Comparative Transcriptome Analysis of Bt Resistant and Susceptible Strains in Ostrinia furnacalis (Guenée) (Lepidoptera: Crambidae)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioassay and Resistance Ratio
2.2. Midgut Dissection and RNA Extraction
2.3. Library Preparation for Transcriptome Sequencing
2.4. Assembly and Functional Gene Annotation
2.5. Selection of Differentially Expressed Genes among ACB-BtS, ACB-FR and ACB-IeR
2.6. RT-qPCR Analysis
3. Results
3.1. Insect Resistance Levels
3.2. RNA-Seq and Sequence Assembly
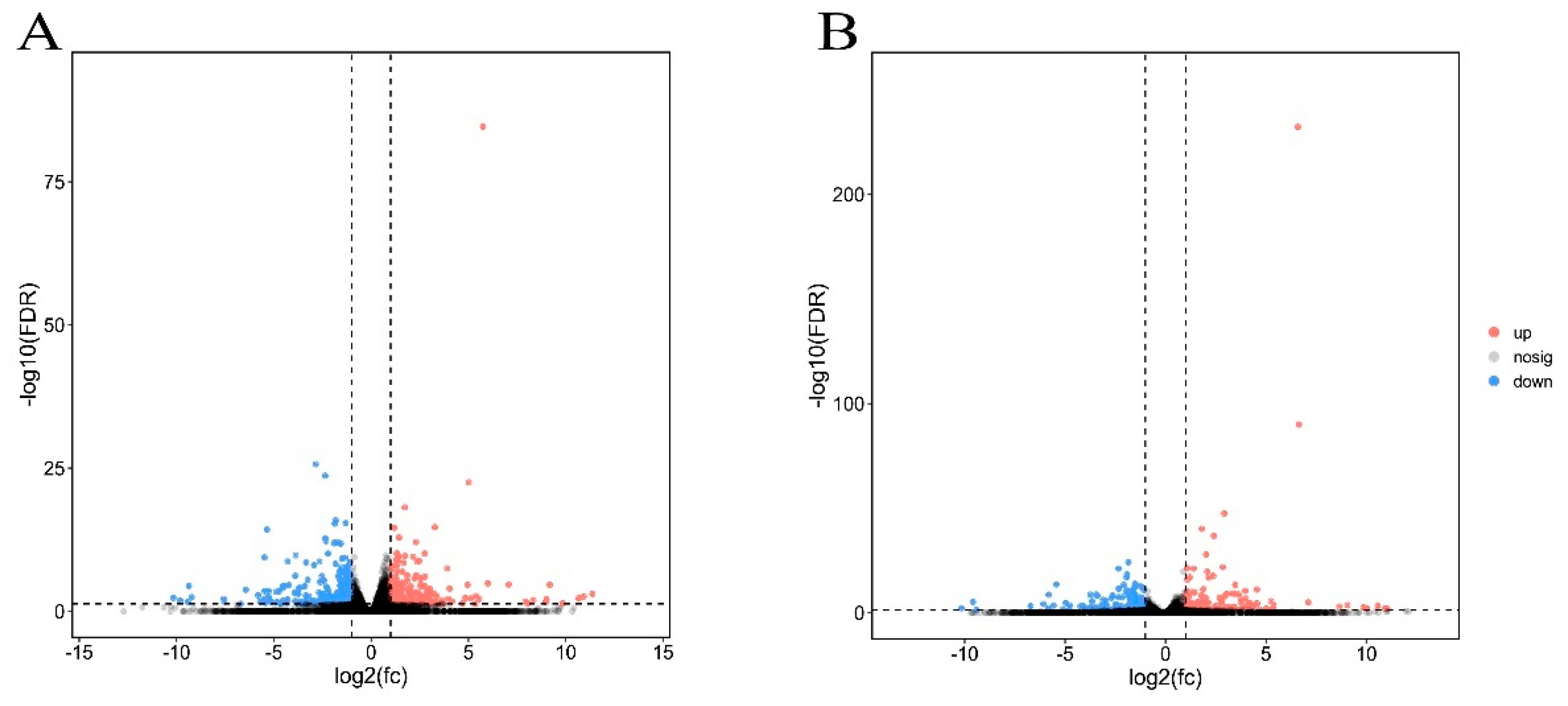
3.3. Differentially Expressed Genes between Cry1F and Cry1Ie-Resistant Strains Compared with Susceptible Strains of O. furnacalis
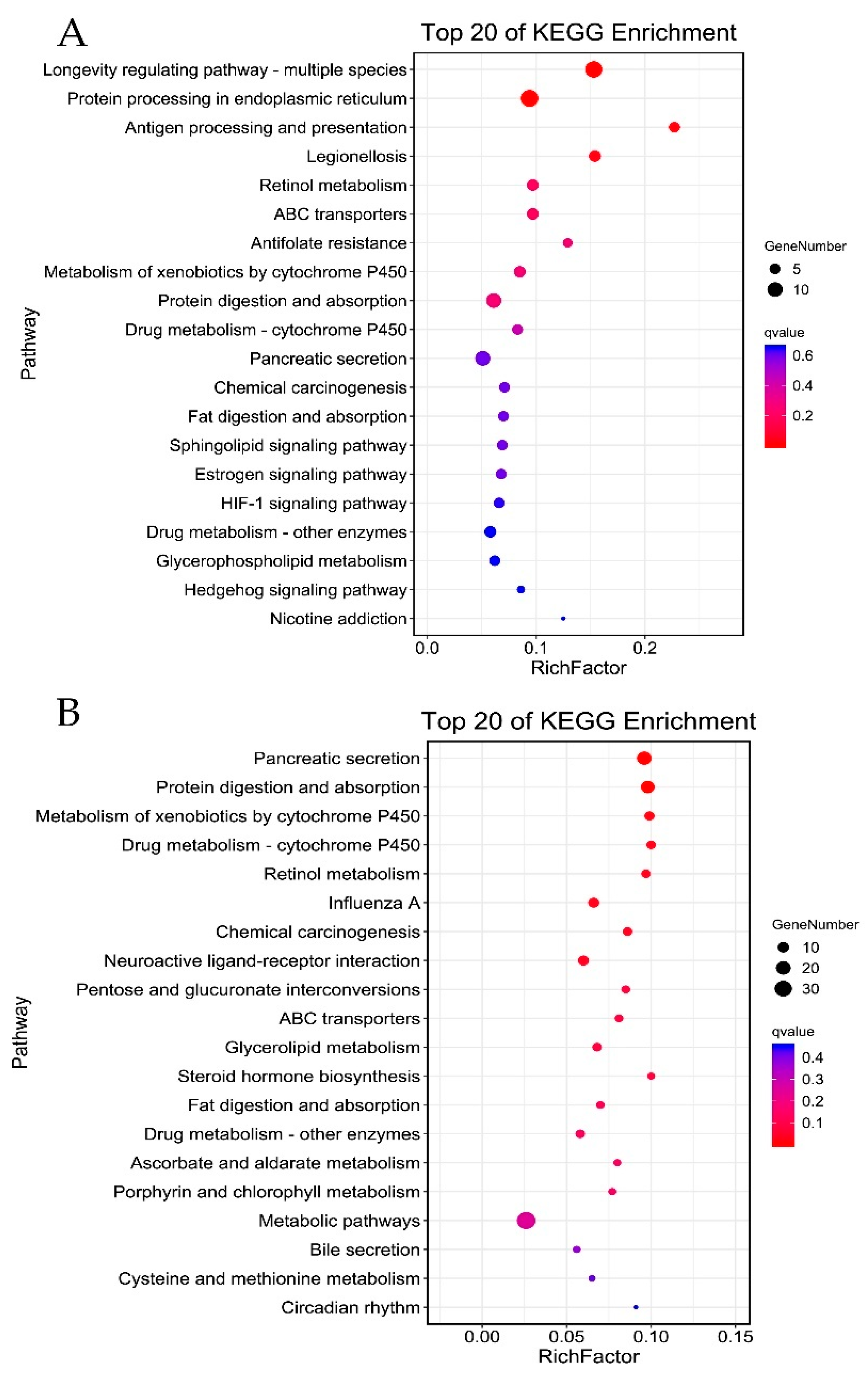
3.4. Gene Ontology and Pathway Enrichment
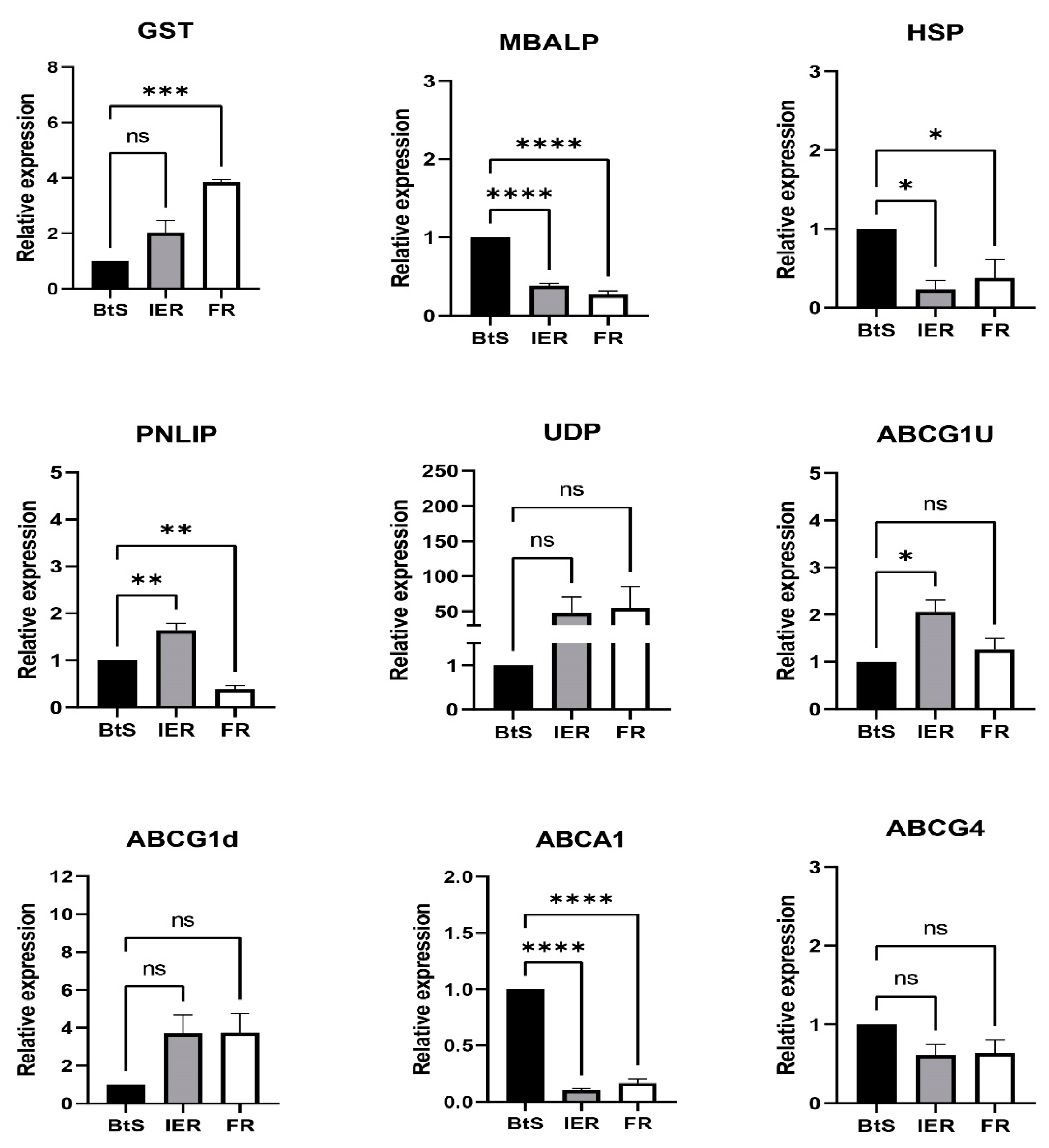
3.5. RT-qPCR Verification of Differentially Expressed Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Wang, Z.; Lu, X.; He, K.; Zhou, D.R. Review of history, present situation and prospect of the Asian maize borer research in China. J. Shenyang Agric. Univ. 2000, 31, 402–412. [Google Scholar]
- Song, L.; Wei, L.; Wang, Z.; He, K.; Cong, B. Effect of infestation by the Asian corn borer together with Fusarium verticillioides on corn yield loss. Acta Phytophy. Sin. 2009, 36, 487–490. [Google Scholar]
- Feng, C.; Huang, J.; Song, Q.; Stanley, D.; Lü, W.; Zhang, Y.; Huang, Y. Parasitization by Macrocentrus cingulum (Hymenoptera: Braconidae) influences expression of prophenoloxidase in Asian corn borer Ostrinia furnacalis. Arch. Insect Biochem. Physiol. 2011, 77, 99–117. [Google Scholar] [CrossRef]
- Patel, S.; Sangeeta, S. Pesticides as the drivers of neuropsychotic diseases, cancers, and teratogenicity among agro-workers as well as general public. Environ. Sci. Pollut. Res. 2019, 26, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Zheng, L.; Quan, Y.; Wei, H. Sublethal pesticide exposure improves resistance to infection in the Asian corn borer. Ecol. Entomol. 2018, 43, 326–331. [Google Scholar] [CrossRef]
