Microbiological and Toxicological Evaluation of Fermented Forages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Microbiological Sample Preparation
2.3. Dry Weight Determination
2.4. Mycotoxin Measurement with HPLC Methods
2.5. Identification of Bacteria Isolated from Fermented Forages by MALDI-TOF MS Method
2.6. Toxin Resistance of Isolated Bacterial Cultures
2.7. Statistical Analysis
3. Results
3.1. Toxicological Characterization of the Fermented Forages
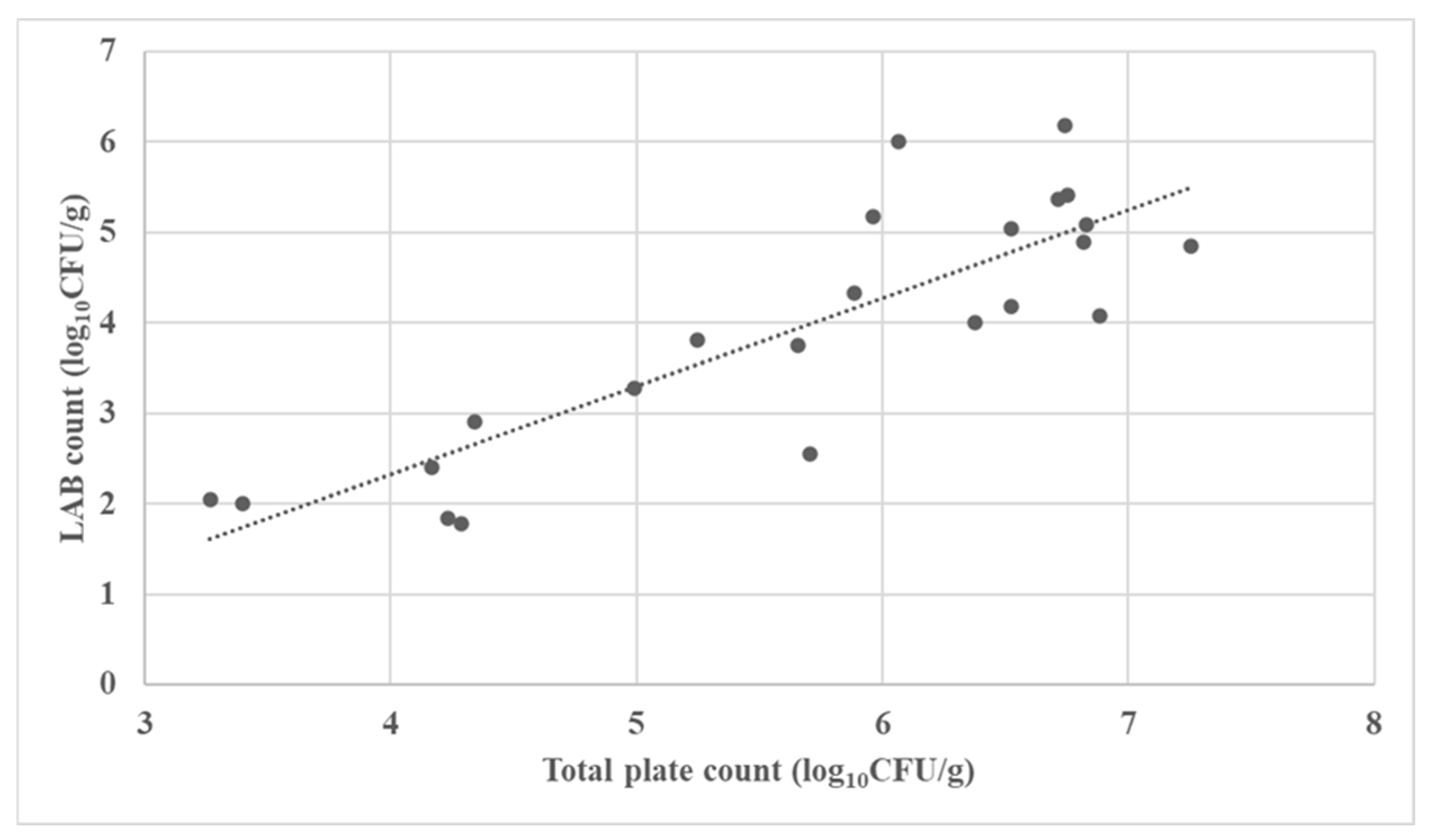
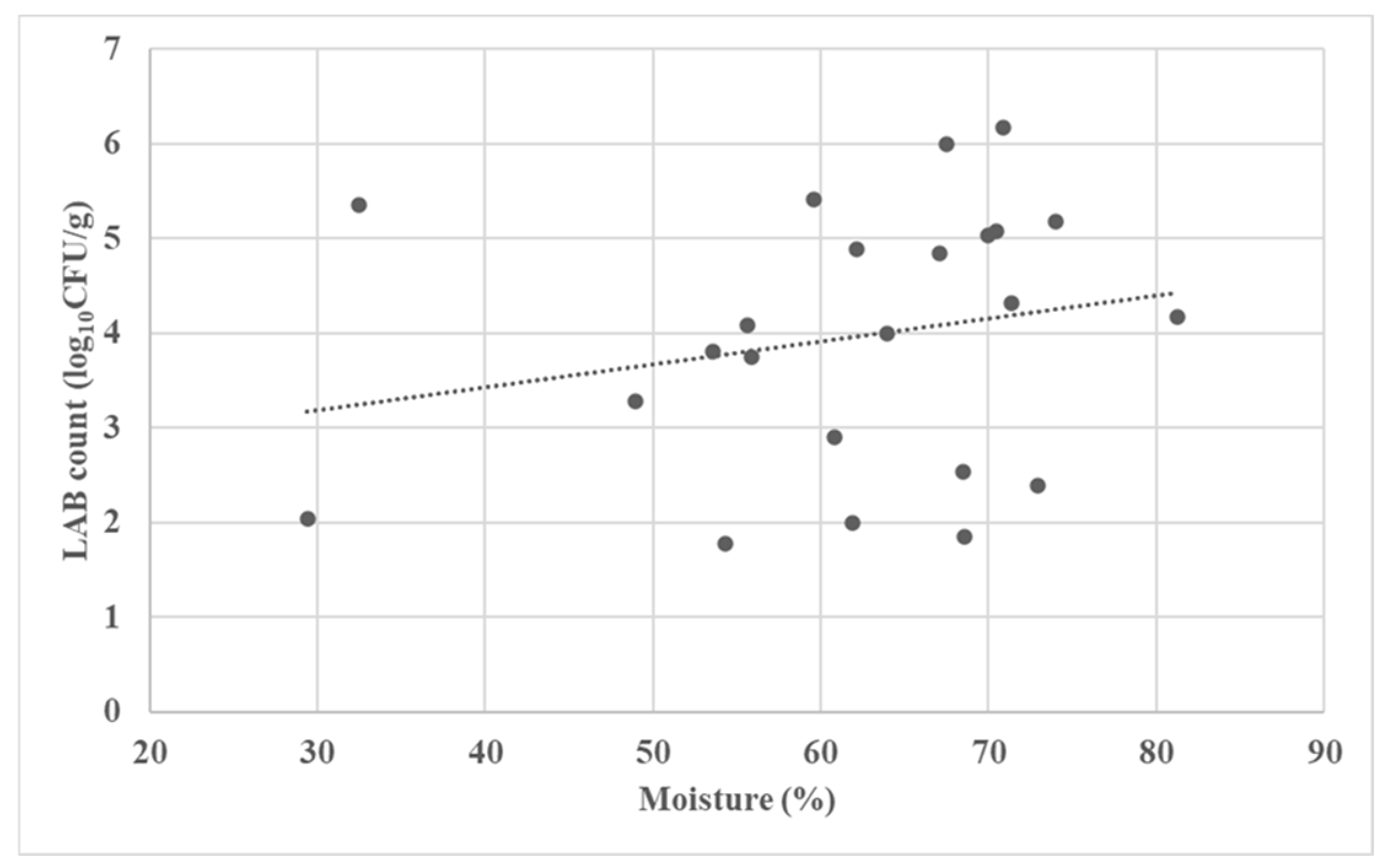
3.2. Microbial Characterization of the Fermented Forages
3.3. Identification of the Isolated Bacteria
3.4. Mycotoxin Resistance of the Isolated Bacteria
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, D.H.; Lee, K.D.; Choi, K.C. Role of LAB in silage fermentation: Effect on nutritional quality and organic acid production—An overview. AIMS Agric. Food 2021, 6, 216–234. [Google Scholar] [CrossRef]
- Lin, C.; Bolsen, K.K.; Brent, B.E.; Fung, D.Y.C. Epiphytic lactic acid bacteria succession during the pre-ensiling and ensiling periods of alfalfa and maize. J. Appl. Bacteriol. 1992, 73, 375–387. [Google Scholar] [CrossRef]
- Adesogan, A.T.; Newman, Y.C. Silage Harvesting, Storing, and Feeding. IFAS Extension, University of Florida, Gainesville. 2014. Available online: http://edis.ifas.ufl.edu/pdf/AG/AG180\AG180-Dz70s07cdf.pdf (accessed on 19 May 2017).
