New Insights into the Use of Rhizobia to Mitigate Soil N2O Emissions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains Used in the Study and Culture Conditions
2.2. Molecular Detection of nosZ Gene Sequences
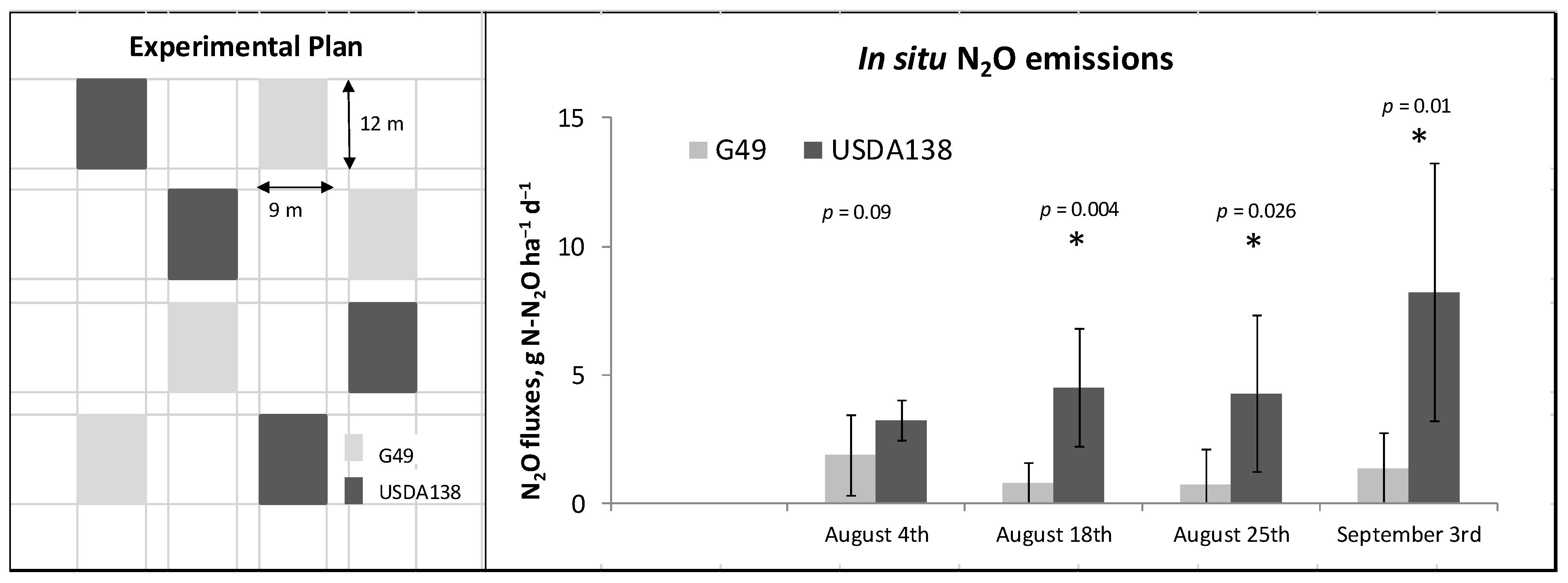
2.3. In Situ N2O Fluxes Measurements
| Species/ Strain Number (Synonyms) | Host Plant of Origin | Reference/Source/Origin |
|---|---|---|
| Ensifer meliloti MIAE00427 (MSDJ848, RCR2011, 2011) | Medicago sativa (alfalfa) | [20]/RCR |
| Rhizobium leguminosarum bv. trifolii MIAE03027(MSDJ134, T117) | Trifolium pratense (clover) | [21] |
| MIAE03034 (MSDJ141, T132) | Trifolium pratense (clover) | [21] |
| Bradyrhizobium lupini MIAE00428 (MSDJ718, LL13) | Lupinus luteus (lupine) | [22] |
| Bradyrhizobium sp. (Lupinus) MIAE00428 (MSDJ718, LL200) | Lupinus luteus (lupine) | [20] |
| Rhizobium leguminosarum bv. viciae MIAE01211 (MSDJ822, FH34) | Vicia faba (faba bean) | [20] |
| MIAE01211 (MSDJ822, FS16) | Vicia faba (faba bean) | [20] |
| Bradyrhizobium diazoefficiens MIAE00426 (MSDJ1996, G49) USDA 110 | Glycine max (soybean) Glycine max (soybean) | [23] USDA |
| Bradyrhizobium japonicum | ||
| USDA138 | Glycine max (soybean) | USDA |
| Ensifer sp. (Trigonella) MIAE06325 (MSDJ3523, 3523) | Trigonella foenum-graecum (fenugreek) | MIAE |
| MIAE06333 (MSDJ3531, 3531) | Trigonella foenum-graecum (fenugreek) | MIAE |
| MIAE01335 (MSDJ3538, 3538) | Trigonella foenum-graecum (fenugreek) | MIAE |
| MIAE06345 (MSDJ3546, 3546) | Trigonella foenum-graecum (fenugreek) | MIAE |
| Bradyrhizobium sp. Arachis MIAE06284 (MSDJ3482, IC7017) MIAE04980 (MSDJ2177, ST2) | Arachis hypogaea (peanut) Arachis hypogaea (peanut) | ICRISAT USA |
2.4. Phenotypic Characterization of Strains Grown as Pure Culture (C) on Growing Medium
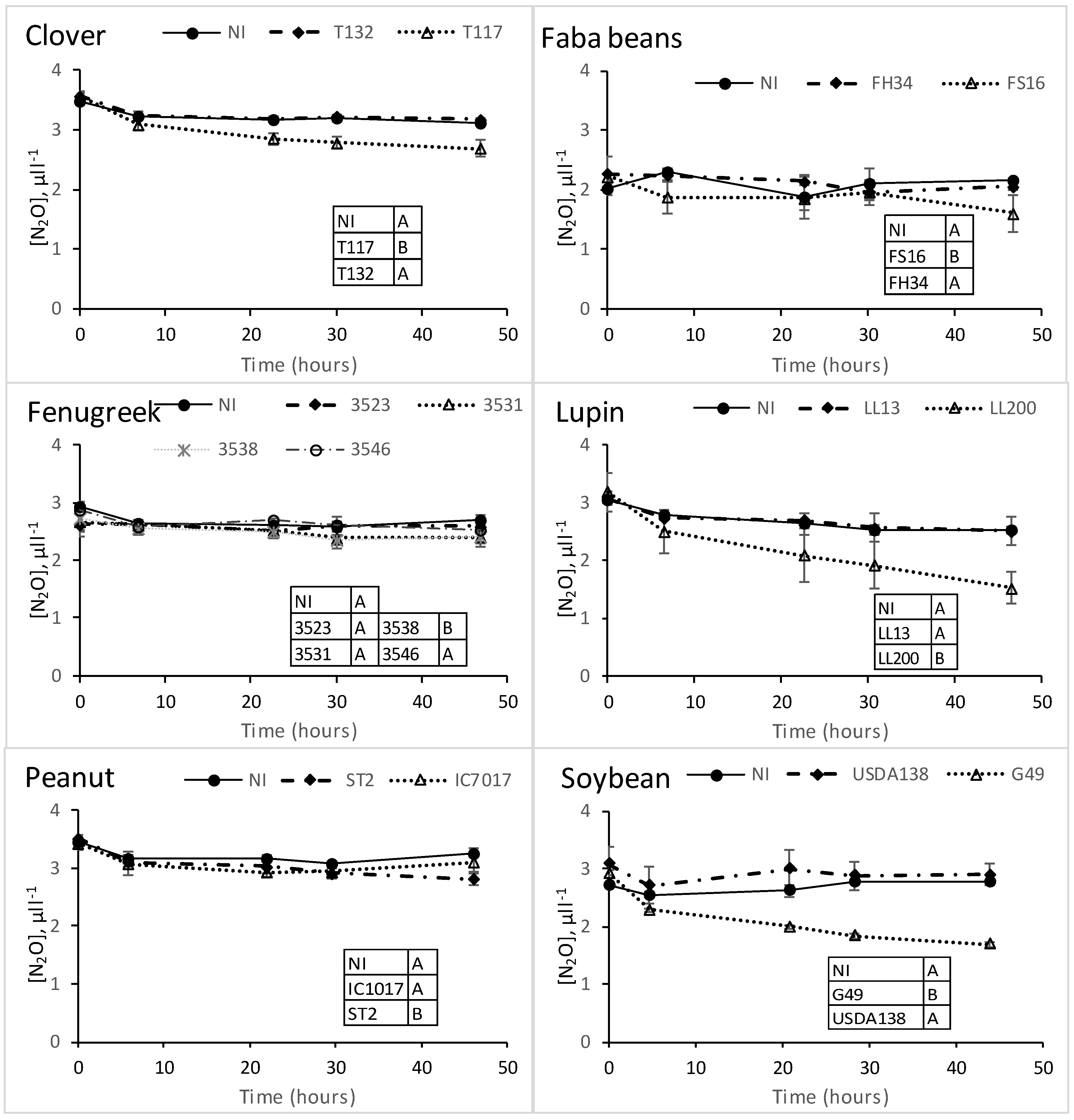
2.5. Characterization of Greenhouse N2O Reduction by the Different Legume–Rhizobia Associations (A)
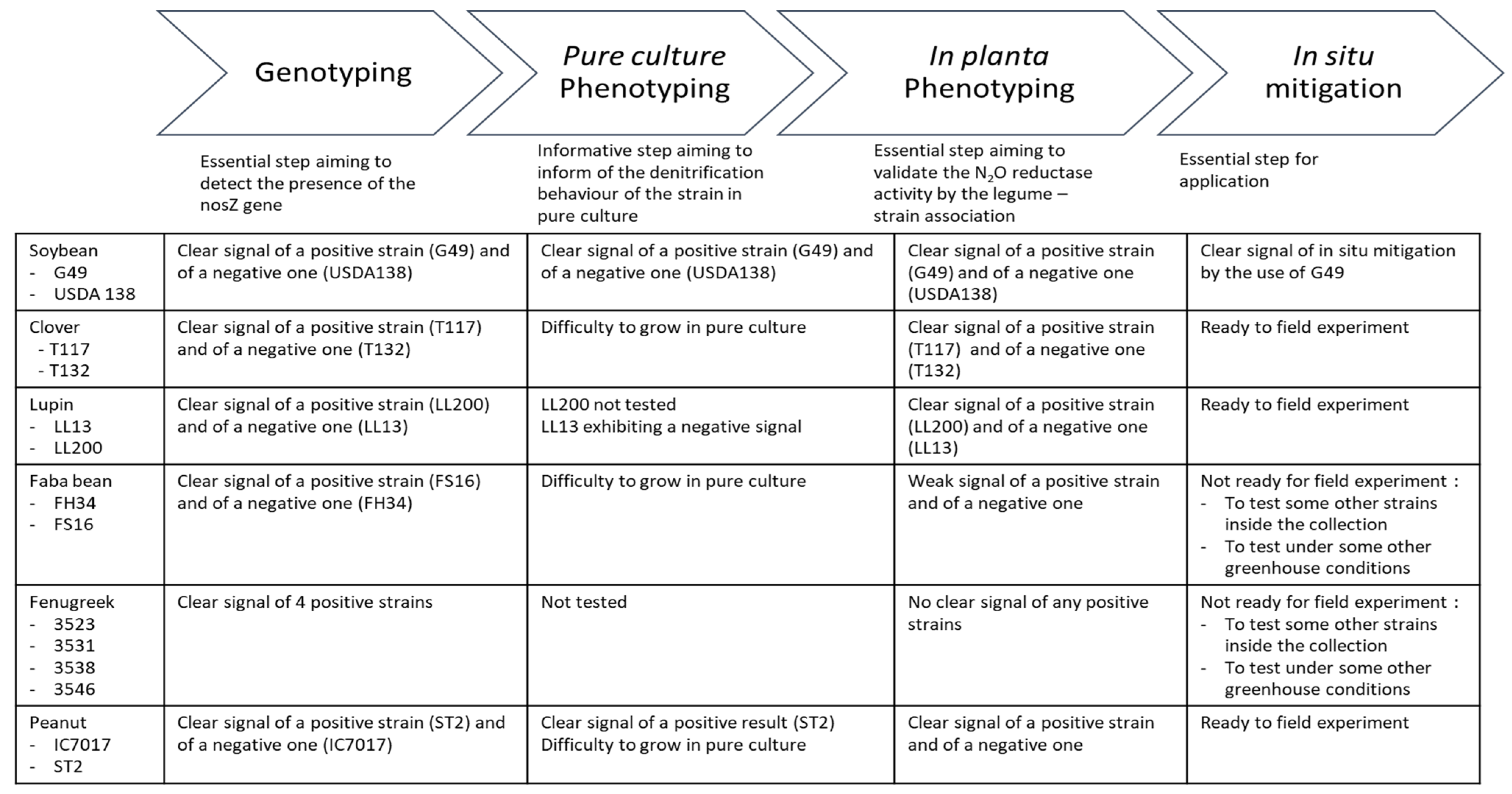
