Morpho-Physiological and Stress-Related Gene Expression of Rice Varieties in Response to Salinity Stress at Early Vegetative Stage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Seed Sterilisation and Treatments
2.2. Germination Observation and Seedlings Physiological Analyses
2.3. Plant Growth and Treatment
2.4. Measurement of Morpho-Physiological Characters
2.4.1. Growth Measurement
2.4.2. Gaseous Exchange
2.4.3. Total Chlorophyll Content
2.4.4. Relative Water Content
2.5. Gene Expression Analyses
2.5.1. RNA Extraction and First-Strand cDNA Synthesis
2.5.2. Semi Quantitative RT-PCR
2.6. Statistical Analyses
3. Results
3.1. Effect of Salinity on MR219 Germination
3.2. Salinity Effects on Plant Morpho-Physiology at Vegetative Stage
3.2.1. Growth Performance
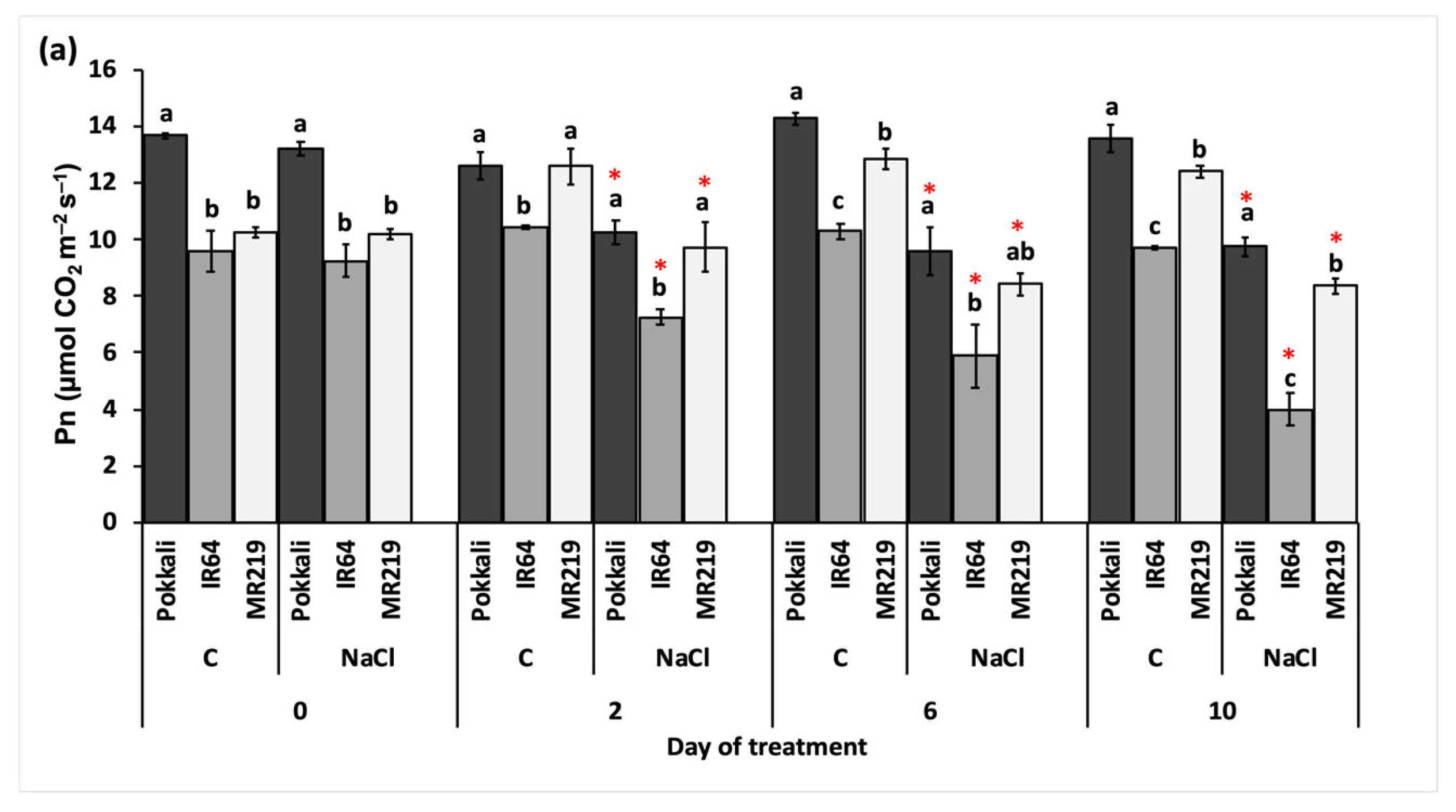
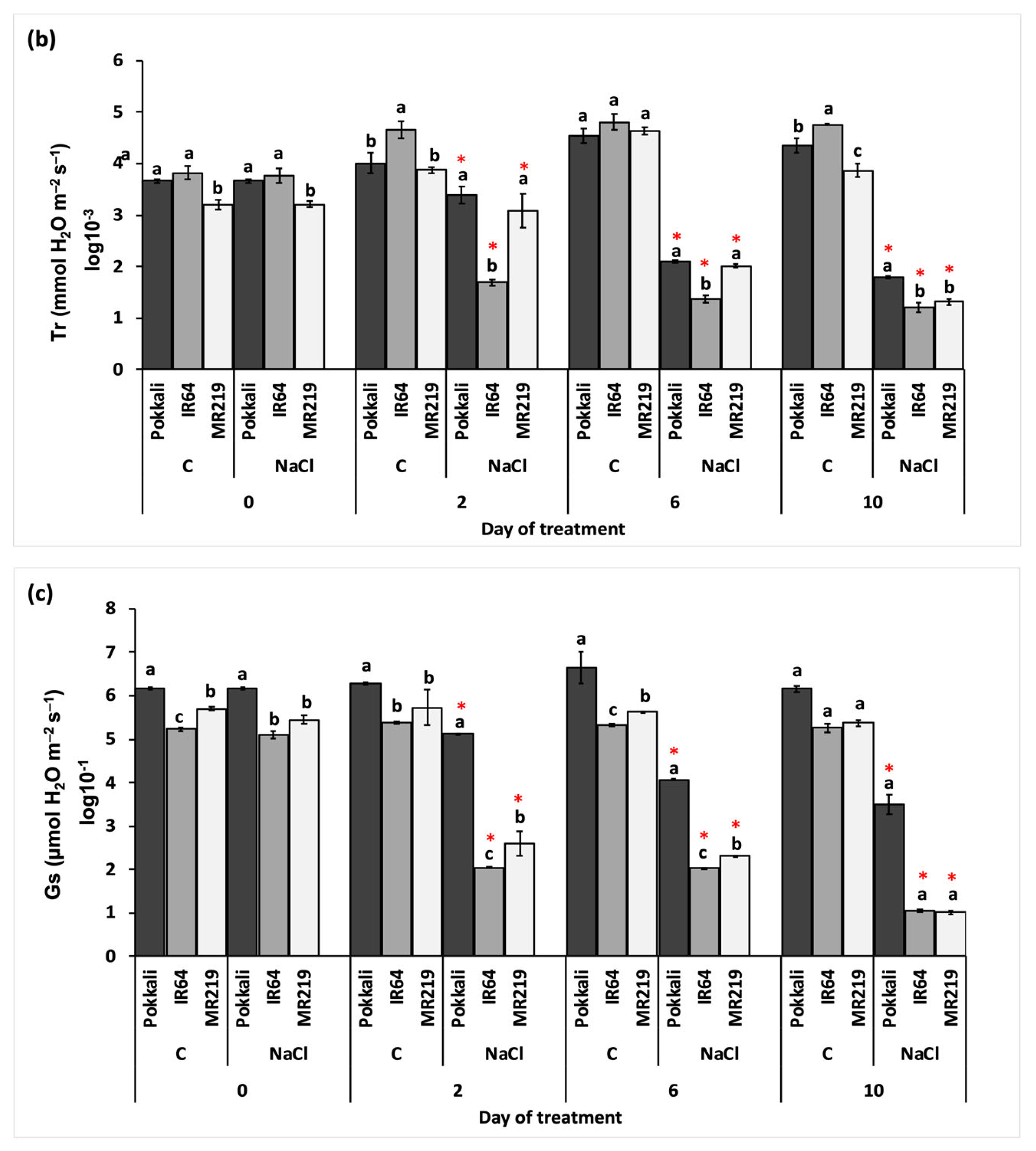
3.2.2. Rate of Gaseous Exchange
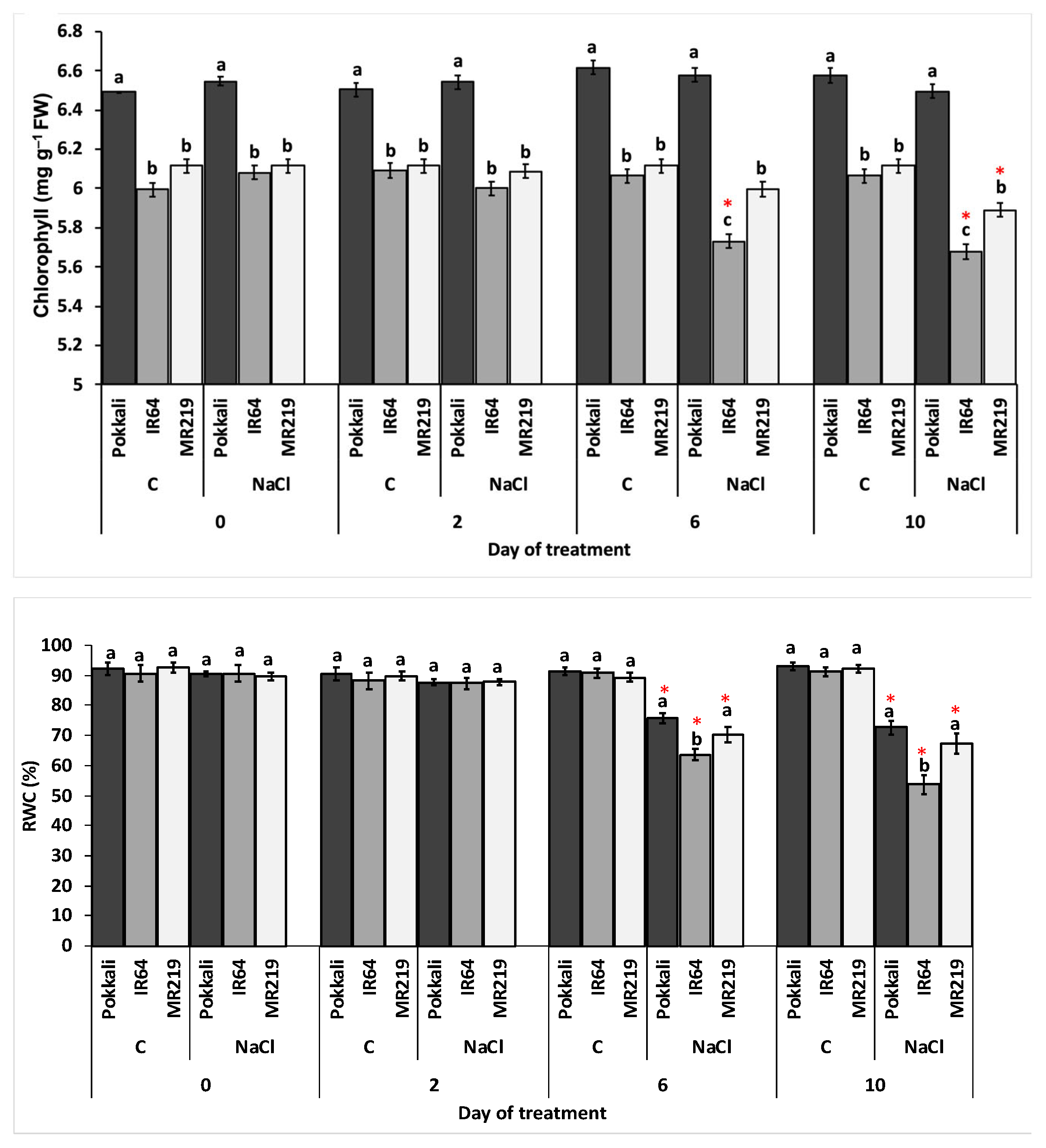
3.2.3. Total Chlorophyll and Relative Water Content
3.2.4. Identification of Phenotypic Correlation Identification Using PCA
3.3. Semi-Quantitative qRT-PCR Analysis of Selected Gene Expression under Salinity Stress
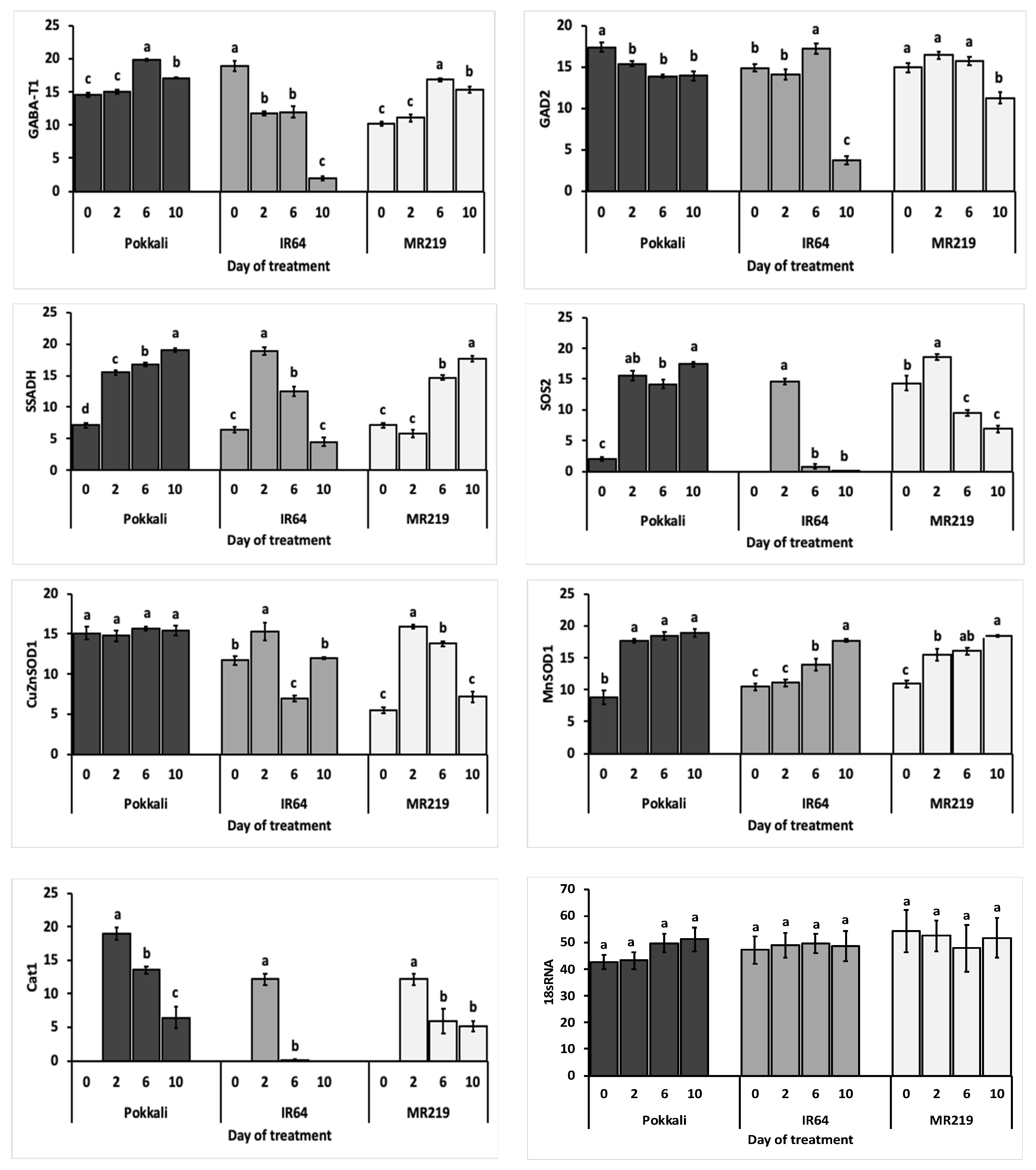
3.3.1. Expression of GABA Shunt Genes
3.3.2. Expression of the Ion Transport Gene
3.3.3. Expression of the Antioxidant Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ghosh, B.; Ali, M.N. Response of rice under salinity stress: A review update. Rice Res. Open Access 2016, 4, 2–9. [Google Scholar] [CrossRef] [Green Version]
- Rathna-Priya, T.S.; Eliazer-Nelson, A.R.L.; Ravichandran, K.; Antony, U. Nutritional and functional properties of coloured rice varieties of South India: A review. J. Ethn. Foods 2019, 6, 11. [Google Scholar] [CrossRef] [Green Version]
- Riaz, M.; Arif, M.S.; Ashraf, M.A.; Mahmood, R.; Yasmeen, T.; Shakoor, M.B.; Shahzad, S.M.; Ali, M.; Saleem, I.; Arif, M.; et al. A comprehensive review on rice responses and tolerance to salt stress. In Advances in Rice Research for Abiotic Stress Tolerance; Woodhead Publishing: Cambridge, UK, 2019; pp. 133–158. [Google Scholar]
- Shrivastava, P.; Kumar, R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J. Biol. Sci. 2015, 22, 123–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.; Sharma, P.K. Soil salinity and food security in India. Front. Sustain. Food Syst. 2020, 4, 533781. [Google Scholar] [CrossRef]
- Machado, R.; Serralheiro, R. Soil salinity: Effect on vegetable crop growth. Management practices to prevent and mitigate soil salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Shaban, M. Effect of water and temperature on seed germination and emergence as a seed hydrothermal time model. Int. J. Adv. Biol. Biomed. Res. 2013, 1, 1686–1691. [Google Scholar]
- Quan, R.; Wang, J.; Hui, J.; Bai, H.; Lyu, X.; Zhu, Y.; Zhang, H.; Zhang, Z.; Li, S.; Huang, R. Improvement of salt tolerance using wild rice genes. Front. Plant Sci. 2018, 8, 2269. [Google Scholar] [CrossRef] [Green Version]
- Sultana, N.; Ikeda, T.; Itoh, R. Effect of NaCl salinity on photosynthesis and dry matter accumulation in developing rice grains. Environ. Exp. Bot. 1999, 42, 211–220. [Google Scholar] [CrossRef]
- Irakoze, W.; Prodjinoto, H.; Nijimbere, S.; Rufyikiri, G.; Lutts, S. NaCl and Na2SO4 salinities have different impact on photosynthesis and yield-related parameters in rice (Oryza sativa L.). Agronomy 2020, 10, 864. [Google Scholar] [CrossRef]
- Hossain, M.S.; Shah, J.S.A. Present scenario of global salt affected soils, its management and Importance of salinity research. Int. Res. J. Biol. Sci. Perspect. 2019, 1, 1–3. [Google Scholar]