- Weidenbörner, M. Encyclopedia of Food Mycotoxins; Springer: Berlin/Heidelberg, Germany, 2001; p. 173. [Google Scholar]
- Rodrigues, I.; Handl, J.; Binder, E.M. Mycotoxin occurrence in commodities, feeds and feed ingredients sourced in the Middle East and Africa. Food Addit. Contam. Part B 2011, 4, 168–179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Döll, S.; Danicke, S. The Fusarium toxins deoxynivalenol (DON) and zearalenone (ZON) in animal feeding. J. Prev. Vet. Med. 2011, 102, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.A.M.; Gonzáles Pereyra, M.L.; Keller, K.M.; Alonso, V.A.; Oliveira, A.A.; Almeida, T.X.; Barbosa, T.S.; Nunes, L.M.T.; Cavaglieri, L.R.; Rosa, C.A.R. Fungal and mycotoxins contamination in corn silage: Monitoring risk before and after fermentation. J. Stored Prod. Res. 2013, 52, 42–47. [Google Scholar] [CrossRef]
- El-Zawahri, M.; Moubasher, A.; Morad, M.; El-Kady, I. Mutagenic effect of aflatoxin B₁. Ann. Nutr. Aliment. 1977, 31, 859–866. [Google Scholar]
- Richard, E.; Heutte, N.; Sage, L.; Pottier, D.; Bouchart, V.; Lebailly, P.; Garon, D. Toxigenic fungi and mycotoxins in mature corn silage. Food Chem. Toxicol. 2007, 45, 2420–2425. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Ghilardelli, F.; Atzori, A.S.; Zara, S.; Novak, B.; Faas, J.; Fancello, F. Co-occurrence of regulated and emerging mycotoxins in corn silage: Relationships with fermentation quality and bacterial communities. Toxins 2021, 13, 232. [Google Scholar] [CrossRef]