3. Results
3.1. Detection of the nosZ Gene Amongst the Strains Tested
3.2. In Situ Proof of Concept of N2O Emission Mitigation by the Soybean–B. diazoefficiens G49 Association
3.3. Phenotypes in Pure Culture
3.4. Greenhouse Phenotypic Characterization on Living Rhizobia-Legume Associations
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- UNEP. Drawing Down N2O to Protect Climate and the Ozone Layer; A UNEP Synthesis Report; United Nations Environment Programme (UNEP): Nairobi, Kenya, 2013. [Google Scholar]
- Butterbach-Bahl, K.; Baggs, E.; Dannenmann, M.; Kiese, R.; Zechmeister-Boltenstern, S. Nitrous oxide emissions from soils: How well do we understand the processes and their controls. Philos. Trans. R. Br. Soc. 2013, 368, 20130122. [Google Scholar] [CrossRef] [PubMed]
- Hénault, C.; Bourennane, H.; Ayzac, A.; Ratié, C.; Saby, N.; Cohan, J.P.; Eglin, T.; Gall, C.L. Management of soil pH promotes nitrous oxide reduction and thus mitigates soil emissions of this greenhouse gas. Sci. Rep. 2019, 9, 20182. [Google Scholar] [CrossRef] [Green Version]
- Bakken, L.R.; Frostegård, Å. Emerging options for mitigating N2O emissions from food production by manipulating the soil microbiota. Curr. Opin. Environ. Sustain. 2020, 47, 89–94. [Google Scholar] [CrossRef]
- Mania, D.; Woliy, K.; Degefu, T.; Frostegård, Å. A common mechanism for efficient N2O reduction in diverse isolates of nodule-forming bradyrhizobia. Environ. Microbiol. 2020, 22, 17–31. [Google Scholar] [CrossRef] [Green Version]
- Itakura, M.; Uchida, Y.; Akiyama, H.; Hoshino, Y.T.; Shimomura, Y.; Morimoto, S.; Tago, K.; Wang, Y.; Hayakawa, C.; Uetake, Y.; et al. Mitigation of nitrous oxide emissions from soils by Bradyrhizobium japonicum inoculation. Nat. Clim. Chang. 2013, 3, 208–212. [Google Scholar] [CrossRef]