- Schnepf, E.; Crickmore, N.; Van Rie, J.; Lereclus, D.; Baum, J.; Feitelson, J.; Zeigler, D.; Dean, D. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 1998, 62, 775–806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jurat-Fuentes, J.L.; Heckel, D.G.; Ferré, J. Mechanisms of resistance to insecticidal proteins from Bacillus thuringiensis. Annu. Rev. Entomol. 2021, 66, 121–140. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.N.; Ferry, N.; Wang, Z.Y.; Zhang, J.; Edwards, M.; Gatehouse, A.; He, K.L. A proteomic approach to study the mechanism of tolerance to Bt toxins in Ostrinia furnacalis larvae selected for resistance to Cry1Ab. Transgenic Res. 2013, 22, 1155–1166. [Google Scholar] [CrossRef] [PubMed]
- Comas, C.; Lumbierres, B.; Pons, X.; Albajes, R. No effects of Bacillus thuringiensis maize on nontarget organisms in the field in southern Europe: A meta-analysis of 26 arthropod taxa. Transgenic Res. 2014, 23, 135–143. [Google Scholar] [CrossRef]
- Morin, S.; Biggs, R.; Sisterson, M.; Shriver, L.; Ellers-Kirk, C.; Higginson, D.; Holley, D.; Gahan, L.; Heckel, D.; Carriere, Y.; et al. Three cadherin alleles associated with resistance to Bacillus thuringiensis in pink bollworm. Proc. Natl. Acad. Sci. USA 2003, 100, 5004–5009. [Google Scholar] [CrossRef] [Green Version]
- Gould, F. Sustainability of Transgenic Insecticidal Cultivars: Integrating Pest Genetics and Ecology. Annu. Rev. Entomol. 1998, 43, 701–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.Q.; Wang, Y.D.; Wang, Z.Y.; Bravo, A.; Soberón, M.; He, K.L. Genetic Basis of Cry1F-Resistance in a Laboratory Selected Asian Corn Borer Strain and Its Cross-Resistance to Other Bacillus thuringiensis Toxins. PLoS ONE 2016, 11, e0161189. [Google Scholar] [CrossRef] [PubMed]
- He, K.L.; Wang, Z.Y.; Zhou, D.R.; Wen, L.P.; Song, Y.Y.; Yao, Z.Y. Evaluation of transgenic Bt corn for resistance to the Asian corn borer (Lepidoptera: Pyralidae). J. Econ. Entomol. 2003, 96, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Gahan, L.; Gould, F.; Heckel, D. Identification of a Gene Associated with Bt Resistance in Heliothis virescens. Science 2001, 293, 857–860. [Google Scholar] [CrossRef] [PubMed]
