescence: Principles and Applications; John Wiley & Sons: Hoboken, NJ, USA, 2012. [Google Scholar]
- Qin, J.; Lu, R. Measurement of the optical properties of fruits and vegetables using spatially resolved hyperspectral diffuse reflectance imaging technique. Postharvest Biol. Technol. 2008, 49, 355–365. [Google Scholar] [CrossRef]
- Bachmann, L.; Zezell, D.M.; Ribeiro, A.D.C.; Gomes, L.; Ito, A.S. Fluorescence Spectroscopy of Biological Tissues—A Review. Appl. Spectrosc. Rev. 2006, 41, 575–590. [Google Scholar] [CrossRef]
- Mitschke, F.; Mitschke, F. Fiber Optics; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Dakin, J.P.; Brown, R. Handbook of Optoelectronics: Concepts, Devices, and Techniques (Volume One); CRC Press: Boca Raton, FL, USA, 2017. [Google Scholar]
- Hoffmann, A.M.; Noga, G.; Hunsche, M. Fluorescence indices for monitoring the ripening of tomatoes in pre- and postharvest phases. Sci. Hortic. 2015, 191, 74–81. [Google Scholar] [CrossRef]
- Karoui, R.; Blecker, C. Fluorescence Spectroscopy Measurement for Quality Assessment of Food Systems—A Review. Food Bioprocess Technol. 2011, 4, 364–386. [Google Scholar] [CrossRef]
- Karim, S.; Zhang, Y.; Laghari, A.A.; Asif, M.R. Image processing based proposed drone for detecting and controlling street crimes. In Proceedings of the 2017 IEEE 17th International Conference on Communication Technology (ICCT), Chengdu, China, 27–30 October 2017; pp. 1725–1730. [Google Scholar]
- Karim, S.; Zhang, Y.; Yin, S.; Laghari, A.A.; Brohi, A.A. Impact of compressed and down-scaled training images on vehicle detection in remote sensing imagery. Multimed. Tools Appl. 2019, 78, 32565–32583. [Google Scholar] [CrossRef]
- Aslan, M.F.; Durdu, A.; Sabanci, K.; Ropelewska, E.; Gültekin, S.S. A Comprehensive Survey of the Recent Studies with UAV for Precision Agriculture in Open Fields and Greenhouses. Appl. Sci. 2022, 12, 1047. [Google Scholar] [CrossRef]
- Laghari, A.A.; He, H.; Shafiq, M.; Khan, A. Assessment of quality of experience (QoE) of image compression in social cloud computing. Multiagent Grid Syst. 2018, 14, 125–143. [Google Scholar] [CrossRef]
- Mitchell, T.M. Machine learning and data mining. Commun. ACM 1999, 42, 30–36. [Google Scholar] [CrossRef]
- Koza, J.R.; Bennett, F.H.; Andre, D.; Keane, M.A. Automated Design of Both the Topology and Sizing of Analog Electrical Circuits Using Genetic Programming. In Artificial Intelligence in Design ’96; Gero, J.S., Sudweeks, F., Eds.; Springer: Dordrecht, The Netherlands, 1996; pp. 151–170. [Google Scholar]
- Kumar, S.D.; Esakkirajan, S.; Bama, S.; Keerthiveena, B. A microcontroller based machine vision approach for tomato grading and sorting using SVM classifier. Microprocess. Microsyst. 2020, 76, 103090. [Google Scholar] [CrossRef]
- Iraji, M.S. Comparison between soft computing methods for tomato quality grading using machine vision. J. Food Meas. Charact. 2019, 13, 1–15. [Google Scholar] [CrossRef]
- Ropelewska, E.; Piecko, J. Discrimination of tomato seeds belonging to different cultivars using machine learning. Eur. Food Res. Technol. 2022, 248, 685–705. [Google Scholar] [CrossRef]
- Slavova, V.; Ropelewska, E.; Sabanci, K.; Aslan, M.F.; Nacheva, E. A comparative evaluation of Bayes, functions, trees, meta, rules and lazy machine learning algorithms for the discrimination of different breeding lines and varieties of potato based on spectroscopic data. Eur. Food Res. Technol. 2022, 248, 1765–1775. [Google Scholar] [CrossRef]
