Allelic Variation of Puroindolines Genes in Iranian Common Wheat Landraces
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Genomic DNA Extraction
2.3. Amplification and Digestion of Pina-D1 and Pinb-D1 Genes
2.4. Cloning of Pinb-D1 Genes
3. Results
3.1. Variation in Grain Hardness
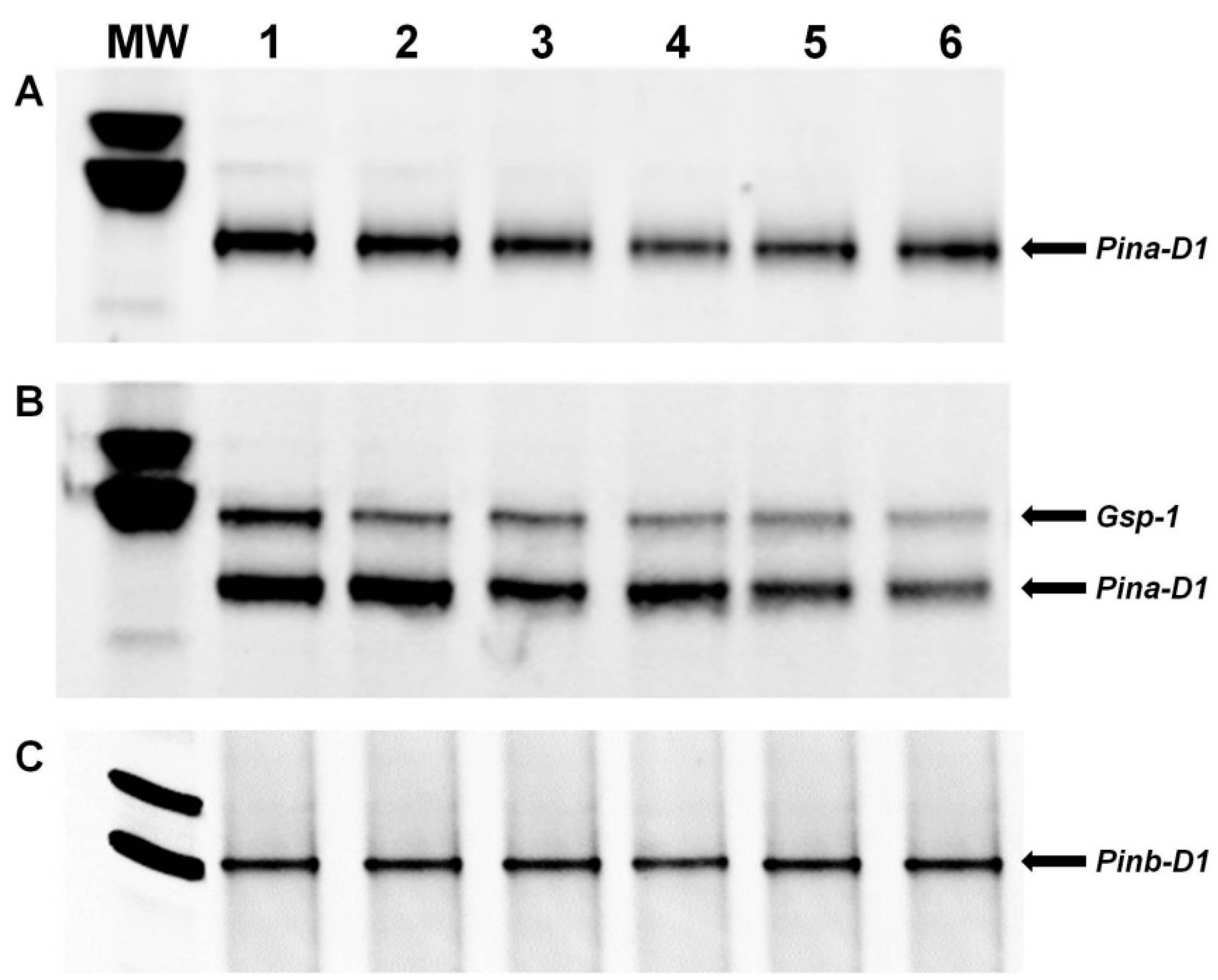
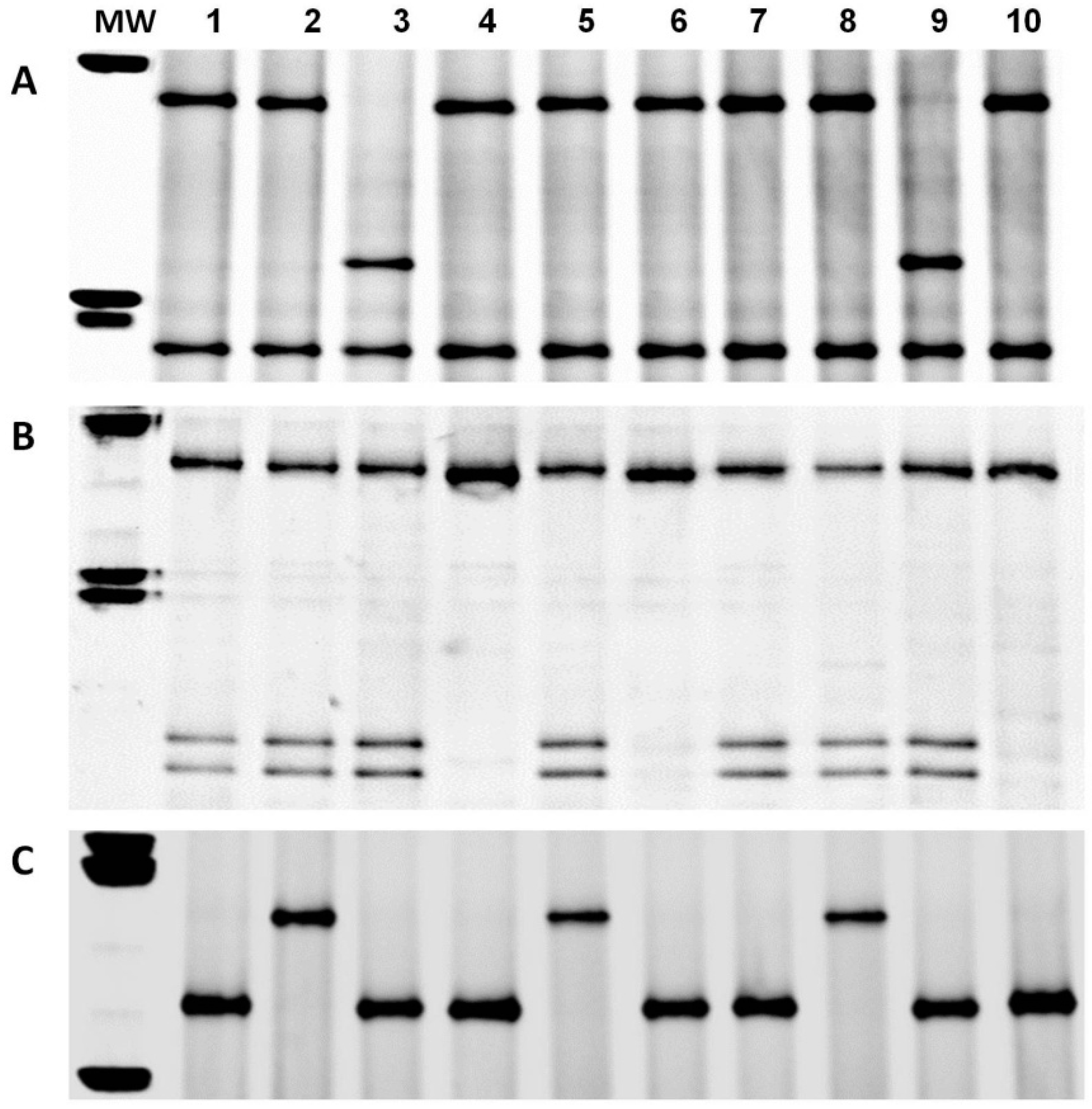
3.2. PCR Analysis of the Pina-D1 and Pinb-D1 Genes
3.3. Enzymatic Digestion and Sequencing of Pinb-D1 Alleles
3.4. Amino Acid Sequence Analysis and Relationship with Grain Hardness
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations FAOSTAT. Available online: http://Www.Fao:Faostat/Es/#data/QC (accessed on 24 June 2022).