- Zachariasova, M.; Dzuman, Z.; Veprikova, Z.; Hajkova, K.; Jiru, M.; Vaclavikova, M.; Zachariasova, A.; Pospichalova, M.; Florian, M.; Hajslova, J. Occurrence of multiple mycotoxins in European feedingstuffs, assessment of dietary intake by farm animals. Anim. Feed Sci. Technol. 2014, 193, 124–140. [Google Scholar] [CrossRef]
- Panasiuk, L.; Jedziniak, P.; Pietruszka, K.; Piatkowska, M.; Bocian, L. Frequency and levels of regulated and emerging mycotoxins in silage in Poland. Mycotoxin Res. 2019, 35, 17–25. [Google Scholar] [CrossRef] [Green Version]
- Peles, F.; Sipos, P.; Kovács, S.; Győri, Z.; Pócsi, I.; Pusztahelyi, T. Biological control and mitigation of aflatoxin contamination in commodities. Toxins 2021, 13, 104. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Yu, M.; Dong, F.; Shi, J.; Xu, J. Esterase activity inspired selection and characterization of zearalenone degrading bacteria Bacillus pumilus ES-21. Food Control 2017, 77, 57–64. [Google Scholar] [CrossRef]
- ISO 4833-1 (2014); Microbiology of the Food Chain. Horizontal Method for the Enumeration of Microorganisms. Part 1: Colony count at 30 Degrees C by the Pour Plate Technique. International Standard Organization: Geneva, Switzerland, 2014.