- Stagnari, F.; Maggio, A.; Galieni, A.; Pisante, M. Multiple benefits of legumes for agriculture sustainability: An overview. Chem. Biol. Technol. Agric. 2017, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Sameshima-Saito, R.; Chiba, K.; Hirayama, J.; Itakura, M.; Mitsui, H.; Eda, S.; Minamisawa, K. Symbiotic Bradyrhizobium japonicum Reduces N2O Surrounding the Soybean Root System via Nitrous Oxide Reductase. Appl. Environ. Microbiol. 2006, 72, 2526–2532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hénault, C.; Revellin, C. Inoculants of leguminous crops for mitigating soil emissions of the greenhouse gas nitrous oxide. Plant Soil 2011, 346, 289–296. [Google Scholar] [CrossRef]
- Akiyama, H.; Hoshino, Y.; Itakura, M.; Shimomura, Y.; Wang, Y.; Yamamoto, A.; Tago, K.; Nakajima, Y.; Minamisawa, K.; Hayatsu, M. Mitigation of soil N2O emission by inoculation with a mixed culture of indigenous Bradyrhizobium diazoefficiens. Sci. Rep. 2016, 6, 32869. [Google Scholar] [CrossRef] [Green Version]
- Obando, M.; Antonelli, C.; Casanave, S.; Maguire, V.; Torres, D.; Pérez, G.; Bailleres, M.; Donadio, F.; Creus, C.; Videla, C.; et al. Evaluation of nitrous oxide emission by soybean inoculated with Bradyrhizobium strains commonly used as inoculants in South America. Plant Soil. 2022. [Google Scholar] [CrossRef]
- Vincent, J.M. A Manual for the Practical Study of Root Nodule Bacteria (Vol. IBP Handbook no. 15); Blackell Scientific Publication: Oxford, UK, 1970. [Google Scholar]
- Beringer, J.E.R. factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 1974, 84, 188–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henry, S.; Bru, D.; Stres, B.; Hallet, S.; Philippot, L. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 2006, 72, 5181–5189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scala, D.J.; Kerkhof, L.J. Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol. Lett. 1998, 162, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Velasco, L.; Mesa, S.; Xu, C.; Delgado, M.J.; Bedmar, E.J. Molecular characterization of nosRZDFYLX genes coding for denitrifying nitrous oxide reductase of Bradyrhizobium japonicum. Antonie Van Leeuwenhoek 2004, 85, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.J.; Rubia, M.I.; de la Peña, T.C.; Pueyo, J.J.; Bedmar, E.J.; Delgado, M.J. Genetic basis for denitrification in Ensifer meliloti. BMC Microbiol. 2014, 14, 142. [Google Scholar] [CrossRef] [Green Version]
- Revellin, C.; Pinochet, X.; Beauclair, P.; Catroux, G. Influence of soil properties and soya bean cropping history on the Bradyrhizobium japonicum population in some French soils. Eur. J. Soil Sci. 1996, 47, 505–510. [Google Scholar] [CrossRef]
- Hutchinson, G.L.; Livingston, G.P. Use of chamber systems to measure trace gas fluxes. In Agricultural Ecosystem Effects on Trace Gases and Global Climate Change; Rolston, D.E., Harper, L.A., Mosier, A.R., Duxbury, J.M., Eds.; ASA Spec. Publ. 55; ASA, CSSA and SSSA: Madison, WI, USA, 1993; pp. 63–78. [Google Scholar]
- Amarger, N. Selection of Rhizobium strains on their competitive ability for nodulation. Soil Biol. Biochem. 1981, 13, 481–486. [Google Scholar] [CrossRef]
- Mazurier, S.I. Diversité de Populations Naturelles Nodulantes de Rhizobium leguminosarum. Ph.D. Thesis, Université Claude Bernard Lyon 1, Lyon, France, 1989. [Google Scholar]
- Laguerre, G.; Geniaux, E.; Mazurier, S.I.; Casartelli, R.R.; Amarger, N. Conformity and diversity among field isolates of Rhizobium leguminosarum bv. viciae, bv. trifolii, and bv. phaseoli revealed by DNA hybridization using chromosome and plasmid probes. Can. J. Microbiol. 1993, 39, 412–419. [Google Scholar] [CrossRef]
- Lagacherie, B.; Hugot, R.; Amarger, N. Selection de souches de Rhizobium japonicum d’après leur compétitivité pour l’infection. Ann. Agron. 1977, 28, 379–389. [Google Scholar]
- Rochette, P.; Eriksen-Hamel, N.S. Chamber Measurements of Soil Nitrous Oxide Flux: Are Absolute Values Reliable? Soil. Sc. Soc. Am. J. 2008, 72, 331–342. [Google Scholar] [CrossRef]
