Metabolomics Analysis of Lettuce (Lactuca sativa L.) Affected by Low Potassium Supply
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Cultivation Procedure
2.2. Fertilization
2.3. Chemical and Metabolomics Analyses
2.4. Statistics
3. Results
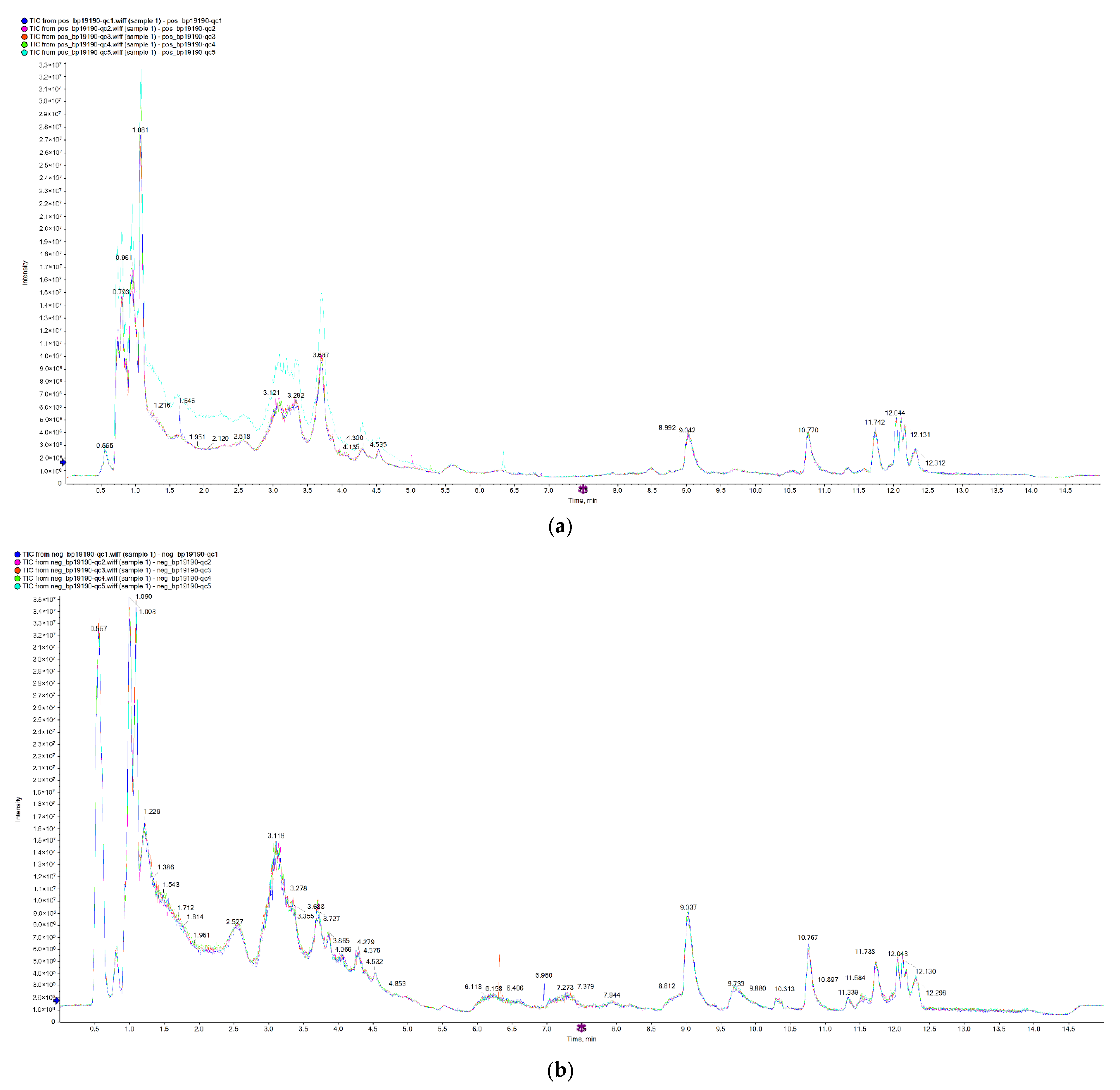
3.1. System Stability Analysis
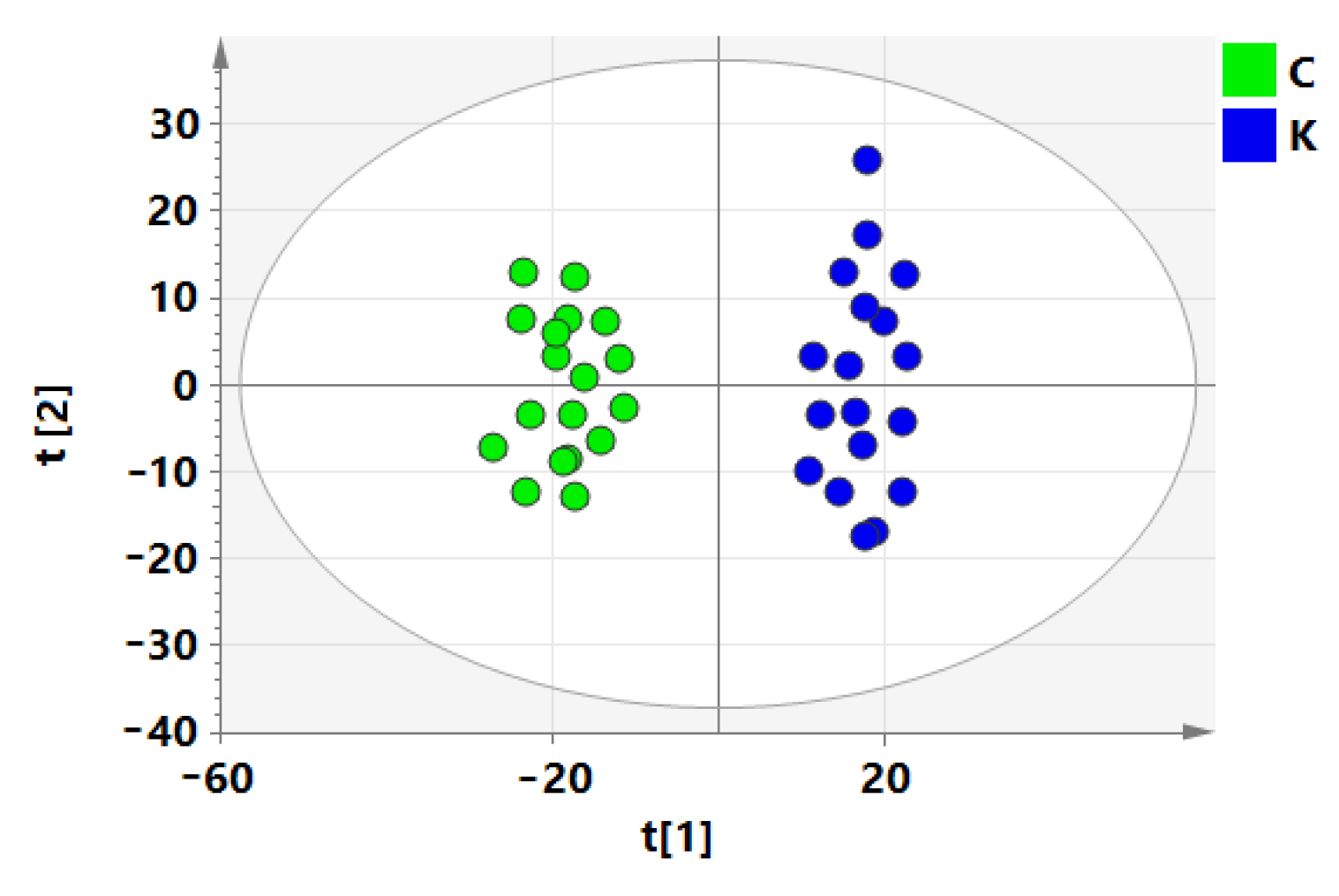
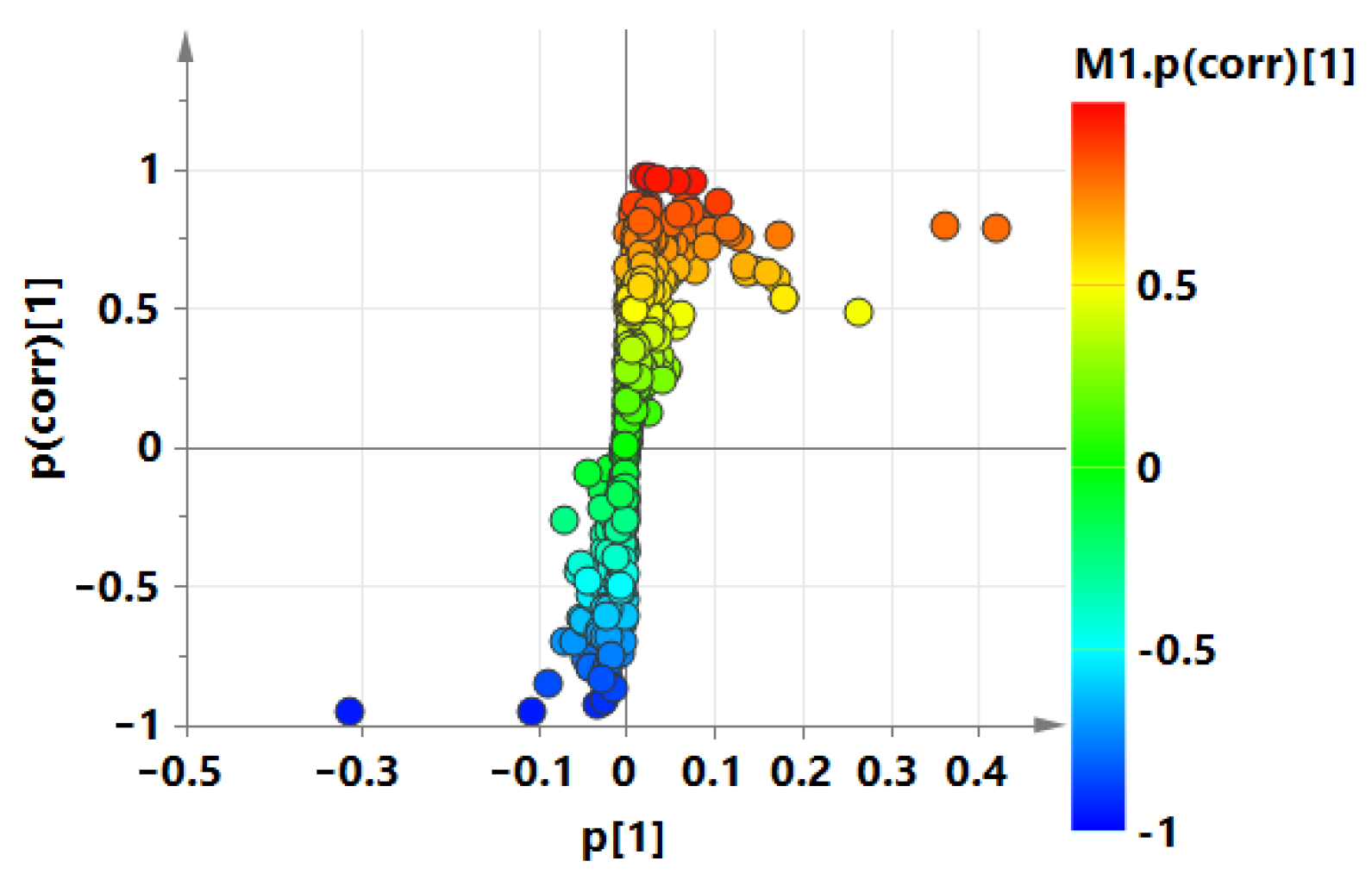
3.2. Screening of Differential Metabolites
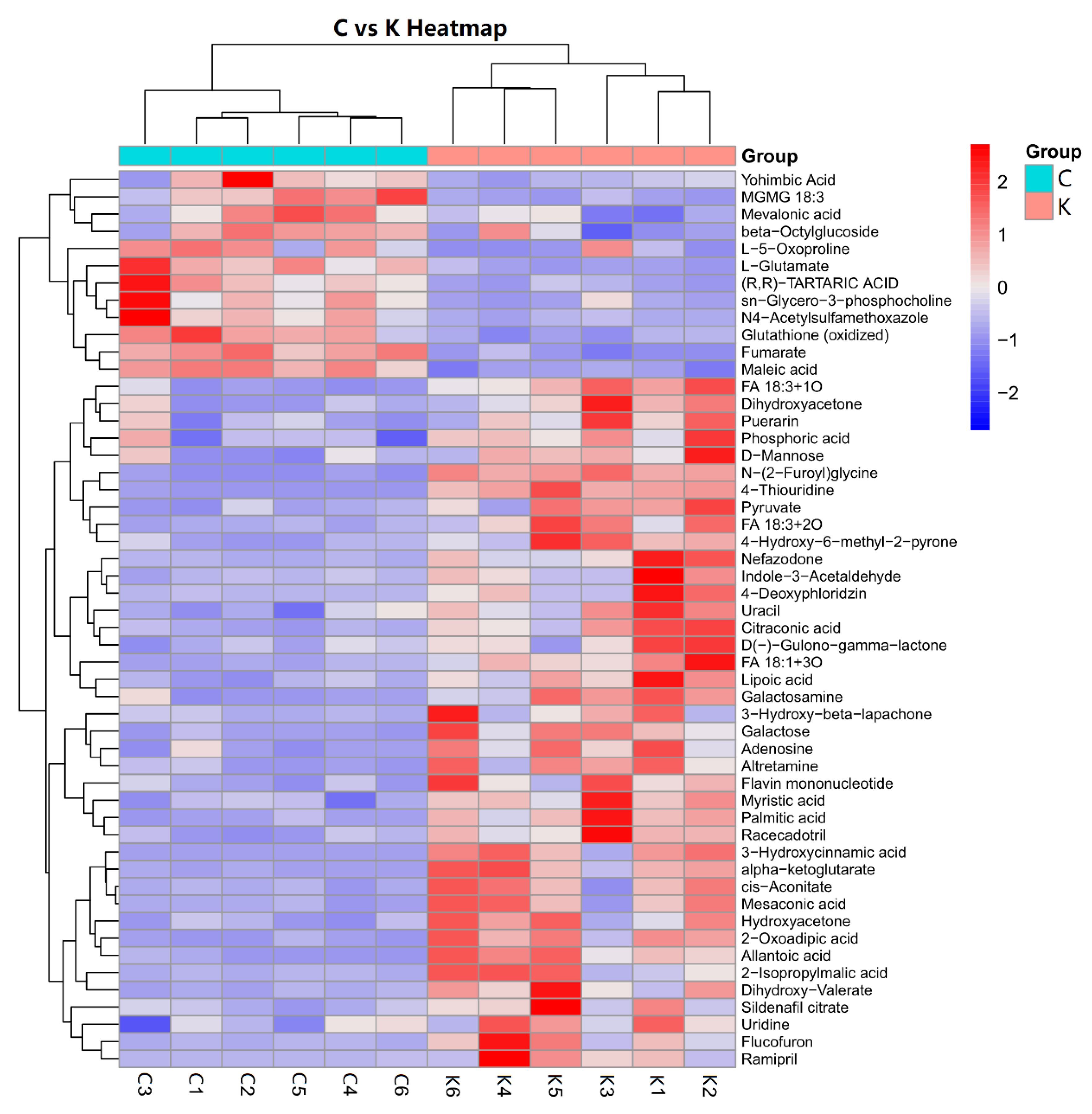
3.3. Hierarchical Clustering for Screening of Differential Metabolites
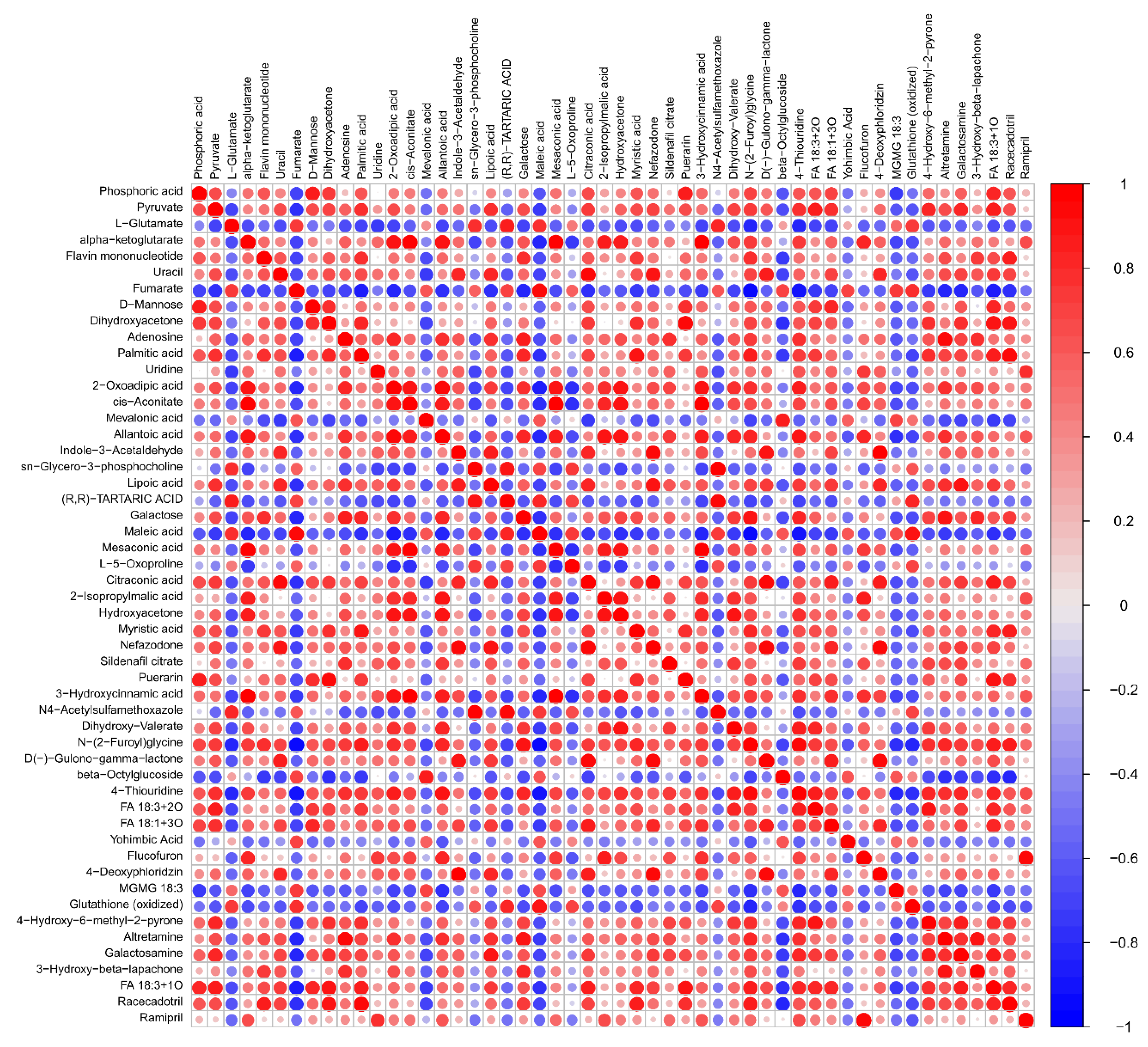
3.4. Metabolite Correlation Analysis
3.5. KEGG Annotation and Pathway Analysis
4. Discussion
4.1. Impact of N, P, K Deficiencies on Lettuce Metabolic Pathways
4.2. Impact of N, P, K Deficiencies on Lettuce Downregulated Metabolic Biomarkers
4.3. Effect of N, P, K Deficiencies on Lettuce Upregulated Metabolic Biomarkers
5. Conclusions and Perspectives
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhou, W.; Liang, X.; Li, K.; Dai, P.; Li, J.; Liang, B.; Sun, C.; Lin, X. Metabolomics analysis reveals potential mechanisms of phenolic accumulation in lettuce (Lactuca sativa L.) induced by low nitrogen supply. Plant Physiol. Biochem. 