- Papageorgiou, G.C.; Murata, N. The unusually strong stabilizing effects of glycine betaine on the structure and function of the oxygen-evolving Photosystem II complex. Photosynth. Res. 1995, 44, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.A.; Sarkhosh, A.; Khan, N.; Balal, R.M.; Ali, S.; Rossi, L.; Gomez, C.; Mattson, N.; Nasim, W.; Garcia-Sanchez, F. Insights into the physiological and biochemical impacts of salt stress on plant growth and development. Agronomy 2020, 10, 938. [Google Scholar] [CrossRef]
- Che-Othman, M.H.; Millar, A.H.; Taylor, N.L. Connecting salt stress signalling pathways with salinity-induced changes in mitochondrial metabolic processes in C3 plants. Plant Cell Environ. 2017, 40, 2875–2905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basu, S.; Roychoudhury, A. Transcript profiling of stress-responsive genes and metabolic changes during salinity in indica and japonica rice exhibit distinct varietal difference. Physiol. Plant 2021, 173, 1434–1447. [Google Scholar] [CrossRef] [PubMed]
- Keisham, M.; Mukherjee, S.; Bhatla, S.C. Mechanisms of sodium transport in plants—progresses and challenges. Int. J. Mol. Sci. 2018, 19, 647. [Google Scholar] [CrossRef] [Green Version]
- Brindha, C.; Vasantha, S.; Raja, A.K.; Tayade, A.S. Characterization of the Salt Overly Sensitive pathway genes in sugarcane under salinity stress. Physiol. Plant 2021, 171, 677–687. [Google Scholar] [CrossRef]
- Tang, J.; Wang, S.Q.; Hu, K.D.; Huang, Z.Q.; Li, Y.H.; Han, Z.; Chen, X.Y.; Hu, L.Y.; Yao, G.F.; Zhang, H. Antioxidative capacity is highly associated with the storage property of tuberous roots in different sweetpotato cultivars. Sci. Rep. 2019, 9, 11141. [Google Scholar] [CrossRef] [Green Version]
- Zuraida, A.R.; Zulkifli, A.S.; Habibuddin, H.; Naziah, B. Regeneration of Malaysian rice variety MR 219 via somatic embryogenesis. J. Trop. Agric. Fd. Sc. 2012, 39, 167–177. [Google Scholar]
- Talei, D.; Valdiani, A.; Maziah, M.; Mohsenkhah, M. Germination response of MR 219 rice variety to different exposure times and periods of 2450 MHz microwave frequency. Sci. World J. 2013, 2013, 408026. [Google Scholar] [CrossRef]
- Ahmed, F.; Rafii, M.Y.; Ismail, M.R.; Juraimi, A.S.; Rahim, H.A.; Latif, M.A.; Hasan, M.M.; Tanweer, F.A. The addition of submergence-tolerant Sub1 gene into high yielding MR219 rice variety and analysis of its BC2F3 population in terms of yield and yield contributing characters to select advance lines as a variety. Biotechnol. Biotechnol. Equip. 2016, 30, 853–863. [Google Scholar] [CrossRef] [Green Version]
- Zulkarnain, W.M.; Ismail, M.R.; Ashrafuzzaman, M.; Saud, H.M.; Haroun, I.C. Growth, physiological and biochemical responses of malaysia rice cultivars to water stress. Pertanika J. Trop. Agric. Sci. 2009, 32, 323–333. [Google Scholar]
- Yadav, P.; Singh, P.; Yadav, B.; Pandey, S.; Singh, V.; Dwivedi, D.K. Evaluation of rice genotypes for salinity tolerance at seedling stage. J. Pharmacogn. Phytochem. 2018, 7, 202–205. [Google Scholar]
- Lakra, N.; Kaur, C.; Singla-Pareek, S.L.; Pareek, A. Mapping the ‘early salinity response’ triggered proteome adaptation in contrasting rice genotypes using iTRAQ approach. Rice 2019, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Noorzuraini, A.R.S.; Ramdzan, O.M.; Idayu, A.R.N.; Hafiz, M.S.M. Evaluating the rice germplasm for salinity tolerance based on phenotypic traits. In IOP Conference Series: Earth and Environmental Science, Proceedings of the International Conference on Biodiversity 2020, Melaka, Malaysia, 4–5 November 2020; IOP Publishing: Bristol, UK, 2021; Volume 736. [Google Scholar]