- Koklu, M.; Sarigil, S.; Ozbek, O. The use of machine learning methods in classification of pumpkin seeds (Cucurbita pepo L.). Genet. Resour. Crop Evol. 2021, 68, 2713–2726. [Google Scholar] [CrossRef]
- Ropelewska, E.; Slavova, V.; Sabanci, K.; Aslan, M.F.; Cai, X.; Genova, S. Discrimination of onion subjected to drought and normal watering mode based on fluorescence spectroscopic data. Comput. Electron. Agric. 2022, 196, 106916. [Google Scholar] [CrossRef]
- Yasmin, J.; Lohumi, S.; Ahmed, M.R.; Kandpal, L.M.; Faqeerzada, M.A.; Kim, M.S.; Cho, B.-K. Improvement in Purity of Healthy Tomato Seeds Using an Image-Based One-Class Classification Method. Sensors 2020, 20, 2690. [Google Scholar] [CrossRef]
- Bouckaert, R.R.; Frank, E.; Hall, M.; Kirkby, R.; Reutemann, P.; Seewald, A.; Scuse, D. WEKA Manual for Version 3-9-1; University of Waikato: Hamilton, New Zealand, 2016. [Google Scholar]
- Frank, E.; Hall, M.; Witten, I. Online Appendix for “Data Mining: Practical Machine Learning Tools and Techniques”; The WEKA Workbench; Elsevier: Amsterdam, The Netherlands, 2016. [Google Scholar]
- Witten, I.H.; Frank, E.; Hall, M.A.; Pal, C.J. Practical machine learning tools and techniques. In Proceedings of the DATA MINING, Las Vegas, NV, USA, 20–23 June 2005; p. 4. [Google Scholar]
- Ropelewska, E.; Szwejda-Grzybowska, J. A comparative analysis of the discrimination of pepper (Capsicum annuum L.) based on the cross-section and seed textures determined using image processing. J. Food Process Eng. 2021, 44, e13694. [Google Scholar] [CrossRef]
- Sabanci, K.; Aslan, M.F.; Ropelewska, E.; Unlersen, M.F. A convolutional neural network-based comparative study for pepper seed classification: Analysis of selected deep features with support vector machine. J. Food Process Eng. 2021, 45, e13955. [Google Scholar] [CrossRef]


| Algorithm | Predicted Class (%) | Actual Class | Average Accuracy (%) | TP Rate | Precision | ROC Area | PRC Area | F-Measure | MCC | |
|---|---|---|---|---|---|---|---|---|---|---|
| Control | Inoculated | |||||||||
| trees.HoeffdingTree | 100 | 0 | control | 100 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| 0 | 100 | inoculated | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | ||
| rules.PART | 100 | 0 | control | 95 | 1.000 | 0.909 | 0.950 | 0.909 | 0.952 | 0.905 |
| 10 | 90 | inoculated | 0.900 | 1.000 | 0.950 | 0.950 | 0.947 | 0.905 | ||
| meta.FilteredClassifier | 100 | 0 | control | 95 | 1.000 | 0.909 | 0.950 | 0.909 | 0.952 | 0.905 |
| 10 | 90 | inoculated | 0.900 | 1.000 | 0.950 | 0.950 | 0.947 | 0.905 | ||
| lazy.IBk | 100 | 0 | control | 95 | 1.000 | 0.909 | 0.990 | 0.982 | 0.952 | 0.905 |
| 10 | 90 | inoculated | 0.900 | 1.000 | 0.990 | 0.983 | 0.947 | 0.905 | ||
| functions.Logistic | 100 | 0 | control | 95 | 1.000 | 0.909 | 1.000 | 1.000 | 0.952 | 0.905 |
| 10 | 90 | inoculated | 0.900 | 1.000 | 1.000 | 1.000 | 0.947 | 0.905 | ||
| bayes.BayesNet | 100 | 0 | control | 100 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| 0 | 100 | inoculated | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | ||
| Algorithm | Predicted Class (%) | Actual Class | Average Accuracy (%) | TP Rate | Precision | ROC Area | PRC Area | F-Measure | MCC | |
|---|---|---|---|---|---|---|---|---|---|---|
| Control | Inoculated | |||||||||
| trees.HoeffdingTree | 100 | 0 | control | 100 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| 0 | 100 | inoculated | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | ||