2021, 158, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Saah, K.J.A.; Kaba, J.S.; Abunyewa, A.A. Inorganic nitrogen fertilizer, biochar particle size and rate of application on lettuce (Lactuca sativa L.) nitrogen use and yield. All Life 2022, 15, 624–635. [Google Scholar] [CrossRef]
- Pettigrew, W.T. Potassium influences on yield and quality production for maize, wheat, soybean and cotton. Physiol. Plant 2010, 133, 670–681. [Google Scholar] [CrossRef]
- Cui, J.; Abadie, C.; Carroll, A.; Lamade, E.; Tcherkez, G. Responses to K deficiency and waterlogging interact via respiratory and nitrogen metabolism. Plant Cell Environ. 2019, 42, 647–658. [Google Scholar] [CrossRef]
- Sung, J.; Lee, S.; Lee, Y.; Ha, S.; Song, B.; Kim, T.; Waters, B.M.; Krishnan, H.B. Metabolomic profiling from leaves and roots of tomato (Solanum lycopersicum L.) plants grown under nitrogen, phosphorus or potassium-deficient condition. Plant Sci. 2015, 241, 55–64. [Google Scholar] [CrossRef]
- Lu, Z.; Ren, T.; Pan, Y.; Li, X.; Cong, R.; Lu, J. Differences on photosynthetic limitations between leaf margins and leaf centers under potassium deficiency for Brassica napus L. Sci. Rep. 2016, 6, 21725. [Google Scholar] [CrossRef] [Green Version]
- Chérel, I.; Michard, E.; Platet, N.; Mouline, K.; Thibaud, J.B. Physical and Functional Interaction of the Arabidopsis K+ Channel AKT2 and Phosphatase AtPP2CA. Plant Cell 2002, 14, 1133–1146. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Johkan, M.; Hohjo, M.; Tsukagoshi, S.; Maruo, T. Plant Growth and Photosynthesis Response to Low Potassium Conditions in Three Lettuce (Lactuca sativa) Types. Hortic. J. 2017, 86, 229–237. [Google Scholar] [CrossRef] [Green Version]
- Mirande-Ney, C.; Tcherkez, G.; Balliau, T.; Zivy, M.; Gilard, F.; Cui, J.; Ghashghaie, J.; Lamade, E. Metabolic leaf responses to potassium availability in oil palm (Elaeis guineensis Jacq.) trees grown in the field. Environ. Exp. Bot. 2020, 175, 104062. [Google Scholar] [CrossRef]
- Mutanga, O.; Skidmore, A.K.; Prins, H.H.T. Predicting in situ pasture quality in the Kruger National Park, South Africa, using continuum-removed absorption features. Remote Sens. Environ. 2004, 89, 393–408. [Google Scholar] [CrossRef]