- Polash, M.A.S.; Sakil, M.A.; Tahjib-Ul-Arif, M.; Hossain, M.A. Effect of salinity on osmolytes and relative water content of selected rice genotypes. Trop. Plant Res. 2018, 5, 227–232. [Google Scholar] [CrossRef]
- Hakim, M.A.; Juraimi, A.S.; Begum, M.; Hanafi, M.M.; Ismail, M.R.; Selamat, A. Effect of salt stress on germination and early seedling growth of rice (Oryza sativa L.). Afr. J. Biotechnol. 2010, 9, 1911–1918. [Google Scholar]
- Bado, S.; Forster, B.P.; Ghanim, A.M.A.; Jankowicz-Cieslak, J.; Berthold, G.; Liu, L.X. Protocol for screening for salt tolerance in rice. In Protocols for Pre-Field Screening of Mutants for Salt Tolerance in Rice, Wheat and Barley; Springer: Cham, Switzerland, 2016; pp. 21–31. [Google Scholar]
- Hu, X.; Tanaka, A.; Tanaka, R. Simple extraction methods that prevent the artifactual conversion of chlorophyll to chlorophyllide during pigment isolation from leaf samples. Plant Methods 2013, 9, 19. [Google Scholar] [CrossRef] [Green Version]
- Shibghatallah, M.A.H.; Khotimah, S.N.; Suhandono, S.; Viridi, S.; Kesuma, T. Measuring leaf chlorophyll concentration from its color: A way in monitoring environment change to plantations. In AIP Conference Proceedings, Proceedings of the Padjadjaran International Physics Symposium 2013 (PIPS-2013), Sumedang, Indonesia, 7–9 May 2013; AIP Publishing: Long Island, NY, USA, 2013; Volume 1554, pp. 210–213. [Google Scholar]
- Turner, N.C. Techniques and experimental approaches for the measurement of plant water status. Plant Soil 1981, 58, 339–366. [Google Scholar] [CrossRef]
- Liu, L.; Xia, W.; Li, H.; Zeng, H.; Wei, B.; Han, S.; Yin, C. Salinity inhibits rice seed germination by reducing α-amylase activity via decreased bioactive gibberellin content. Front. Plant Sci. 2018, 9, 275. [Google Scholar] [CrossRef]
- Fogliatto, S.; Serra, F.; Patrucco, L.; Milan, M.; Vidotto, F. Effect of different water salinity levels on the germination of imazamox-resistant and sensitive weedy rice and cultivated rice. Agronomy 2019, 9, 658. [Google Scholar] [CrossRef] [Green Version]
- Panuccio, M.R.; Jacobsen, S.E.; Akhtar, S.S.; Muscolo, A. Effect of saline water on seed germination and early seedling growth of the halophyte quinoa. AoB Plants 2014, 6, plu047. [Google Scholar] [CrossRef]
- Zhang, H.; Irving, L.J.; McGill, C.; Matthew, C.; Zhou, D.; Kemp, P. The effects of salinity and osmotic stress on barley germination rate: Sodium as an osmotic regulator. Annals Bot. 2010, 106, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.R.; Tahjib-Ul-Arif, M.; Polash, M.A.S.; Hossen, M.Z.; Hossain, M.A. Physiological mechanisms of exogenous calcium on alleviating salinity-induced stress in rice (Oryza sativa L.). Physiol. Mol. Biol. Plants 2019, 25, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Singh, A.; Pandey, A.K.; Singh, A.K.; Singh, R.; Singh, A.; Yadav, R.K. Effect salinity on germination percentage (%) and seed vigour index of rice (Oryza sativa L.). J. Pharmacogn. Phytochem. 2020, 9, 1130–1133. [Google Scholar] [CrossRef]
- Singh, R.K.; Kota, S.; Flowers, T.J. Salt tolerance in rice: Seedling and reproductive stage QTL mapping come of age. Theor. Appl. Genet. 2021, 134, 3495–3533. [Google Scholar] [CrossRef]
- Prajapati, K.; Modi, H.A. The importance of potassium in plant growth—A review. Indian J. Plant Sci. 2012, 1, 2319–3824. [Google Scholar]