- Heckel, D.G. How do toxins from Bacillus thuringiensis kill insects? An evolutionary perspective. Arch. Insect Biochem. Physiol. 2020, 104, e21673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vadlamudi, R.; Weber, E.; Ji, I.; Ji, T.; Bulla, L. Cloning and Expression of a Receptor for an Insecticidal Toxin of Bacillus thuringiensis. J. Biol. Chem. 1995, 270, 5490–5494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knight, P.; Crickmore, N.; Ellar, D. The receptor for Bacillus thuringiensis CrylA(c) delta-endotoxin in the brush border membrane of the lepidopteran Manduca sexta is aminopeptidase N. Mol. Microbiol. 1994, 11, 429–436. [Google Scholar] [CrossRef]
- Bravo, A.; Gill, S.S.; Soberon, M. Mode of action of Bacillus thuringiensis Cry and Cyt toxins and their potential for insect control. Toxicon 2007, 49, 423–435. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Dong, X.; Liu, J.; Hu, M.; Zhong, G.; Geng, P.; Yi, X. Molecular Cloning, Expression and Molecular Modeling of Chemosensory Protein from Spodoptera litura and Its Binding Properties with Rhodojaponin III. PLoS ONE 2012, 7, e47611. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.T.; Coates, B.; Wang, Y.Q.; Wang, Y.D.; Bai, S.X.; Wang, Z.Y.; He, K.L. Down-regulation of aminopeptidase N and ABC transporter subfamily G transcripts in Cry1Ab and Cry1Ac resistant Asian corn borer, Ostrinia furnacalis (Lepidoptera: Crambidae). Int. J. Biol. Sci. 2017, 13, 835–851. [Google Scholar] [CrossRef] [Green Version]
- Shabbir, M.Z.; Zhang, T.T.; Wang, Z.Y.; He, K.L. Transcriptome and proteome alternation with resistance to Bacillus thuringiensis Cry1Ah toxin in Ostrinia furnacalis. Front. Physiol. 