- Pomeranz, Y.; Williams, P.C. Wheat Hardness. Its genetic, structural, and biochemical background, measurement, and significance. Adv. Cereal Sci. Technol. 1990, 10, 471–544. [Google Scholar]
- Guttieri, M.J.; Bowen, D.; Gannon, D.; O’Brien, K.; Souza, E. Solvent retention capacities of irrigated soft white spring wheat flours. Crop Sci. 2001, 41, 1054–1061. [Google Scholar] [CrossRef]
- Morris, C.F. Puroindolines: The molecular genetic basis of wheat grain hardness. Plant Mol. Biol. 2002, 48, 633–647. [Google Scholar] [CrossRef]
- Bhave, M.; Morris, C.F. Molecular genetics of puroindolines and related genes: Allelic diversity in wheat and other grasses. Plant Mol. Biol. 2008, 66, 205–219. [Google Scholar] [CrossRef]
- Giroux, M.J.; Morris, C.F. A glycine to serine change in puroindoline b is associated with wheat grain hardness and low levels of starch-surface friabilin. Theor. Appl. Genet. 1997, 95, 857–864. [Google Scholar] [CrossRef]
- Gautier, M.F.; Aleman, M.E.; Marion, D.; Joudrier, P. Triticum aestivum puroindolines, two basic cystine-rich seed proteins: Cdna sequence analy.sis and developmental gene expression. Plant Mol. Biol. 1994, 25, 43–57. [Google Scholar] [CrossRef]
- Chantret, N.; Salse, J.; Sabot, F.; Rahman, S.; Bellec, A.; Laubin, B.; Dubois, I.; Dossat, C.; Sourdille, P.; Joudrier, P.; et al. Molecular basis of evolutionary events that shaped the hardness locus in diploid and polyploid wheat species (Triticum and Aegilops). Plant Cell 2005, 17, 1033–1045. [Google Scholar] [CrossRef] [Green Version]
- Morris, C.F.; Geng, H.; Beecher, B.S.; Ma, D. A review of the occurrence of grain softness protein-1 genes in wheat (Triticum aestivum L.). Plant Mol. Biol. 2013, 83, 507–521. [Google Scholar] [CrossRef]
- Chen, F.; Yu, Y.; Xia, X.; He, Z. Prevalence of a novel puroindoline b allele in Yunnan endemic wheats (Triticum aestivum ssp. yunnanense King). Euphytica 2007, 156, 39–46. [Google Scholar] [CrossRef]
- Giroux, M.J.; Talbert, L.; Habernicht, D.K.; Lanning, S.; Hemphill, A.; Martin, J.M. Association of puroindoline sequence type and grain hardness in hard red spring wheat. Crop Sci. 2000, 40, 370–374. [Google Scholar] [CrossRef]
- Martin, J.M.; Frohberg, R.C.; Morris, C.F.; Talbert, L.E.; Giroux, M.J. Milling and bread baking traits associated with puroindoline sequence type in hard red spring wheat. Crop Sci. 2001, 41, 228–234. [Google Scholar] [CrossRef] [Green Version]
- Hammer, K.; Knupffer, H.; Xhuveli, L. Estimating genetic erosion in landraces. Genet. Resour. Crop Evol. 1996, 43, 329–336. [Google Scholar] [CrossRef]
- Lillemo, M.; Morris, C.F. A leucine to proline mutation in puroindoline b frequently present in Northen Europe. Theor. Appl. Genet. 2000, 100, 1100–1107. [Google Scholar] [CrossRef]
- Ayala, M.; Guzmán, C.; Alvarez, J.B.; Peña, R.J. Characterization of genetic diversity of puroindoline genes in Mexican wheat landraces. Euphytica 2013, 190, 53–63. [Google Scholar] [CrossRef]
- Ma, X.; Sajjad, M.; Wang, J.; Yang, W.; Sun, J.; Li, X.; Zhang, A.; Liu, D. Diversity, distribution of puroindolines genes and their effect on kernel hardness in a diverse panel of Chinese wheat germplasm. Plant Biol. 2017, 17, 158. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, H.; Morris, C.F.; Haruna, M.; Tsujimoto, H. Prevalence of puroindoline alleles in wheat varieties from Eastern Asia including the discovery of a new SNP in puroindoline b. Plant Genet. Resour. 2008, 6, 142–152. [Google Scholar] [CrossRef]
- Kumar, R.; Arora, S.; Singh, K.; Garg, M. Puroindoline allelic diversity in Indian wheat germplasm and identification of new allelic variants. Breed. Sci. 2015, 65, 319–326. [Google Scholar] [CrossRef] [Green Version]