- Xiong, Y.T.; Ohashi, S.; Nakano, K.; Jiang, W.Z.; Takizawa, K.; Iijima, K.; Maniwara, P. Quantification of potassium concentration with Vis-SWNIR spectroscopy in fresh lettuce. J. Innov. Opt. Health Sci. 2020, 13, 2050029. [Google Scholar] [CrossRef]
- Liu, D.S.; Jin, L.S.; Yang, Z.H.; Han, X.R.; Lin, G.L. Rapid Determination of Potassium Content in Flue-cured Tobacco Leaves by Near Infrared Reflectance Spectroscopy. Chin. J. Soil Sci. 2010, 41, 417–419. [Google Scholar]
- Xu, Z.; Guo, X.; Zhu, A.; He, X.; Zhao, X.; Han, Y.; Subedi, R. Using Deep Convolutional Neural Networks for Image-Based Diagnosis of Nutrient Deficiencies in Rice. Comput. Intell. Neurosci. 2020, 2020, 7307252. [Google Scholar] [CrossRef]
- Chen, L.S.; Huang, S.H.; Sun, Y.Y.; Zhu, E.Y.; Wang, K. Rapid Identification of Potassium Nutrition Stress in Rice Based on Machine Vision and Object-Oriented Segmentation. J. Spectro. 2019, 2019, 4623545. [Google Scholar] [CrossRef]
- Weksler, S.; Rozenstein, O.; Haish, N.; Moshelion, M.; Wallach, R.; Ben-Dor, E. Detection of Potassium Deficiency and Momentary Transpiration Rate Estimation at Early Growth Stages Using Proximal Hyperspectral Imaging and Extreme Gradient Boosting. Sensors 2021, 21, 958. [Google Scholar] [CrossRef]