2019, 10, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.T.; He, M.X.; Gatehouse, A.; Wang, Z.Y.; Edwards, M.; He, K.L. Inheritance Patterns, Dominance and Cross-Resistance of Cry1Ab- and Cry1Ac-Selected Ostrinia furnacalis (Guenée). Toxins 2014, 6, 2694–2707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, D.D.; Jin, M.H.; Yang, Y.C.; Zhang, J.F.; Yang, Y.B.; Liu, K.Y.; Soberón, M.; Bravo, A.; Xiao, Y.T.; Wu, K.M. Synergistic resistance of Helicoverpa armigera to Bt toxins linked to cadherin and ABC transporters mutations. Insect Biochem. Mol. Biol. 2021, 137, 103635. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Yang, J.; Quan, Y.D.; Wang, Z.Y.; Cai, W.Z.; He, K.L. Characterization of Asian corn borer resistance to Bt toxin Cry1Ie. Toxins 2017, 9, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, M.X.; He, K.L.; Wang, Z.Y.; Wang, X.Y.; Li, Q. Selection for Cry1Ie resistance and cross-resistance of the selected strain to other Cry toxins in the Asian corn borer, Ostrinia furnacalis (Lepidoptera: Crambidae). Acta Entomol. Sin. 2013, 56, 1135–1142. [Google Scholar]
- Pereira, E.J.G.; Lang, B.A.; Storer, N.P.; Siegfried, B.D. Selection for Cry1F resistance in the European corn borer and cross-resistance to other Cry toxins. Entomol. Exp. Appl. 2008, 126. [Google Scholar] [CrossRef]
- Hua, G.; Masson, L.; Jurat-Fuentes, J.L.; Schwab, G.; Adang, M.J. Binding analyses of Bacillus thuringiensis Cry δ-endotoxins using brush border membrane vesicles of Ostrinia nubilalis. Appl. Environ. Microbiol. 2001, 67, 872–879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siqueira, H.A.; Moellenbeck, D.; Spencer, T.; Siegfried, B.D. Cross-resistance of Cry1Ab-selected Ostrinia nubilalis (Lepidoptera: Crambidae) to Bacillus thuringiensis δ-endotoxins. J. Econ. Entomol. 2004, 97, 1049–1057. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Jin, F.; Hu, Z.; Chen, H.; Yin, F.; Li, Z.; Dong, X.; Zhang, D.; Ren, S.; Zhang, Z. Transcriptome Analysis of Chlorantraniliprole Resistance Development in the Diamondback Moth Plutella xylostella. PLoS ONE 2013, 8, e72314. [Google Scholar] [CrossRef] [Green Version]
- Lei, Y.Y.; Zhu, X.; Xie, W.; Wu, Q.J.; Wang, S.L.; Guo, Z.J.; Xu, B.Y.; Li, X.C.; Zhou, X.G.; Zhang, Y.J. Midgut transcriptome response to a Cry toxin in the diamondback moth, Plutella xylostella (Lepidoptera: Plutellidae). Gene 2014, 533, 180–187. [Google Scholar] [CrossRef]