- ISO 15214 (1998); Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of Mesophilic Lactic Acid Bacteria—Colony-Count Technique at 30 °C. International Standard Organization: Geneva, Switzerland, 1998.
- ISO 7954 (1999); Microbiology. General Guidance for Enumeration of Yeasts and Moulds. Colony Count Technique at 25 °C. International Standard Organization: Geneva, Switzerland, 1999.
- ISO 6496 (1999); Animal Feeding Stuffs—Determination of Moisture and Other Volatile Matter Content. International Standard Organization: Geneva, Switzerland, 1999.
- Garon, D.; Richard, E.; Sage, L.; Bouchart, V.; Pottier, D.; Lebailly, P. Mycoflora and multimycotoxin detection in corn silage: Experimental study. J. Agric. Food Chem. 2006, 54, 3479–3484. [Google Scholar] [CrossRef]
- Ogunade, I.M.; Martinez-Tuppia, C.; Queiroz, O.C.M.; Jiang, Y.; Drouin, P.; Wu, F.; Vyas, D.; Adesogan, A.T. Silage review: Mycotoxins in silage: Occurrence, effects, prevention, and mitigation. J. Dairy Sci. 2018, 101, 4034–4059. [Google Scholar] [CrossRef] [PubMed]
- Jensen, T.; de Boevre, M.; Preußke, N.; de Saeger, S.; Birr, T.; Verreet, J.; Sönnichsen, F. Evaluation of high-resolution mass spectrometry for the quantitative analysis of mycotoxins in complex feed matrices. Toxins 2019, 11, 531. [Google Scholar] [CrossRef] [Green Version]
- Boudra, H.; Morgavi, D.P. Reduction in fusarium toxin levels in corn silage with low dry matter and storage time. J. Agric. Food Chem. 2008, 56, 4523–4528. [Google Scholar] [CrossRef]
- Driehuis, F.; Spanjer, M.C.; Scholten, J.M.; Te Giffel, M.C. Occurrence of mycotoxins in maize, grass and wheat silage for dairy cattle in the Netherlands. Food Addit. Contam. Part B Surveill. 2008, 1, 41–50. [Google Scholar] [CrossRef]
- Cavallarin, L.; Tabacco, E.; Antoniazzi, S.; Borreani, G. Aflatoxin accumulation in whole crop maize silage as a result of aerobic exposure. J. Sci. Food Agric. 2011, 91, 2419–2425. [Google Scholar] [CrossRef]
- Huerta-Treviño, A.; Dávila-Aviña, J.E.; Sánchez, E.; Heredia, N.; García, S. Occurrence of mycotoxins in alfalfa (Medicago sativa L.), sorghum [Sorghum bicolor (L.) Moench], and grass (Cenchrus ciliaris L.) retailed in the state of Nuevo León, México. Agrociencia 2016, 50, 825–836. [Google Scholar]
- Kosicki, R.; Błajet-Kosicka, A.; Grajewski, A.; Twaruzek, M. Multiannual mycotoxin survey in feed materials and feeding stuffs. Anim. Feed Sci. Technol. 2016, 215, 165–180. [Google Scholar] [CrossRef]