- Cui, J.; Nieves-Cordones, M.; Rubio, F.; Tcherkez, G. Involvement of salicylic acid in the response to potassium deficiency revealed by metabolomics. Plant Physiol. Biochem. 2021, 163, 201–204. [Google Scholar] [CrossRef]
- Alexandersson, E.; Jacobson, D.; Vivier, M.A.; Weckwerth, W.; Andreasson, E. Field-omics-understanding large-scale molecular data from field crops. Front Plant Sci. 2014, 5, 286. [Google Scholar] [CrossRef] [Green Version]
- Yoshio, T.; Tetsuya, M.; Ryo, N.; Makoto, K.; Kazuki, S.; Seiichi, O.; Wang, N.; Miyako, K. Metabolomic Evaluation of the Quality of Leaf Lettuce Grown in Practical Plant Factory to Capture Metabolite Signature. Front. Plant Sci. 2018, 9, 665. [Google Scholar]
- Cui, J.; Davanture, M.; Zivy, M.; Lamade, E.; Tcherkez, G. Metabolic responses to potassium availability and waterlogging reshape respiration and carbon use efficiency in oil palm. New Phytol. 2019, 223, 310–322. [Google Scholar] [CrossRef]
- Cui, J.; Pottosin, I.; Lamade, E.; Tcherkez, G. What is the role of putrescine accumulated under potassium deficiency? Plant Cell Environ. 2020, 43, 1331–1347. [Google Scholar] [CrossRef]
- Jing, C.A.; El, B.; Gt, A. Potassium deficiency reconfigures sugar export and induces catecholamine accumulation in oil palm leaves. Plant Sci. 2020, 300, 110628. [Google Scholar]
- Takahashi, H.; Imamura, T.; Miyagi, A.; Uchimiya, H. Comparative metabolomics of developmental alterations caused by mineral deficiency during in vitro culture of Gentiana triflora. Metabolomics 2012, 8, 154–163. [Google Scholar] [CrossRef]
- Gao, H.Y.; Mao, H.P.; Ullah, I. Analysis of Metabolomic Changes in Lettuce Leaves under Low Nitrogen and Phosphorus Deficiencies Stresses. Agric.-Basel 2020, 10, 406. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, X.M.; Wang, X.C.; Liu, J.H.; Huang, B.Y.; Guo, X.Y.; Xiong, S.P.; La, G.X. UPLC-QTOF analysis reveals metabolomic changes in the flag leaf of wheat (Triticum aestivum L.) under low-nitrogen stress. Plant Physiol. Biochem. 2017, 111, 30–38. [Google Scholar] [CrossRef]
- MetaboAnalyst 4.0. Available online: https://www.metaboanalyst.ca/ (accessed on 15 December 2021).