- Cheneby, D.; Hartmann, A.; Hénault, C.; Topp, E.; Germon, J.C. Diversity of denitrifying microflora and ability to reduce N2O in two soils. Biol. Fert. Soils. 1998, 28, 19–26. [Google Scholar] [CrossRef]
- Castaldo, D.A.; Harroon, M.; Schrader, L.E.; Youngs, V.L. Rapid colorimetric determination of nitrate in plant-tissue by nitration of salycilic acid. Commun. Soil Sci. Plant Anal. 1975, 6, 71–80. [Google Scholar] [CrossRef]
- Yoshinari, T.; Hynes, R.; Knowles, R. Acetylene inhibition of nitrous oxide reduction and measurement of denitrification and nitrogen fixation in soil. Soil Biol. Biochem. 1977, 9, 177–1833. [Google Scholar] [CrossRef]
- Green, S.J.; Prakash, O.; Gihring, T.M.; Akob, D.M.; Jasrotia, P.; Jardine, P.M.; Watson, D.B.; Brown, S.D.; Palumbo, A.V.; Kostka1, J.E. Denitrifying Bacteria Isolated from Terrestrial Subsurface Sediments Exposed to Mixed-Waste Contamination. Appl. Environ. Microbiol. 2010, 76, 3244–3254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catroux, G. Inoculant quality standards and controls in France. In Expert Consultation on Legume Inoculant Production and Quality Control; Thompson, V.A., Ed.; FAO: Rome, Italy, 1991; Volume 19–21, pp. 113–120. [Google Scholar]
- Mahne, I.; Tiedje, J.M. Criteria and methodology for identifying respiratory denitrifiers. Appl. Environ. Microbiol. 1995, 61, 1110–1115. [Google Scholar] [CrossRef] [Green Version]
- Woliy, K.; Degefu, T.; Frostegard, A. Host Range and Symbiotic Effectiveness of N2O Reducing Bradyrhizobium Strains. Front. Microbiol. 2019, 10, 10. [Google Scholar] [CrossRef]
- Obando, M.; Correa-Galeote, D.; Castellano-Hinojosa, A.; Gualp, J.L.; Hidalgo, A.; Alche, J.; Bedmar, E.J.; Cassan, F.D. Analysis of the denitrification pathway and greenhouse gases emissions in Bradyrhizobium sp. strains used as biofertilizers in South America. J. Appl. Microbiol. 2019, 127, 739–749. [Google Scholar] [CrossRef]