- Borreani, G.; Tabacco, E.; Schmidt, R.J.; Holmes, B.J.; Muck, R.E. Silage review: Factors affecting dry matter and quality losses in silages. J. Dairy Sci. 2018, 101, 3952–3979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porto, M.C.W.; Kuniyoshi, T.M.; Azevedo, P.O.S.; Vitolo, M.; Oliveira, R.P.S. Pediococcus spp.: An important genus of lactic acid bacteria and pediocin producers. Biotechnol. Adv. 2017, 35, 361–374. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.S.; Yang, P.; Wu, A.B.; Zuo, D.Y.; He, W.J.; Guo, M.W.; Huang, T.; Li, H.P.; Liao, Y.C. Variation in the microbiome, trichothecenes, and aflatoxins in stored wheat grains in Wuhan, China. Toxins 2018, 10, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, H.; Shuai, Y.; Ran, Q.; Yan, Y.; Wang, X.; Li, D.; Cai, Y.; Zhang, X. The microbiome and metabolome of Napier grass silages prepared with screened lactic acid bacteria during ensiling and aerobic exposure. Anim. Feed Sci. Technol. 2020, 269, 114673. [Google Scholar] [CrossRef]
- Fabiszewska, A.U.; Zielińska, K.J.; Wróbel, B. Trends in designing microbial silage quality by biotechnological methods using lactic acid bacteria inoculants: A minireview. World J. Microbiol. Biotechnol. 2019, 35, 76. [Google Scholar] [CrossRef] [Green Version]
- Zielińska, K.J.; Fabiszewska, A.U.; Stefańska, I. Different aspects of Lactobacillus inoculants on the improvement of quality and safety of alfalfa silage. Chil. J. Agric. Res. 2015, 75, 298–306. [Google Scholar] [CrossRef] [Green Version]
- Queiroz, O.C.M.; Kim, S.C.; Adesogan, A.T. Effect of treatment with a mixture of bacteria and fibrolytic enzymes on the quality and safety of corn silage infested with different levels of rust. J. Dairy Sci. 2012, 95, 5285–5291. [Google Scholar] [CrossRef] [Green Version]
- Iglesias, C.; Bach, A.; Devant, M.; Adelantado, C.; Calvo, M.A. The effect of Lactobacillus buchneri inoculation on corn silages conservation. In Proceedings of the XI Jornadas sobre Producción Animal, Gobierno de Aragón, Servicio de Investigación Agroalimentaria, Zaragoza, Spain, 11–12 May 2005; pp. 611–613. [Google Scholar]
- Schnürer, J.; Magnusson, J. Antifungal lactic acid bacteria as biopreservatives. Trends Food Sci. Technol. 2005, 16, 70–78. [Google Scholar] [CrossRef]
- Dogi, C.A.; Fochesato, A.; Armando, R.; Pribull, B.; de Souza, M.M.; da Silva Coelho, I.; Araújo de Melo, D.; Dalcero, A.; Cavaglieri, L. Selection of lactic acid bacteria to promote an efficient silage fermentation capable of inhibiting the activity of Aspergillus parasiticus and Fusarium gramineraum and mycotoxin production. J. Appl. Microbiol. 2013, 114, 1650–1660. [Google Scholar] [CrossRef]