- Shen, T.H.; Xiong, Q.Q.; Zhong, L.; Shi, X.; Cao, C.H.; He, H.H.; Chen, X.R. Analysis of main metabolisms during nitrogen deficiency and compensation in rice. Acta Physiol. Plant. 2019, 41, 68. [Google Scholar] [CrossRef]
- Chen, J.; Wang, W.; Kong, J.; Yue, Y.; Dong, Y.; Zhang, J.; Liu, L. Application of UHPLC-Q-TOF MS based untargeted metabolomics reveals variation and correlation amongst different tissues of Eucommia ulmoides Oliver. Microchem. J. 2022, 172, 106919. [Google Scholar] [CrossRef]
- Kralova, K.; Jampilek, J. Responses of Medicinal and Aromatic Plants to Engineered Nanoparticles. Appl. Sci.-Basel 2021, 11, 1813. [Google Scholar] [CrossRef]
- Yue, H.F.; Zhou, M.; Hou, X.K.; Yang, X.; Zhao, H.; Xu, M.S.; Wang, L.; Liu, H.M.; Wuyun, T.N.; Zhu, G.P.; et al. Effect of elements nitrogen, phosphorus and potassium on phenotype, photosynthesis and biomass accumulation in juvenile phase of Prunus armeniaca × Sibirica. Pak. J. Bot. 2022, 54, 577–588. [Google Scholar] [CrossRef]
- Cao, Y.W.; Qu, R.J.; Tang, X.Q.; Sun, L.Q.; Chen, Q.Q.; Miao, Y.J. UPLC-Triple TOF-MS/MS based metabolomics approach to reveal the influence of nitrogen levels on Isatis indigotica seedling leaf. Sci. Hortic. 2020, 266, 109280. [Google Scholar] [CrossRef]
- Chen, J.; Zeng, H.; Zhang, X. Integrative transcriptomic and metabolomic analysis of D-leaf of seven pineapple varieties differing in N-P-K% contents. BMC Plant Biol. 2021, 21, 550. [Google Scholar] [CrossRef]
- Groher, T.; Schmittgen, S.; Noga, G.; Hunsche, M. Limitation of mineral supply as tool for the induction of secondary metabolites accumulation in tomato leaves. Plant Physiol. Biochem. 2018, 130, 105–111. [Google Scholar] [CrossRef]
- Jan, R.; Asaf, S.; Numan, M.; Lubna; Kim, K.M. Plant Secondary Metabolite Biosynthesis and Transcriptional Regulation in Response to Biotic and Abiotic Stress Conditions. Agron.-Basel 2021, 11, 968. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, W.H. Regulation of potassium transport and signaling in plants. Curr. Opin. Plant Biol. 2017, 39, 123–128. [Google Scholar] [CrossRef]
- Armengaud, P.; Sulpice, R.; Miller, A.J.; Stitt, M.; Amtmann, A.; Gibon, Y. Multilevel analysis of primary metabolism provides new insights into the role of potassium nutrition for glycolysis and nitrogen assimilation in Arabidopsis roots. Plant Physiol. 2009, 150, 772–785. [Google Scholar] [CrossRef] [Green Version]
- Patra, B.; Schluttenhofer, C.; Wu, Y.; Pattanaik, S.; Ling, Y. Transcriptional regulation of secondary metabolite biosynthesis in plants-ScienceDirect. Biochim. Et Biophys. Acta (BBA)-Gene Regul. Mech. 2013, 1829, 1236–1247. [Google Scholar] [CrossRef]
- Jiang, J.; Wang, Y.P.; Yang, Y.H.; Yu, M.X.; Wang, C.; Yan, J.H. Interactive effects of nitrogen and phosphorus additions on plant growth vary with ecosystem type. Plant Soil 2019, 440, 523–537. [Google Scholar] [CrossRef]
- Li, Y.; Niu, S.; Yu, G. Aggravated phosphorus limitation on biomass production under increasing nitrogen loading: A meta-analysis. Glob. Change Biol. 2016, 22, 934–943. [Google Scholar] [CrossRef]
- Coskun, D.; Britto, D.T.; Kronzucker, H.J. Regulation and mechanism of potassium release from barley roots: An in planta 42K+ analysis. New Phytol. 2010, 188, 1028–1038. [Google Scholar] [CrossRef]
- Coskun, D.; Britto, D.T.; Li, M.; Oh, S.; Kronzucker, H.J. Capacity and plasticity of potassium channels and high-affinity transporters in roots of barley and Arabidopsis. Plant Physiol. 2013, 162, 496–511. [Google Scholar] [CrossRef] [Green Version]