- Kakar, N.; Jumaa, S.H.; Redoña, E.D.; Warburton, M.L.; Reddy, K.R. Evaluating rice for salinity using pot-culture provides a systematic tolerance assessment at the seedling stage. Rice 2019, 12, 57. [Google Scholar] [CrossRef] [Green Version]
- Asch, F.; Wopereis, M.C.S. Responses of field-grown irrigated rice cultivars to varying levels of floodwater salinity in a semi-arid environment. Field Crop Res. 2001, 70, 127–137. [Google Scholar] [CrossRef]
- Albacete, A.; Ghanem, M.E.; Martínez-Andújar, C.; Acosta, M.; Sánchez-Bravo, J.; Martínez, V.; Lutts, S.; Dodd, I.C.; Perez-Alfocea, F. Hormonal changes in relation to biomass partitioning and shoot growth impairment in salinized tomato (Solanum lycopersicum L.) plants. J. Exp. Bot. 2008, 59, 4119–4131. [Google Scholar] [CrossRef]
- Sánchez-Blanco, M.J.; Rodríguez, P.; Olmos, E.; Morales, M.A.; Torrecillas, A. Differences in the effects of simulated sea aerosol on water relations, salt content, and leaf ultrastructure of Rock-Rose plants. J. Environ. Qual. 2004, 33, 1369–1375. [Google Scholar] [CrossRef] [Green Version]
- Acosta-Motos, J.R.; Ortuño, M.F.; Bernal-Vicente, A.; Diaz-Vivancos, P.; Sanchez-Blanco, M.J.; Hernandez, J.A. Plant responses to salt stress: Adaptive mechanisms. Agronomy 2017, 7, 18. [Google Scholar] [CrossRef] [Green Version]
- Dalton, F.N.; Maggio, A.; Piccinni, G. Simulation of shoot chloride accumulation: Separation of physical and biochemical processes governing plant salt tolerance. Plant Soil 2000, 219, 1–11. [Google Scholar] [CrossRef]
- Kazemi, S.; Eshghizadeh, H.R.; Zahedi, M. Responses of four rice varieties to elevated CO2 and different salinity levels. Rice Sci. 2018, 25, 142–151. [Google Scholar] [CrossRef]
- Siregar, M.P.A.; Hanum, C.; Siregar, L.A.M.; Tistama, R. Morphological and physiological performance of brown rice (Oryza nivara L.) under salinity stress. SABRAO J. Breed. Genet. 2021, 53, 228–239. [Google Scholar]
- Seo, D.H.; Seomun, S.; Choi, Y.D.; Jang, G. Root development and stress tolerance in rice: The key to improving stress tolerance without yield penalties. Int. J. Mol. Sci. 2020, 21, 1807. [Google Scholar] [CrossRef] [Green Version]
- Safitri, H.; Purwoko, B.S.; Dewi, I.S.; Ardie, S.W. Salinity tolerance of several rice genotypes at seedling stage. Indones. J. Agric. Sci. 2018, 18, 63. [Google Scholar] [CrossRef] [Green Version]
- Maggio, A.; Raimondi, G.; Martino, A.; De Pascale, S. Salt stress response in tomato beyond the salinity tolerance threshold. Environ. Exp. Bot. 2007, 59, 276–282. [Google Scholar] [CrossRef]
- Pattanagul, W.; Thitisaksakul, M. Effect of salinity stress on growth and carbohydrate metabolism in three rice (Oryza sativa L.) cultivars differing in salinity tolerance. Indian J. Exp. Biol. 2008, 46, 736–742. [Google Scholar]
- Song, X.J.; Matsuoka, M. Bar the windows: An optimized strategy to survive drought and salt adversities. Genes Dev. 2009, 23, 1709–1713. [Google Scholar] [CrossRef] [Green Version]
- Safdar, H.; Amin, A.; Shafiq, Y.; Ali, A.; Yasin, R. A review: Impact of salinity on plant growth. Nat. Sci. 2019, 17, 34–40. [Google Scholar]
- Xu, N.; Chu, Y.; Chen, H.; Li, X.; Wu, Q.; Jin, L.; Wang, G.; Huang, J. Rice transcription factor OsMADS25 modulates root growth and confers salinity tolerance via the ABA–mediated regulatory pathway and ROS scavenging. PLoS Genet. 2018, 14, e100766. [Google Scholar] [CrossRef] [Green Version]