- Wu, Y.D. Detection and Mechanisms of Resistance Evolved in Insects to Cry Toxins from Bacillus thuringiensis. Adv. Insect Physiol. 2014, 47, 297–342. [Google Scholar]
- Sayed, A.; Nekl, E.; Siqueira, H.; Wang, H.; Ffrench-Constant, R.; Bagley, M.; Siegfried, B. A novel cadherin-like gene from western corn rootworm, Diabrotica virgifera virgifera (Coleoptera: Chrysomelidae), larval midgut tissue. Insect Mol. Biol. 2007, 16, 591–600. [Google Scholar] [CrossRef] [Green Version]
- Bravo, A.; Likitvivatanavong, S.; Gill, S.; Soberón, M. Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 2011, 41, 423–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabrick, J.; Ponnuraj, J.; Singh, A.; Tanwar, R.; Unnithan, G.; Yelich, A.; Li, X.C.; Carriere, Y.; Tabashnik, B. Alternative Splicing and Highly Variable Cadherin Transcripts Associated with Field-Evolved Resistance of Pink Bollworm to Bt Cotton in India. PLoS ONE 2014, 9, e97900. [Google Scholar] [CrossRef]
- Zhang, H.N.; Tian, W.; Zhao, J.; Jin, L.; Yang, J.; Liu, C.H.; Yang, Y.H.; Wu, S.W.; Wu, K.M.; Cui, J.J.; et al. Diverse genetic basis of field-evolved resistance to Bt cotton in cotton bollworm from China. Proc. Natl. Acad. Sci. USA 2012, 109, 10275–10280. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhang, H.N.; Wang, H.D.; Zhao, S.; Zuo, Y.Y.; Yang, Y.H.; Wu, Y.D. Functional validation of cadherin as a receptor of Bt toxin Cry1Ac in Helicoverpa armigera utilizing the CRISPR/Cas9 system. Insect Biochem. Mol. Biol. 2016, 76, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.X.; Fu, S.; Ma, X.L.; Baxter, S.W.; Vasseur, L.; Xiong, L.; Huang, Y.P.; Yang, G.; You, S.J.; You, M.S. Resistance to Bacillus thuringiensis Cry1Ac toxin requires mutations in two Plutella xylostella ATP-binding cassette transporter paralogs. PLoS Pathog. 2020, 16, e1008697. [Google Scholar] [CrossRef]