- Sansaloni, C.; Franco, J.; Santos, B.; Percival-Alwyn, L.; Singh, S.; Petroli, C.; Campos, J.; Dreher, K.; Payne, T.; Marshall, D.; et al. Diversity analysis of 80,000 wheat accessions reveals consequences and opportunities of selection footprints. Nat. Commun. 2020, 11, 4572. [Google Scholar] [CrossRef]
- Vikram, P.; Franco, J.; Burgueño, J.; Li, H.; Sehgal, D.; Saint-Pierre, C.; Ortiz, C.; Singh, V.K.; Sneller, C.; Sharma, A.; et al. Strategic use of Iranian bread wheat landrace accessions for genetic improvement: Core set formulation and validation. Plant Breed. 2021, 140, 87–99. [Google Scholar] [CrossRef]
- American Association of Cereal Chemists. Approved Methods of Analysis, AACC Method 55-30.01, 11th ed.; Association of Cereal Chemists: St Paul, MN, USA, 2000. [Google Scholar]
- Stacey, J.; Isaac, P.G. Isolation of DNA from plants. In Protocols for Nucleic Acid Analysis by Nonradioactive Probes; Isaac, P.G., Ed.; Humana Press: Totowa, NJ, USA, 1994; pp. 9–15. [Google Scholar]
- Lillemo, M.; Chen, F.; Xia, X.; William, M.; Peña, R.J.; Trethowan, R.; He, Z. Puroindoline grain hardness alleles in CIMMYT bread wheat germplasm. J. Cereal Sci. 2006, 44, 86–92. [Google Scholar] [CrossRef]
- Massa, A.N.; Morris, C.F.; Gill, B.S. Sequence diversity of puroindoline-a, puroindoline-b, and the grain softness protein genes in Aegilops tauschii Coss. Crop Sci. 2004, 44, 1808–1816. [Google Scholar] [CrossRef]
- Chen, F.; He, Z.H.; Xia, X.C.; Xia, L.Q.; Zhang, X.Y.; Lillemo, M.; Morris, C.F. Molecular and biochemical characterization of puroindoline a and b alleles in Chinese landraces and historical cultivars. Theor. Appl. Genet. 2006, 112, 400–409. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, G.; Xia, X.; He, Z.; Mu, P. Molecular characterization of Pina and Pinb allelic variations in Xinjiang landraces and commercial wheat cultivars. Euphytica 2008, 164, 745–752. [Google Scholar] [CrossRef]
- Li, G.; He, Z.; Lillemo, M.; Sun, Q.; Xia, X. Molecular characterization of allelic variations at Pina and Pinb loci in Shandong wheat landraces, historical and current cultivars. J. Cereal Sci. 2008, 47, 510–517. [Google Scholar] [CrossRef]
- Choi, Y.; Sims, G.E.; Murphy, S.; Miller, J.R.; Chan, A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE 2012, 7, e46688. [Google Scholar] [CrossRef] [Green Version]
- Guzmán, C.; Caballero, L.; Martín, M.A.; Alvarez, J.B. Molecular characterization and diversity of the Pina and Pinb genes in cultivated and wild diploid wheat. Mol. Breed. 2012, 30, 69–78. [Google Scholar] [CrossRef]
- Xia, L.; Chen, F.; Chen, X.; Morris, F. Occurrence of puroindoline alleles in Chinese winter wheats. Cereal Chem. 2005, 82, 38–43. [Google Scholar] [CrossRef] [Green Version]
- Morris, C.F.; Lillemo, M.; Simeone, M.C.; Giroux, M.J.; Babb, S.L.; Kidwell, K.K. Prevalence of puroindoline grain hardness genotypes among historically significant North American spring and winter wheats. Crop Sci. 2001, 41, 218–228. [Google Scholar] [CrossRef]
- Vikram, P.; Franco, J.; Burgueño-Ferreira, J.; Li, H.; Sehgal, D.; Saint Pierre, C.; Ortiz, C.; Sneller, C.; Tattaris, M.; Guzmán, C.; et al. Unlocking the genetic diversity of Creole wheats. Sci. Rep. 2016, 6, 23092. [Google Scholar] [CrossRef] [Green Version]
- Maryami, Z.; Azimi, M.R.; Guzmán, C.; Dreisigacker, S.; Najafian, G. Puroindoline (Pina-D1 and Pinb-D1) and waxy (Wx-1) genes in Iranian bread wheat (Triticum aestivum L.) landraces. Biotechnol. Biotechnol. Equip. 2020, 34, 1019–1027. [Google Scholar] [CrossRef]
- Cane, K.; Spackman, M.; Eagles, H. Puroindoline genes and their effects on grain quality traits in southern Australian wheat cultivars. Aust. J. Agric. Res. 2004, 55, 89–95. [Google Scholar] [CrossRef]