- Kerstiens, G.; Tych, W.; Robinson, M.F.; Mansfield, T.A. Sodium-related partial stomatal closure and salt tolerance of Aster tripolium. New Phytol. 2002, 153, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Y.; Yin, Z.; Jiang, J.; Zhang, M.; Guo, X.; Ye, Z.; Zhao, Y.; Xiong, H.; Zhang, Z.; et al. OsASR5 enhances drought tolerance through a stomatal closure pathway associated with ABA and H2O2 signalling in rice. Plant Biotechnol. J. 2017, 15, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Sharma, P.K. Effect of NaCl on chlorophyll fluorescence and thylakoid membrane proteins in leaves of salt sensitive and tolerant rice (Oryza sativa L) varieties. J. Stress Physiol. Biochem. 2021, 17, 35–44. [Google Scholar]
- Wang, X.; Wang, W.; Huang, J.; Peng, S.; Xiong, D. Diffusional conductance to CO2 is the key limitation to photosynthesis in salt-stressed leaves of rice (Oryza sativa). Physiol. Plant 2018, 163, 45–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahman, A.; Nahar, K.; Al-Mahmud, J.; Hasanuzzaman, M.; Hossain, M.S.; Fujita, M. Salt stress tolerance in rice: Emerging role of exogenous phytoprotectants. In Advances in International Rice Research; IntechOpen: Rijeka, Croatia, 2017; pp. 139–174. [Google Scholar]
- Jogawat, A. Osmolytes and their role in abiotic stress tolerance in plants. In Molecular Plant Abiotic Stress: Biology and Biotechnology; Wiley: Hoboken, NJ, USA, 2019; pp. 91–104. [Google Scholar]
- Razzaque, S.; Elias, S.M.; Haque, T.; Biswas, S.; Jewel, G.M.N.A.; Rahman, S.; Weng, X.; Ismail, A.M.; Walia, H.; Juenger, T.E.; et al. Gene Expression analysis associated with salt stress in a reciprocally crossed rice population. Sci. Rep. 2019, 9, 8249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheteiwy, M.S.; Shao, H.; Qi, W.; Hamoud, Y.A.; Shaghaleh, H.; Khan, N.U.; Yang, R.; Tang, B. GABA-alleviated oxidative injury induced by salinity, osmotic stress and their combination by regulating cellular and molecular signals in rice. Int. J. Mol. Sci. 2019, 20, 5709. [Google Scholar] [CrossRef] [Green Version]
- Challabathula, D.; Analin, B.; Mohanan, A.; Bakka, K. Differential modulation of photosynthesis, ROS and antioxidant enzyme activities in stress-sensitive and -tolerant rice cultivars during salinity and drought upon restriction of COX and AOX pathways of mitochondrial oxidative electron transport. J. Plant Physiol. 2022, 268, 153583. [Google Scholar] [CrossRef]
- Che-Othman, M.H.; Jacoby, R.P.; Millar, A.H.; Taylor, N.L. Wheat mitochondrial respiration shifts from the tricarboxylic acid cycle to the GABA shunt under salt stress. New Phytol. 2020, 225, 1166–1180. [Google Scholar] [CrossRef]
- Renault, H.; Roussel, V.; El Amrani, A.; Arzel, M.; Renault, D.; Bouchereau, A.; Deleu, C. The Arabidopsis pop2-1mutant reveals the involvement of GABA transaminase in salt stress tolerance. BMC Plant Biol. 2010, 10, 20. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Liu, Z.; Fan, Y.; Liu, C.; Wang, H.; Li, Y.; Xin, Y.; Gai, Y.; Ji, Z. Characterization of GABA-transaminase gene from mulberry (Morus multicaulis) and its role in salt stress tolerance. Genes 2022, 13, 501. [Google Scholar] [CrossRef]