- Bretschneider, A.; Heckel, D.; Pauchet, Y. Three toxins, two receptors, one mechanism: Mode of action of Cry1A toxins from Bacillus thuringiensis in Heliothis virescens. Insect Biochem. Mol. Biol. 2016, 76, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Tay, W.; Mahon, R.; Heckel, D.; Walsh, T.; Downes, S.; James, W.; Lee, S.; Reineke, A.; Williams, A.; Gordon, K. Insect Resistance to Bacillus thuringiensis Toxin Cry2Ab Is Conferred by Mutations in an ABC Transporter Subfamily A Protein. PLoS Genet. 2015, 11, e1005534. [Google Scholar] [CrossRef]
- Badenes-Pérez, F.; Morrison, A.; Vogel, H.; Crickmore, N.; Kain, W.; Wang, P.; Heckel, D.; Jiggins, C. Parallel Evolution of Bacillus thuringiensis Toxin Resistance in Lepidoptera. Genetics 2011, 189, 675–679. [Google Scholar]
- Guo, Z.J.; Kang, S.; Zhu, X.; Xia, J.X.; Wu, Q.J.; Wang, S.; Xie, W.; Zhang, Y.J. Down-regulation of a novel ABC transporter gene (Pxwhite) is associated with Cry1Ac resistance in the diamondback moth, Plutella xylostella (L.). Insect Biochem. Mol. Biol. 2015, 59, 30–40. [Google Scholar] [CrossRef]
- Li, H.; Oppert, B.; Higgins, R.; Huang, F.; Buschman, L.; Gao, J.; Zhu, K. Characterization of cDNAs encoding three trypsin-like proteinases and mRNA quantitative analysis in Bt-resistant and-susceptible strains of Ostrinia nubilalis. Insect Biochem. Mol. Biol. 2005, 35, 847–860. [Google Scholar] [CrossRef] [PubMed]
- Tabashnik, B.; Liu, Y.; Unnithan, D.; Carrière, Y.; Dennehy, T.; Morin, S. Shared genetic basis of resistance to Bt toxin Cry1Ac in independent strains of pink bollworm. J. Econ. Entomol. 2004, 97, 721–726. [Google Scholar] [CrossRef]
- Zhang, M.; Wei, J.Z.; Ni, X.Z.; Zhang, J.; Jurat-Fuentes, J.; Fabrick, J.; Carrière, Y.; Tabashnik, B.; Li, X.C. Decreased Cry1Ac activation by midgut proteases associated with Cry1Ac resistance in Helicoverpa zea. Pest Manag. Sci. 2019, 75, 1099–1106. [Google Scholar] [CrossRef]
- Tabashnik, B.; Finson, N.; Johnson, M.; Heckel, D. Cross-resistance to Bacillus thuringiensis toxin CryIF in the diamondback moth (Plutella xylostella). Appl. Environ. Microbiol. 1994, 60, 4627–4629. [Google Scholar] [CrossRef] [Green Version]