| rules.PART | 100 | 0 | control | 100 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| 0 | 100 | inoculated | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | ||
| meta.FilteredClassifier | 100 | 0 | control | 95 | 1.000 | 0.909 | 0.950 | 0.909 | 0.952 | 0.905 |
| 10 | 90 | inoculated | 0.900 | 1.000 | 0.950 | 0.950 | 0.947 | 0.905 | ||
| lazy.IBk | 100 | 0 | control | 100 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 |
| 0 | 100 | inoculated | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | 1.000 | ||
| functions.Logistic | 100 | 0 | control | 95 | 1.000 | 0.909 | 0.950 | 0.909 | 0.952 | 0.905 |
| 10 | 90 | inoculated | 0.900 | 1.000 | 1.000 | 1.000 | 0.947 | 0.905 | ||
| bayes.BayesNet | 100 | 0 | control | 95 | 1.000 | 0.909 | 1.000 | 1.000 | 0.952 | 0.905 |
| 10 | 90 | inoculated | 0.900 | 1.000 | 1.000 | 1.000 | 0.947 | 0.905 | ||
| Algorithm | Predicted Class (%) | Actual Class | Average Accuracy (%) | TP Rate | Precision | ROC Area | PRC Area | F-Measure | MCC | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | Inoculated | ||||||||||
| trees.HoeffdingTree | 90 | 10 | control | 85 | 0.900 | 0.818 | 0.880 | 0.826 | 0.857 | 0.704 | |
| 20 | 80 | inoculated | 0.800 | 0.889 | 0.880 | 0.923 | 0.842 | 0.704 | |||
| rules.PART | 90 | 10 | control | 80 | 0.900 | 0.750 | 0.830 | 0.775 | 0.818 | 0.612 | |
| 30 | 70 | inoculated | 0.700 | 0.875 | 0.830 | 0.813 | 0.778 | 0.612 | |||
| meta.FilteredClassifier | 90 | 10 | control | 90 | 0.900 | 0.900 | 0.960 | 0.967 | 0.900 | 0.800 | |
| 10 | 90 | inoculated | 0.900 | 0.900 | 0.960 | 0.962 | 0.900 | 0.800 | |||
| lazy.KStar | 100 | 0 | control | 95 | 1.000 | 0.909 | 0.935 | 0.882 | 0.952 | 0.905 | |
| 10 | 90 | inoculated | 0.900 | 1.000 | 0.900 | 0.950 | 0.947 | 0.905 | |||
| functions.Logistic | 90 | 10 | control | 90 | 0.900 | 0.900 | 0.960 | 0.967 | 0.900 | 0.800 | |
| 10 | 90 | inoculated | 0.900 | 0.900 | 0.960 | 0.962 | 0.900 | 0.800 | |||
| bayes.BayesNet | 90 | 10 | control | 85 | 0.900 | 0.818 | 0.890 | 0.834 | 0.857 | 0.704 | |
| 20 | 80 | inoculated | 0.800 | 0.889 | 0.890 | 0.932 | 0.842 | 0.704 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ropelewska, E.; Slavova, V.; Sabanci, K.; Aslan, M.F.; Masheva, V.; Petkova, M. Differentiation of Yeast-Inoculated and Uninoculated Tomatoes Using Fluorescence Spectroscopy Combined with Machine Learning. Agriculture 2022, 12, 1887. https://doi.org/10.3390/agriculture12111887
Ropelewska E, Slavova V, Sabanci K, Aslan MF, Masheva V, Petkova M. Differentiation of Yeast-Inoculated and Uninoculated Tomatoes Using Fluorescence Spectroscopy Combined with Machine Learning. Agriculture. 2022; 12(11):1887. https://doi.org/10.3390/agriculture12111887
Chicago/Turabian StyleRopelewska, Ewa, Vanya Slavova, Kadir Sabanci, Muhammet Fatih Aslan, Veselina Masheva, and Mariana Petkova. 2022. "Differentiation of Yeast-Inoculated and Uninoculated Tomatoes Using Fluorescence Spectroscopy Combined with Machine Learning" Agriculture 12, no. 11: 1887. https://doi.org/10.3390/agriculture12111887
APA StyleRopelewska, E., Slavova, V., Sabanci, K., Aslan, M. F., Masheva, V., & Petkova, M. (2022). Differentiation of Yeast-Inoculated and Uninoculated Tomatoes Using Fluorescence Spectroscopy Combined with Machine Learning. Agriculture, 12(11), 1887. https://doi.org/10.3390/agriculture12111887