- Ragoubi, C.; Quintieri, L.; Greco, D.; Mehrez, A.; Maatouk, I.; D’Ascanio, V.; Landoulsi, A.; Avantaggiato, G. Mycotoxin removal by Lactobacillus spp. and their application in animal liquid feed. Toxins 2021, 13, 185. [Google Scholar] [CrossRef]
- Adebo, O.A.; Njobeh, P.B.; Mavumengwana, V. Degradation and detoxification of AFB1 by Staphylocococcus warneri, Sporosarcina sp. and Lysinibacillus fusiformis. Food Control 2016, 68, 92–96. [Google Scholar] [CrossRef]
| No. | Fermented Forages | AF (µg/kg) | OTA (µg/kg) | DON (mg/kg) | ZEA (mg/kg) |
|---|---|---|---|---|---|
| 1 | Corn silage | <0.01 | <0.5 | <0.1 | <0.1 |
| 2 | Corn silage | <0.01 | <0.5 | <0.1 | <0.1 |
| 3 | Corn silage | <0.01 | <0.5 | <0.1 | <0.1 |
| 4 | Corn silage | <0.01 | <0.5 | <0.1 | <0.1 |
| 5 | Corn silage | B1 1.15 | <0.5 | 3.25 | <0.1 |
| 6 | Corn silage | <0.01 | <0.5 | 1.07 | <0.1 |
| 7 | Corn silage | <0.01 | <0.5 | 1.35 | <0.1 |
| 8 | Corn silage | <0.01 | <0.5 | 0.47 | <0.1 |
| 9 | Corn silage | <0.01 | <0.5 | 1.03 | <0.1 |
| 10 | Corn silage | <0.01 | <0.5 | 2.57 | 0.42 |
| 11 | Corn silage | <0.01 | <0.5 | 1.19 | <0.1 |
| 12 | Corn silage | <0.01 | <0.5 | 1.24 | <0.1 |
| 13 | Corn silage | G2 0.045 B1 < 0.01 | <0.5 | <0.1 | <0.1 |
| 14 | Corn silage | <0.01 | <0.5 | <0.1 | <0.1 |
| 15 | Alfalfa silage | <0.01 | <0.5 | 0.45 | <0.1 |
| 16 | Alfalfa silage | <0.01 | 0.80 | <0.1 | <0.1 |
| 17 | Alfalfa haylage | <0.01 | 2.52 | <0.1 | <0.1 |
| 18 | Alfalfa haylage | G2 24.23 B1 < 0.01 | 27.57 | <0.1 | <0.1 |
| 19 | Rye silage | <0.01 | <0.5 | <0.1 | <0.1 |
| 20 | Rye haylage | G2 0.301 B1 < 0.01 | <0.5 | <0.1 | <0.1 |
| 21 | Triticale haylage | <0.01 | <0.5 | <0.1 | <0.1 |
| 22 | Triticale haylage | <0.01 | <0.5 | <0.1 | <0.1 |
| No. | Fermented Forages | Total Aerobic Plate Count (log10 CFU/g) | LAB Count (log10 CFU/g) | Mold Count (log10 CFU/g) |
|---|---|---|---|---|
| 1 | Corn silage | 3.265 | 2.041 | 2.204 |
| 2 | Corn silage | 6.068 | 6.000 | 1.000 |
| 3 | Corn silage | 6.740 | 6.176 | 1.000 |
| 4 | Corn silage | 6.526 | 4.176 | 1.000 |
| 5 | Corn silage | 3.397 | 2.000 | 1.000 |
| 6 | Corn silage | 5.247 | 3.813 | 1.000 |
| 7 | Corn silage | 5.656 | 3.756 | 3.477 |
| 8 | Corn silage | 6.830 | 5.079 | 1.602 |
| 9 | Corn silage | 4.991 | 3.279 | 1.000 |
| 10 | Corn silage | 6.755 | 5.415 | 6.079 |
| 11 | Corn silage | 6.884 | 4.079 | 1.699 |
| 12 | Corn silage | 4.341 | 2.908 | 1.000 |
| 13 | Corn silage | 4.235 | 1.845 | 1.000 |
| 14 | Corn silage | 5.965 | 5.176 | 4.146 |
| 15 | Alfalfa silage | 6.522 | 5.041 | 3.176 |
| 16 | Alfalfa silage | 7.254 | 4.851 | 1.000 |
| 17 | Alfalfa haylage | 6.377 | 4.000 | 1.301 |
| 18 | Alfalfa haylage | 4.289 | 1.778 | 1.000 |