- Jackson, R.; Gould, F.; Bradley, J.; Van Duyn, J. Genetic variation for resistance to Bacillus thuringiensis toxins in Helicoverpa zea (Lepidoptera: Noctuidae) in eastern North Carolina. J. Econ. Entomol. 2006, 99, 1790–1797. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Du, L.X.; Liu, C.X.; Gong, L.; Han, L.Z.; Peng, Y.F. RNAi in the striped stem borer, Chilo suppressalis, establishes a functional role for aminopeptidase N in Cry1Ab intoxication. J. Invertebr. Pathol. 2017, 143, 1–10. [Google Scholar] [CrossRef]
- Pavlidi, N.; Vontas, J.; Van Leeuwen, T. The role of glutathione S-transferases (GSTs) in insecticide resistance in crop pests and disease vectors. Curr. Opin. Insect Sci. 2018, 27, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.X.; Xiao, Y.T.; Li, X.C.; Oppert, B.; Tabashnik, B.; Wu, K.M. Cis-mediated down-regulation of a trypsin gene associated with Bt resistance in cotton bollworm. Sci. Rep. 2014, 4, 7219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, Z.; Wang, G.; Xie, Z.; Wang, J.; Zhang, C.; Dong, F.; Chen, C. Identification of novel and differentially expressed microRNAs of dairy goat mammary gland tissues using Solexa sequencing and bioinformatics. PLoS ONE 2012, 7, e49463. [Google Scholar] [CrossRef] [PubMed]
- Gahan, L.J.; Pauchet, Y.; Vogel, H.; Heckel, D.G. An ABC Transporter Mutation Is Correlated with Insect Resistance to Bacillus thuringiensis Cry1Ac toxin. PLoS Genet. 2010, 6, e1001248. [Google Scholar] [CrossRef] [Green Version]
- Feyereisen, R. Insect P450 enzymes. Annu. Rev. Entomol. 1999, 44, 507–533. [Google Scholar] [CrossRef]
- Berge, J.B.; Feyereisen, R.; Amichot, M. Cytochrome P450 Monooxygenases and Insecticide Resistance in Insects. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1998, 353, 1701–1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, F.; Guo, Q.; Shen, B.; Zhu, C. A cluster of CYP6 gene family associated with the major quantitative trait locus is responsible for the pyrethroid resistance in Culex pipiens pallen. Insect Mol. Biol. 2019, 28, 528–536. [Google Scholar] [PubMed]