- Su, N.; Wu, Q.; Chen, J.; Shabala, L.; Mithöfer, A.; Wang, H.; Qu, M.; Yu, M.; Cui, J.; Shabala, S. GABA operates upstream of H+-ATPase and improves salinity tolerance in Arabidopsis by enabling cytosolic K+ retention and Na+ exclusion. J. Exp. Bot. 2019, 70, 6349–6361. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Kang, Z.; Zhu, K.; Zhao, D.; Yuan, Y.; Yang, S.; Zhen, W.; Hu, X. RBOH1-dependent H2O2 mediates spermine-induced antioxidant enzyme system to enhance tomato seedling tolerance to salinity–alkalinity stress. Plant Physiol. Biochem. 2021, 164, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Golnari, S.; Vafaee, Y.; Nazari, F.; Ghaderi, N. Gamma-aminobutyric acid (GABA) and salinity impacts antioxidative response and expression of stress-related genes in strawberry cv. Aromas. Braz. J. Bot. 2021, 44, 639–651. [Google Scholar] [CrossRef]
- Bobrovskikh, A.; Zubairova, U.; Kolodkin, A.; Doroshkov, A. Subcellular compartmentalization of the plant antioxidant system: An integrated overview. PeerJ 2020, 8, e9451. [Google Scholar] [CrossRef]
- Corpas, F.J.; González-Gordo, S.; Palma, J.M. Plant peroxisomes: A factory of reactive species. Front. Plant Sci. 2020, 11, 853. [Google Scholar] [CrossRef]
- Tounsi, S.; Feki, K.; Kamoun, Y.; Saïdi, M.N.; Jemli, S.; Ghorbel, M.; Alcon, C.; Brini, F. Highlight on the expression and the function of a novel MnSOD from diploid wheat (T. monococcum) in response to abiotic stress and heavy metal toxicity. Plant Physiol. Biochem. 2019, 142, 384–394. [Google Scholar] [CrossRef]
- Shafi, A.; Mir, M.A.; Zahoor, I. Effect of salinity stress on germination, tolerance and antioxidant response in Arabidopsis thaliana overexpressing Cu/Zn-SOD. Int. J. Life Sci. Res. 2017, 5, 69–78. [Google Scholar]
- Lee, H.-J.; Yang, H.Y.; Choi, J.I. Study of functional verification to abiotic stress through antioxidant gene transformation of Pyropia yezoensis (Bangiales, Rhodophyta) APX and MnSOD in Chlamydomonas. J. Microbiol. Biotechnol. 2018, 28, 1217–1224. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S.; Mukherjee, A.; Rastogi, R.P.; Verma, J.P. Salt-tolerant plant growth-promoting Bacillus pumilus strain JPVS11 to enhance plant growth attributes of rice and improve soil health under salinity stress. Microbiol. Res. 2021, 242, 126616. [Google Scholar] [CrossRef]
- Ighodaro, O.; Akinloye, O. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 2017, 54, 287–293. [Google Scholar] [CrossRef] [Green Version]
- Poli, Y.; Nallamothu, V.; Balakrishnan, D.; Ramesh, P.; Desiraju, S.; Mangrauthia, S.K.; Violeti, S.R.; Neelamraju, S. Increased catalase activity and maintenance of Photosystem II distinguishes high-yield mutants from low-yield mutants of rice var. Nagina22 under low-phosphorus stress. Front. Plant Sci. 2018, 9, 1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarifth Shafika, K.; Noraziyah, A.A.S.; Muhd Hafiz, C.O.; Shakri, T.; Tan, L.W.; Noor Liyana, S.; Isa, N.M.; Rahman, Z.A.; Zainal, Z. Morpho-physiology and antioxidant enzyme activities of transgenic rice plant overexpressing ABP57 under reproductive stage drought condition. Agronomy 2020, 10, 1530. [Google Scholar]
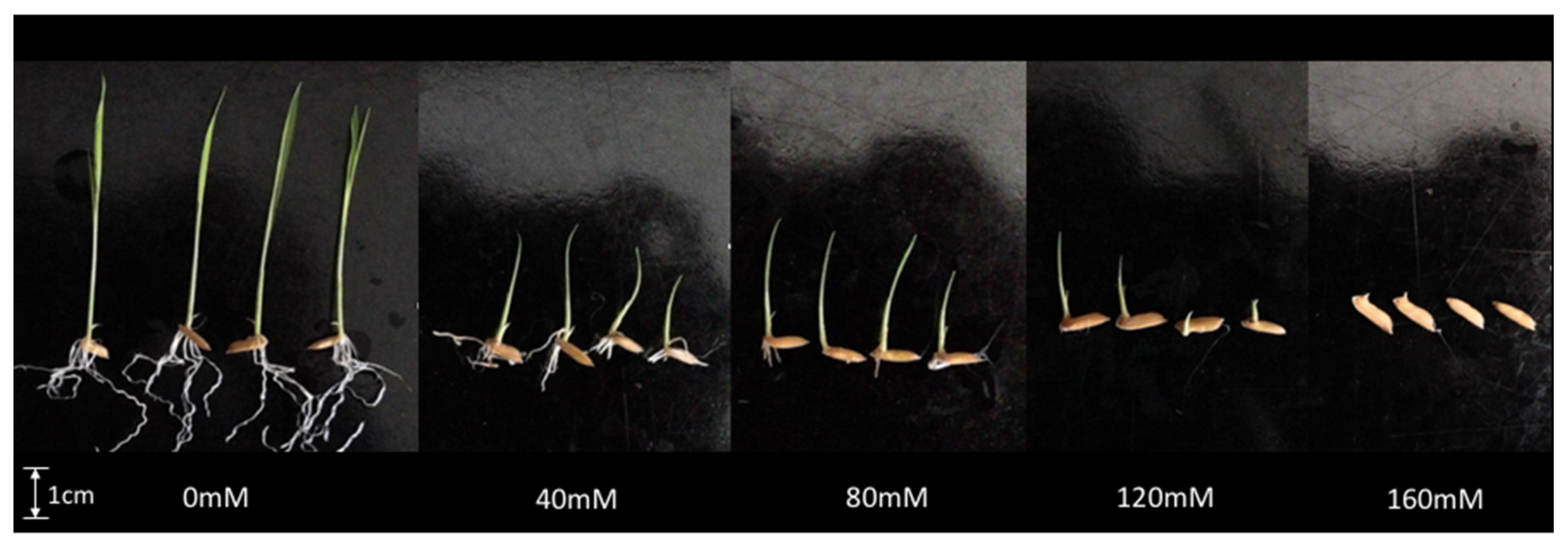

| NaCl (mM) | SG (d−1) | GE (%) | FGP (%) | Shoot (cm) | Root (cm) | RWC (%) |
|---|---|---|---|---|---|---|
| 0 | 5.07 (±1.12) a | 73.33 (±15.28) b | 73.33 (±15.28) b | 5.0 (±1.6) a | 5.5 (±1.0) a | 82.80 (±3.68) a,b |
| 40 | 5.93 (±0.46) a | 86.67 (±5.77) a | 90.00 (±0.00) a | 2.0 (±0.9) b | 5.5 (±1.1) a | 91.93 (±8.63) a |
| 80 | 5.75 (±0.15) a | 85.00 (±5.00) a,b | 90.00 (±0.00) a | 1.9 (±0.6) b | 2.0 (±0.2) b | 88.11 (±2.12) a |
| 120 | 3.97 (±0.38) b | 56.67 (±11.55) c | 66.67 (±5.77) b | 1.3 (±0.9) b,c | 0.5 (±0.1) c | 77.84 (±6.71) b |
| 160 | 0.33 (±0.31) c | 3.33 (±3.33) d | 6.67 (±5.77) c | 0.1 (±0.0) c | 0.0 (±0.0) c | 77.49 (±6.09) b |
| Mean | 4.21 | 61.00 | 65.33 | 2.1 | 2.7 | 86.63 |