| 19 | Rye silage | 5.886 | 4.322 | 1.000 |
| 20 | Rye haylage | 4.170 | 2.398 | 1.477 |
| 21 | Tritikale haylage | 6.715 | 5.362 | 2.903 |
| 22 | Tritikale haylage | 5.705 | 2.544 | 1.000 |
| Source | Identified Organism | Highest Score Value per Isolate | Growth Inhibition by AFB1 | Growth Inhibition by OTA |
|---|---|---|---|---|
| corn silage | Lactiplantibacillus plantarum | 2.292 | 0% | 43% |
| 2.261 | 0% | 45% | ||
| 2.317 | 0% | 66% | ||
| 2.295 | 0% | 67% | ||
| 2.343 | 0% | 33% | ||
| 2.297 | 0% | 48% | ||
| 2.242 | 0% | 69% | ||
| 2.379 | 0% | 52% | ||
| 2.382 | 0% | 64% | ||
| corn silage | Lactiplantibacillus pentosus | 2.377 | 0% | 64% |
| 2.211 | 0% | 61% | ||
| 2.235 | 0% | 58% | ||
| 2.181 | 0% | 47% | ||
| 2.271 | 0% | 57% | ||
| 2.283 | 0% | 65% | ||
| alfalfa silage | Lactiplantibacillus pentosus | 2.273 | 0% | 61% |
| 2.338 | 0% | 80% | ||
| 2.391 | 0% | 39% | ||
| corn silage | Latilactobacillus curvatus | 2.327 | 13% | 77% |
| corn silage | Lacticaseibacillus paracasei | 2.473 | 0% | 41% |
| corn silage | Levilactobacillus brevis | 2.226 | 0% | 57% |
| 2.226 | 24% | 25% | ||
| corn silage | Pediococcus pentosaceus | 2.214 | 0% | 57% |
| rye silage | Pediococcus acidilactici | 2.21 | 0% | 53% |
| 2.207 | 0% | 53% | ||
| corn silage | Loigolactobacillus coryniformis | 2.239 | 12% | 64% |
| corn silage | Klebsiella pneumoniae | 2.39 | 0% | 86% |
| corn silage | Bacillus thuringiensis | 2.137 | 0% | 65% |
| corn silage | Lysinibacillus fusiformis | 1.959 | 0% | 73% |
| alfalfa haylage | Rummeliibacillus suwonensis | 1.956 | 0% | 54% |
| alfalfa haylage | Lysinibacillus boronitolerans | 2.258 | 0% | 46% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adácsi, C.; Kovács, S.; Pócsi, I.; Győri, Z.; Dombrádi, Z.; Pusztahelyi, T. Microbiological and Toxicological Evaluation of Fermented Forages. Agriculture 2022, 12, 421. https://doi.org/10.3390/agriculture12030421
Adácsi C, Kovács S, Pócsi I, Győri Z, Dombrádi Z, Pusztahelyi T. Microbiological and Toxicological Evaluation of Fermented Forages. Agriculture. 2022; 12(3):421. https://doi.org/10.3390/agriculture12030421
Chicago/Turabian StyleAdácsi, Cintia, Szilvia Kovács, István Pócsi, Zoltán Győri, Zsuzsanna Dombrádi, and Tünde Pusztahelyi. 2022. "Microbiological and Toxicological Evaluation of Fermented Forages" Agriculture 12, no. 3: 421. https://doi.org/10.3390/agriculture12030421
APA StyleAdácsi, C., Kovács, S., Pócsi, I., Győri, Z., Dombrádi, Z., & Pusztahelyi, T. (2022). Microbiological and Toxicological Evaluation of Fermented Forages. Agriculture, 12(3), 421. https://doi.org/10.3390/agriculture12030421