| Species | Collection Name | Detection of the nosZ Gene | Genbank Accession Numbers of Sequences | |
|---|---|---|---|---|
| Primers nosZ1F/nosZ2R | Primers nos661F*/nos1773R* | |||
| E. meliloti | 2011 | + | NT | |
| B. diazoefficiens | G49 | + | + | OL741415 |
| B. diazoefficiens | USDA 110 | + | + | |
| B. japonicum | USDA 138 | − | − | |
| R. leguminosarum bv. trifolii | T132 | − | − | |
| R. leguminosarum bv. trifolii | T117 | + | + | OL691940 |
| B. lupini | LL13 | − | − | |
| Bradyrhizobium sp. | LL200 | + | + | |
| R. leguminosarum bv. viciae | FH34 | − | − | |
| R. leguminosarum bv. viciae | FS16 | + | + | OL691939 |
| Ensifer sp. | 3523 | + | + | |
| Ensifer sp. | 3531 | + | + | |
| Ensifer sp. | 3538 | + | + | |
| Ensifer sp. | 3546 | + | + | |
| Bradyrhizobium sp. arachis | IC7017 | − | − | |
| Bradyrhizobium sp. arachis | ST2 | + | + | OL741416 |
| Species | Without Acetylene | With Acetylene | |||
|---|---|---|---|---|---|
| Collection Name | Nitrate Consumption % of Added Nitrate | N2O Accumulation % of Consumed Nitrate | Nitrate Consumption% of Added Nitrate | N2O Accumulation% of Consumed Nitrate | |
| E. meliloti | 2011 | 62 | 0.4 (low) | 14 | 34 (high) |
| R. legumin. bv. trifolii | T117 | 100 | 0 (very low) | 0 | 0 (very low) |
| B. lupini | LL13 | 100 | 100 (very high) | NT | NT |
| R. legumin. bv. viciae | FH34 | 0 | 0 (very low) | NT | NT |
| R. legumin. bv. viciae | FS16 | 32 | 0 (very low) | 2 | 0 (very low) |
| B. diazoefficiens | G49 | 100 | 0 (very low) | 100 | 100 (very high) |
| B. diazoefficiens | USDA 110 | 94 | 0 (very low) | 26 | 100 (very high) |
| B. japonicum | USDA 138 | 96 | 84 (very high) | NT | NT |
| B. sp. arachis | IC7017 | 0 | 0 (very low) | NT | NT |
| B. sp. arachis | ST2 | 100 | 0 (very low) | 71 | 100 (very high) |
| Genotype | Phenotypic Characterization | |||||
|---|---|---|---|---|---|---|
| Species (Abbreviated) | Collection Name | Strain in Growth Medium with Anaerobic Conditions | Strain in Symbiosis with Its Host Plant | |||