| CV (%) | 51.95 | 53.86 | 49.72 | 95.90 | 93.15 | 9.38 |
| Source of Variation | df | Mean Square | |||||
|---|---|---|---|---|---|---|---|
| PH (cm) | SR | B(g) | LA (cm2) | LN | NN | ||
| Replication (R) | 3 | 1.84 | 0.21 | 0.01 | 0.72 | 0.04 | 0.09 |
| Variety (V) | 2 | 6692.61 * | 11.43 * | 0.24 * | 2876.16 * | 3.82 * | 4.51 * |
| Day of treatment (T) | 3 | 2431.19 * | 2.69 * | 0.51 * | 972.27 * | 35.20 * | 2.12 |
| Salinity (S) | 1 | 602.00 * | 0.10 | 0.10 | 667.82 * | 4.59 | 3.76 |
| V×T | 6 | 127.57 * | 0.14 | 0.06 * | 91.11 * | 0.35 | 0.50 |
| V×S | 2 | 5.18 | 0.26 | 0.06 | 10.83 | 0.78 | 0.32 |
| T×S | 3 | 190.21 * | 0.03 | 0.07 * | 228.86 * | 0.59 | 0.59 |
| V×T×S | 6 | 11.41 | 0.08 | 0.04 * | 9.26 | 0.36 | 0.11 |
| Error | 69 | 6.45 | 0.04 | 0.01 | 2.24 | 0.33 | 0.27 |
| Day | Variety | PH (cm) | SR | B (g) | LA (cm2) | LN | NN | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | S | C | S | C | S | C | S | C | S | C | S | ||
| 0 | Pokkali | 64.6 (±1.2) a | 66.1 (±2.9) a | 2.64 (±0.21) a | 2.72 (±0.21) a | 0.23 (±0.02) a | 0.22 (±0.03) a | 18.6 (±0.6) a | 19.3 (±1.4) a | 5.5 (±0.6) a | 5.5 (±0.6) a | 6.5 (±0.6) a | 6.5 (±0.6) a |
| IR64 | 46.4 (±1.5) b | 45.6 (±1.9) b | 1.68 (±0.24) b | 1.71 (±0.16) b | 0.19 (±0.02) a | 0.19 (±0.03) a | 9.5 (±0.7) b | 9.1 (±1.0) b | 5.8 (±0.5) a | 5.8 (±0.5) a | 7.0 (±0.0) a | 7.0 (±0.0) a | |
| MR219 | 46.0 (±1.9) b | 45.7 (±1.5) b | 1.90 (±0.17) b | 1.91 (±0.45) b | 0.21 (±0.05) a | 0.23 (±0.04) a | 9.3 (±0.9) b | 9.1 (±0.8) b | 5.8 (±0.5) a | 5.5 (±0.6) a | 6.5 (±0.6) a | 6.5 (±0.6) a | |
| Mean | 52.3 | 52.4 | 2.07 | 2.10 | 0.21 | 0.21 | 12.4 | 12.5 | 5.7 | 5.6 | 6.7 | 6.7 | |
| CV (%) | 17.50 | 19.56 | 22.65 | 25.12 | 16.03 | 16.62 | 36.84 | 41.09 | 8.69 | 9.22 | 7.39 | 7.39 | |
| 2 | Pokkali | 67.4 (±1.2) a | 67.3 (±1.7) a | 2.58 (±0.19) a | 2.69 (±0.19) a | 0.27 (±0.02) a | 0.25 (±0.03) a | 24.9 (±0.7) a | 24.9 (±1.0) a | 7.5 (±0.6) a | 7.0 (±0.0) b | 6.5 (±0.6) a | 6.0 (±0.0) a |
| IR64 | 48.2 (±1.5) b | 47.1 (±1.9) b | 1.75 (±0.25) b | 1.71 (±0.14) b | 0.23 (±0.02) a | 0.22 (±0.03) a | 10.4 (±0.7) b | 9.8 (±1.0) b | 8.0 (±0.0) a | 8.0 (±0.0) a | 7.0 (±0.0) a | 7.0 (±0.0) a | |
| MR219 | 47.5 (±1.9) b | 47.3 (±1.5) b | 1.91 (±0.17) b | 1.92 (±0.44) b | 0.25 (±0.05) a | 0.26 (±0.04) a | 10.2 (±0.9) b | 9.9 (±0.8) b | 7.8 (±0.5) a | 7.3 (±0.5) b | 6.8 (±0.5) a | 6.3 (±0.5) a | |
| Mean | 54.4 | 53.9 | 2.08 | 2.11 | 0.25 | 0.24 | 15.1 | 14.9 | 7.8 | 7.4 | 6.8 | 6.42 | |
| CV (%) | 17.71 | 18.59 | 20.17 | 24.22 | 13.43 | 14.54 | 47.83 | 50.04 | 5.84 | 6.94 | 6.70 | 8.02 | |
| 6 | Pokkali | 93.1 (±3.7) a | 80.3 (±2.6) a* | 3.45 (±0.07) a | 3.50 (±0.28) a | 0.59 (±0.18) a | 0.51 (±0.04) a | 40.5 (±2.9) a | 30.2 (±2.1) a* | 8.0 (±0.0) a | 7.0 (±0.0) b* | 7.0 (±0.0) a | 6.0 (±0.0) b* |
| IR64 | 61.5 (±2.9) c | 53.9 (±1.5) b* | 2.31 (±0.18) b | 2.05 (±0.13) c | 0.27 (±0.04) b | 0.30 (±0.08) b | 21.4 (±1.7) b | 13.2 (±0.6) b* | 8.5 (±0.6) a | 7.8 (±0.5) a | 7.5 (±0.6) a | 6.8 (±0.5) a | |
| MR219 | 61.6 (±2.3) b | 56.4 (±2.1) b | 2.14 (±0.14) b | 2.46 (±0.20) b* | 0.35 (0.05) b | 0.29 (±0.02) b* | 21.4 (±1.4) b | 14.5 (±1.0) b* | 8.3 (±0.5) a | 7.5 (±0.6) a,b | 7.0 (±0.0) a | 6.5 (±0.6) a,b | |
| Mean | 72.0 | 63.5 | 2.63 | 2.67 | 0.40 | 0.36 | 27.8 | 19.3 | 8.3 | 7.4 | 7.2 | 6.4 | |
| CV (%) | 21.90 | 19.75 | 23.59 | 24.85 | 43.80 | 31.80 | 34.57 | 42.35 | 5.48 | 6.94 | 5.43 | 8.02 | |