- Ayala, M.; Guzmán, C.; Peña, R.J.; Alvarez, J.B. genetic diversity and molecular characterization of puroindoline genes (Pina-D1 and Pinb-D1) in bread wheat landraces from Andalusia (Southern Spain). J. Cereal Sci. 2016, 71, 61–65. [Google Scholar] [CrossRef]
- Przyborowski, M.; Gasparis, S.; Kala, M.; Orczyk, W.; Nadolska-Orczyk, A. The variability of puroindoline-encoding alleles and their influence on grain hardness in modern wheat cultivars cultivated in Poland, breeding lines and Polish old landraces (Triticum aestivum L.). Agronomy 2020, 10, 1075. [Google Scholar] [CrossRef]
- Khurshid, M.; Ahmad, M. Prevalence of puroindoline genes and their impact on quality traits in a diverse germplasm of wheat genotypes. J. Agric. Sci. 2021, 43, 454–465. [Google Scholar] [CrossRef]
- Pan, Z.; Song, W.; Meng, F.; Xu, L.; Liu, B.; Zhu, J. Characterization of genes encoding wheat grain hardness from Chinese cultivar GaoCheng 8901. Cereal Chem. 2004, 81, 287–289. [Google Scholar] [CrossRef]
- Corona, V.; Gazza, L.; Boggini, G.; Pogna, N.E. Variation in friabilin composition as determined by A-PAGE fractionation and PCR amplification, and its relationship to grain hardness in bread wheat. J. Cereal Sci. 2001, 34, 243–250. [Google Scholar] [CrossRef]
- Feiz, L.; Beecher, B.S.; Martin, J.M.; Giroux, M.J. In planta mutagenesis determines the functional regions of the wheat puroindoline proteins. Genetics 2009, 183, 853–860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douliez, J.P.; Michon, T.; Elmorjani, K.; Marion, D. Mini review: Structure, biological and technological functions of lipid transfer proteins and indolines, the major lipid binding proteins from cereal kernels. J. Cereal Sci. 2000, 32, 1–20. [Google Scholar] [CrossRef]
- Wang, J.; Sun, J.; Liu, D.; Yang, W.; Wang, D.; Tong, Y.; Zhang, A. Analysis of Pina and Pinb alleles in the micro-core collections of Chinese wheat germplasm by Ecotilling and identification of a novel Pinb allele. J. Cereal Sci. 2008, 48, 836–842. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; He, Z.; Xia, X.; Lillemo, M.; Morris, C. A new puroindoline b mutation present in Chinese winter wheat cultivar Jingdong 11. J. Cereal Sci. 2005, 42, 267–269. [Google Scholar] [CrossRef]



| Allele | Mutation | N | Reference |
|---|---|---|---|
| Pinb-D1b | Gly/Ser at position 46 | 11 | [6,26] |
| Pinb-D1ab | Gln/Stop at position 382 | 175 | [17,26] |
| Pinb-D1p | Lsy/Asn at position 210 | 80 | [27,30] |
| Pinb-D1e | Trp/Stop at position 39 | 0 | [26,31] |
| Pinb-D1c | Leu/Pro at position 60 | 0 | [14] |
| Puroindoline Composition | N | PSI (% ± s.d.) | Range |
|---|---|---|---|
| Pina-D1a/Pinb-D1b | 11 | 45.91 ± 3.58 AB* | 40–50 |
| Pina-D1a/Pinb-D1ab | 175 | 47.13 ± 2.70 A | 40–50 |
| Pina-D1a/Pinb-D1p | 80 | 47.48 ± 2.64 A | 41–50 |
| Pina-D1a/Pinb-D1ak | 5 | 44.20 ± 2.93 B | 41–49 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huertas-García, A.B.; Guzmán, C.; Tabbita, F.; Alvarez, J.B. Allelic Variation of Puroindolines Genes in Iranian Common Wheat Landraces. Agriculture 2022, 12, 1196. https://doi.org/10.3390/agriculture12081196
Huertas-García AB, Guzmán C, Tabbita F, Alvarez JB. Allelic Variation of Puroindolines Genes in Iranian Common Wheat Landraces. Agriculture. 2022; 12(8):1196. https://doi.org/10.3390/agriculture12081196
Chicago/Turabian StyleHuertas-García, Ana B., Carlos Guzmán, Facundo Tabbita, and Juan B. Alvarez. 2022. "Allelic Variation of Puroindolines Genes in Iranian Common Wheat Landraces" Agriculture 12, no. 8: 1196. https://doi.org/10.3390/agriculture12081196