| Host Plant | Varieties | Efficient Reduction of N2O Observed on Living Inoculated Plants | ||||
| E. meliloti | 2011 | nosZ+ | C-N2OR+ | NT | ||
| R. legumin. trif. | T132 | nosZ− | NT | Clover | HUIA | A-N2OR− |
| R. legumin. trif. | T117 | nosZ+ | NNC* | Clover | HUIA | A-N2OR+ |
| B. lupini | LL13 | nosZ− | CN2OR− | Lupin | LUBLANC | A-N2OR− |
| B. sp. (Lup.) | LL200 | nosZ+ | NT | Lupin | LUBLANC | A-N2OR+ |
| R. legumin. vic. | FH34 | nosZ− | NNC | Faba beans | EXPRESSO | A-N2OR− |
| R. legumin. vic. | FS16 | nosZ+ | NNC* | Faba beans | EXPRESSO | A-N2OR− |
| B. diaz. | G49 | nosZ+ | C-N2OR+ | Soybean | NAVIGATOR | A-N2OR+ |
| B. diaz. | USDA 110 | nosZ+ | C-N2OR+ | NT | ||
| B. japon. | USDA 138 | nosZ− | C-N2OR− | Soybean | NAVIGATOR | A-N2OR− |
| E. sp. (Trigon.) | 3523 | nosZ+ | NT | Fenugreek | HANKA | A-N2OR− |
| E. sp. (Trigon.) | 3531 | nosZ+ | NT | Fenugreek | HANKA | A-N2OR− |
| E. sp. (Trigon.) | 3538 | nosZ+ | NT | Fenugreek | HANKA | A-N2OR+/− |
| E. sp. (Trigon.) | 3546 | nosZ+ | NT | Fenugreek | HANKA | A-N2OR− |
| B. sp. (Arachis) | IC7017 | nosZ− | NNC | Peanut | Unknown | A-N2OR− |
| B. sp. (Arachis) | ST2 | nosZ+ | C-N2OR+ | Peanut | Unknown | A-N2OR+ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hénault, C.; Barbier, E.; Hartmann, A.; Revellin, C. New Insights into the Use of Rhizobia to Mitigate Soil N2O Emissions. Agriculture 2022, 12, 271. https://doi.org/10.3390/agriculture12020271
Hénault C, Barbier E, Hartmann A, Revellin C. New Insights into the Use of Rhizobia to Mitigate Soil N2O Emissions. Agriculture. 2022; 12(2):271. https://doi.org/10.3390/agriculture12020271
Chicago/Turabian StyleHénault, Catherine, Elodie Barbier, Alain Hartmann, and Cécile Revellin. 2022. "New Insights into the Use of Rhizobia to Mitigate Soil N2O Emissions" Agriculture 12, no. 2: 271. https://doi.org/10.3390/agriculture12020271