- Cui, J.; Tcherkez, G. Potassium dependency of enzymes in plant primary metabolism. Plant Physiol. Biochem. 2021, 166, 522–530. [Google Scholar] [CrossRef]
- Coskun, D.; Britto, D.T.; Kronzucker, H.J. The nitrogen-potassium intersection: Membranes, metabolism, and mechanism. Plant Cell Environ. 2017, 40, 2029–2041. [Google Scholar] [CrossRef]





| Metabolite Name | ESI Mode | Adduct | VIP | m/z | RT (min) |
|---|---|---|---|---|---|
| Adenosine | ESI+ | [M+H]+ | 11.07 | 268.10 | 3.96 |
| FA 18:1+3O | ESI− | [M-H]− | 9.51 | 329.23 | 3.22 |
| Uridine | ESI− | [M-H]− | 4.81 | 243.06 | 3.73 |
| cis-Aconitate | ESI− | [M-H]− | 4.76 | 173.01 | 10.54 |
| D(-)-Gulono-gamma-lactone | ESI− | [M-H]− | 4.28 | 177.04 | 2.55 |
| 2-Isopropylmalic acid | ESI− | [M-H]− | 3.99 | 175.06 | 6.95 |
| Dihydroxyacetone | ESI− | (2M-H)− | 3.96 | 179.06 | 6.17 |
| D-Mannose | ESI− | [M-H]− | 3.91 | 179.06 | 7.17 |
| Mesaconic acid | ESI− | [M-H]− | 3.57 | 129.02 | 10.55 |
| Galactosamine | ESI+ | [M+NH4]+ | 3.08 | 180.09 | 6.09 |
| Dihydroxy-Valerate | ESI− | [M-H]− | 3.06 | 133.05 | 1.96 |
| 2-Oxoadipic acid | ESI− | (M-H2O-H)− | 2.71 | 141.02 | 9.83 |
| Myristic acid | ESI− | [M-H]− | 2.65 | 227.20 | 1.12 |
| Palmitic acid | ESI− | [M-H]− | 2.65 | 255.23 | 1.13 |
| Hydroxyacetone | ESI− | (M+CH3COO)− | 2.50 | 133.05 | 2.02 |
| Phosphoric acid | ESI− | [M-H]− | 2.16 | 96.96 | 10.34 |
| Pyruvate | ESI− | (M-H)− | 2.13 | 87.01 | 2.34 |
| N-(2-Furoyl)glycine | ESI− | [M-H]− | 2.08 | 168.03 | 9.72 |
| 3-Hydroxycinnamic acid | ESI− | [M-H]− | 2.02 | 163.04 | 1.19 |
| Citraconic acid | ESI− | (M-H)− | 1.88 | 129.02 | 2.51 |
| Uracil | ESI− | [M-H]− | 1.88 | 111.02 | 3.74 |
| Allantoic acid | ESI− | [M-H]− | 1.80 | 175.05 | 8.54 |
| FA 18:3+1O | ESI+ | [M+H]+ | 1.64 | 295.23 | 3.14 |
| alpha-ketoglutarate | ESI− | (M-H)− | 1.56 | 145.01 | 8.68 |
| Galactose | ESI+ | [M+NH4]+ | 1.55 | 198.10 | 9.02 |
| 4-Deoxyphloridzin | ESI− | [M-H]− | 1.52 | 419.13 | 3.81 |
| 4-Thiouridine | ESI− | [M-H]− | 1.51 | 261.06 | 9.80 |
| Lipoic acid | ESI− | [M-H]− | 1.50 | 205.04 | 1.39 |
| Nefazodone | ESI+ | [M+H]+ | 1.47 | 470.23 | 3.09 |
| Indole-3-Acetaldehyde | ESI− | [M-H]− | 1.43 | 240.04 | 2.56 |
| Flavin mononucleotide | ESI− | (M-H)− | 1.40 | 455.10 | 0.56 |
| 3-Hydroxy-beta-lapachone | ESI+ | [M+H]+ | 1.37 | 259.10 | 1.00 |
| Altretamine | ESI+ | (M+NH4)+ | 1.36 | 228.19 | 0.89 |
| Racecadotril | ESI+ | [M+H]+ | 1.30 | 386.14 | 6.14 |
| Flucofuron | ESI− | [M-H]− | 1.15 | 414.98 | 9.73 |
| 4-Hydroxy-6-methyl-2-pyrone | ESI+ | [M+H]+ | 1.10 | 127.04 | 6.17 |
| Ramipril | ESI+ | [M+H]+ | 1.08 | 417.24 | 5.91 |
| Puerarin | ESI− | [M-H]− | 1.00 | 415.11 | 9.10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, H.; Gong, L.; Ni, J.; Li, Q. Metabolomics Analysis of Lettuce (Lactuca sativa L.) Affected by Low Potassium Supply. Agriculture 2022, 12, 1153. https://doi.org/10.3390/agriculture12081153
Gao H, Gong L, Ni J, Li Q. Metabolomics Analysis of Lettuce (Lactuca sativa L.) Affected by Low Potassium Supply. Agriculture. 2022; 12(8):1153. https://doi.org/10.3390/agriculture12081153
Chicago/Turabian StyleGao, Hongyan, Liyan Gong, Jiheng Ni, and Qinglin Li. 2022. "Metabolomics Analysis of Lettuce (Lactuca sativa L.) Affected by Low Potassium Supply" Agriculture 12, no. 8: 1153. https://doi.org/10.3390/agriculture12081153
APA StyleGao, H., Gong, L., Ni, J., & Li, Q. (2022). Metabolomics Analysis of Lettuce (Lactuca sativa L.) Affected by Low Potassium Supply. Agriculture, 12(8), 1153. https://doi.org/10.3390/agriculture12081153