- Schama, R.; Pedrini, N.; Juárez, M.P.; Nelson, D.R.; Torres, A.Q.; Valle, D.; Mesquita, R.D. Rhodnius prolixus supergene families of enzymes potentially associated with insecticide resistance. Insect Biochem. Mol. Biol. 2016, 69, 91–104. [Google Scholar] [CrossRef] [PubMed]

| Bt Toxin | Insect Strain | LC50 (95% CI) 1 μg/g | Resistance Ratio |
|---|---|---|---|
| Cry1F | ACB-BtS | 0.52 (0.36–0.70) | / |
| ACB-FR | >500.00 | >961.00 | |
| Cry1Ie | ACB-BtS | 1.16 (0.84–1.66) | / |
| ACB-IeR | >1000.00 | >862.07 |
| Samples | Raw Reads | Clean Reads | Q20 (%) | Q30 (%) | GC (%) | Total Mapped |
|---|---|---|---|---|---|---|
| ACB-BtS | 39,687,801 | 39,127,741 | 97.63 | 93.31 | 48.99 | 31,285,151 (79.96%) |
| ACB-FR | 47,222,879 | 46,637,357 | 97.89 | 93.90 | 49.72 | 39,798,320 (85.34%) |
| ACB-IeR | 50,484,709 | 49,893,262 | 98.06 | 94.30 | 49.95 | 41,890,268 (83.96%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Y.; Gao, Q.; Wang, Y.; Wang, Z.; He, K.; Shang, S.; Zhang, T. Comparative Transcriptome Analysis of Bt Resistant and Susceptible Strains in Ostrinia furnacalis (Guenée) (Lepidoptera: Crambidae). Agriculture 2022, 12, 298. https://doi.org/10.3390/agriculture12020298
Lin Y, Gao Q, Wang Y, Wang Z, He K, Shang S, Zhang T. Comparative Transcriptome Analysis of Bt Resistant and Susceptible Strains in Ostrinia furnacalis (Guenée) (Lepidoptera: Crambidae). Agriculture. 2022; 12(2):298. https://doi.org/10.3390/agriculture12020298
Chicago/Turabian StyleLin, Yaling, Qing Gao, Yueqin Wang, Zhenying Wang, Kanglai He, Suqin Shang, and Tiantao Zhang. 2022. "Comparative Transcriptome Analysis of Bt Resistant and Susceptible Strains in Ostrinia furnacalis (Guenée) (Lepidoptera: Crambidae)" Agriculture 12, no. 2: 298. https://doi.org/10.3390/agriculture12020298
APA StyleLin, Y., Gao, Q., Wang, Y., Wang, Z., He, K., Shang, S., & Zhang, T. (2022). Comparative Transcriptome Analysis of Bt Resistant and Susceptible Strains in Ostrinia furnacalis (Guenée) (Lepidoptera: Crambidae). Agriculture, 12(2), 298. https://doi.org/10.3390/agriculture12020298