| 10 | Pokkali | 100.6 (±0.4) a | 88.3 (±4.1) a* | 3.26 (±0.05) a | 3.71 (±0.23) a* | 1.02 (±0.16) a | 0.45 (±0.07) a* | 49.5 (±0.3) a | 32.6 (±2.9) a* | 8.5 (±0.6) a | 7.0 (±0.0) b* | 7.0 (±0.0) a | 6.0 (±0.0) b* |
| IR64 | 63.8 (±2.1) c | 54.5 (±4.6) b* | 2.45 (±0.30) b | 2.17 (±0.10) c | 0.48 (±0.12) b | 0.41 (±0.11) a | 20.8 (±1.3) c | 11.5 (±2.3) b* | 8.8 (±1.5) a | 9.0 (±0.8) a | 7.8 (±1.5) a | 7.5 (±0.6) a | |
| MR219 | 70.6 (±1.8) b | 58.9 (±5.3) b* | 2.11 (±0.07) c | 2.45 (±0.16) b | 0.43 (±0.17) b | 0.40 (±0.12) a | 24.8 (±1.1) b | 13.7 (±2.7) b* | 8.8 (±0.5) a | 8.5 (±0.6) a | 7.8 (±0.5) a | 7.5 (±0.6) a | |
| Mean | 78.3 | 67.2 | 2.61 | 2.77 | 0.64 | 0.42 | 31.7 | 19.3 | 8.7 | 8.2 | 7.5 | 7.00 | |
| CV (%) | 21.44 | 24.17 | 20.31 | 25.86 | 48.39 | 23.31 | 41.95 | 52.67 | 10.24 | 12.61 | 12.06 | 12.18 | |
| Source of Variation | df | Mean Square | ||
|---|---|---|---|---|
| Pn (µmol CO2 m−2 s−1) | Tr (mmol H2O m−2 s−1) | Gs (µmol H2O m−2 s−1) | ||
| Replication (R) | 3 | 0.05 | 0.21 | 0.12 |
| Variety (V) | 2 | 118.32 * | 0.68 * | 22.86 * |
| Day of treatment (T) | 3 | 8.04 | 2.10 * | 15.31 * |
| Salinity (S) | 1 | 219.80 * | 77.76 * | 134.80 * |
| V×T | 6 | 4.86 | 0.43 * | 0.57 * |
| V×S | 2 | 0.93 | 3.00 * | 3.57 * |
| T×S | 3 | 24.06 * | 11.08 * | 14.87 * |
| V×T×S | 6 | 0.58 | 0.56 * | 0.50 * |
| Error | 69 | 1.00 | 3.77 | 5.37 |
| Source of Variation | df | Mean Square | |
|---|---|---|---|
| Chl (mg g−1 FW) | RWC (%) | ||
| Replication (R) | 3 | 0.01 | 37.89 |
| Variety (V) | 2 | 3.06 * | 176.07 * |
| Day of treatment (T) | 3 | 0.04 | 923.98 * |
| Salinity (S) | 1 | 0.20 * | 4010.89 * |
| V×T | 6 | 0.03 * | 45.54 |
| V×S | 2 | 0.06 * | 81.69 |
| T×S | 3 | 0.09 * | 1055.62 * |
| V×T×S | 6 | 0.02 | 50.68 |
| Error | 69 | 0.35 | 15.22 |
| Source of Variations | df | Mean Square | |||||||
|---|---|---|---|---|---|---|---|---|---|
| GABA-T1 | SSADH | GAD2 | SOS2 | CuZn SOD1 | MnSOD1 | CAT1 | 18sRNA | ||
| Replication (R) | 2 | 0.15 | 0.14 | 1.51 | 1.06 | 2.14 | 8.94 | 0.62 | 21.76 |
| Variety (V) | 2 | 91.04 * | 54.93 * | 23.86 * | 286.15 * | 72.05 * | 22.86 * | 135.01 * | 73.69 |
| Day of treatment (T) | 3 | 39.81 * | 113.85 * | 78.92 * | 196.79 * | 36.21 * | 109.98 * | 335.78 * | 11.55 |
| V×T | 6 | 77.22 * | 97.92 * | 26.29 * | 92.15 * | 36.35 * | 10.50 * | 27.55 * | 35.52 |
| Error | 22 | 0.80 | 1.11 | 1.05 | 1.41 | 1.27 | 1.02 | 3.19 | 31.63 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shakri, T.; Che-Othman, M.H.; Md Isa, N.; Sukiran, N.L.; Zainal, Z. Morpho-Physiological and Stress-Related Gene Expression of Rice Varieties in Response to Salinity Stress at Early Vegetative Stage. Agriculture 2022, 12, 638. https://doi.org/10.3390/agriculture12050638
Shakri T, Che-Othman MH, Md Isa N, Sukiran NL, Zainal Z. Morpho-Physiological and Stress-Related Gene Expression of Rice Varieties in Response to Salinity Stress at Early Vegetative Stage. Agriculture. 2022; 12(5):638. https://doi.org/10.3390/agriculture12050638
Chicago/Turabian StyleShakri, Tasneem, Muhammad Hafiz Che-Othman, Nurulhikma Md Isa, Noor Liyana Sukiran, and Zamri Zainal. 2022. "Morpho-Physiological and Stress-Related Gene Expression of Rice Varieties in Response to Salinity Stress at Early Vegetative Stage" Agriculture 12, no. 5: 638. https://doi.org/10.3390/agriculture12050638
APA StyleShakri, T., Che-Othman, M. H., Md Isa, N., Sukiran, N. L., & Zainal, Z. (2022). Morpho-Physiological and Stress-Related Gene Expression of Rice Varieties in Response to Salinity Stress at Early Vegetative Stage. Agriculture, 12(5), 638. https://doi.org/10.3390/agriculture12050638






