Automated Wheat Diseases Classification Framework Using Advanced Machine Learning Technique
Abstract
:1. Introduction
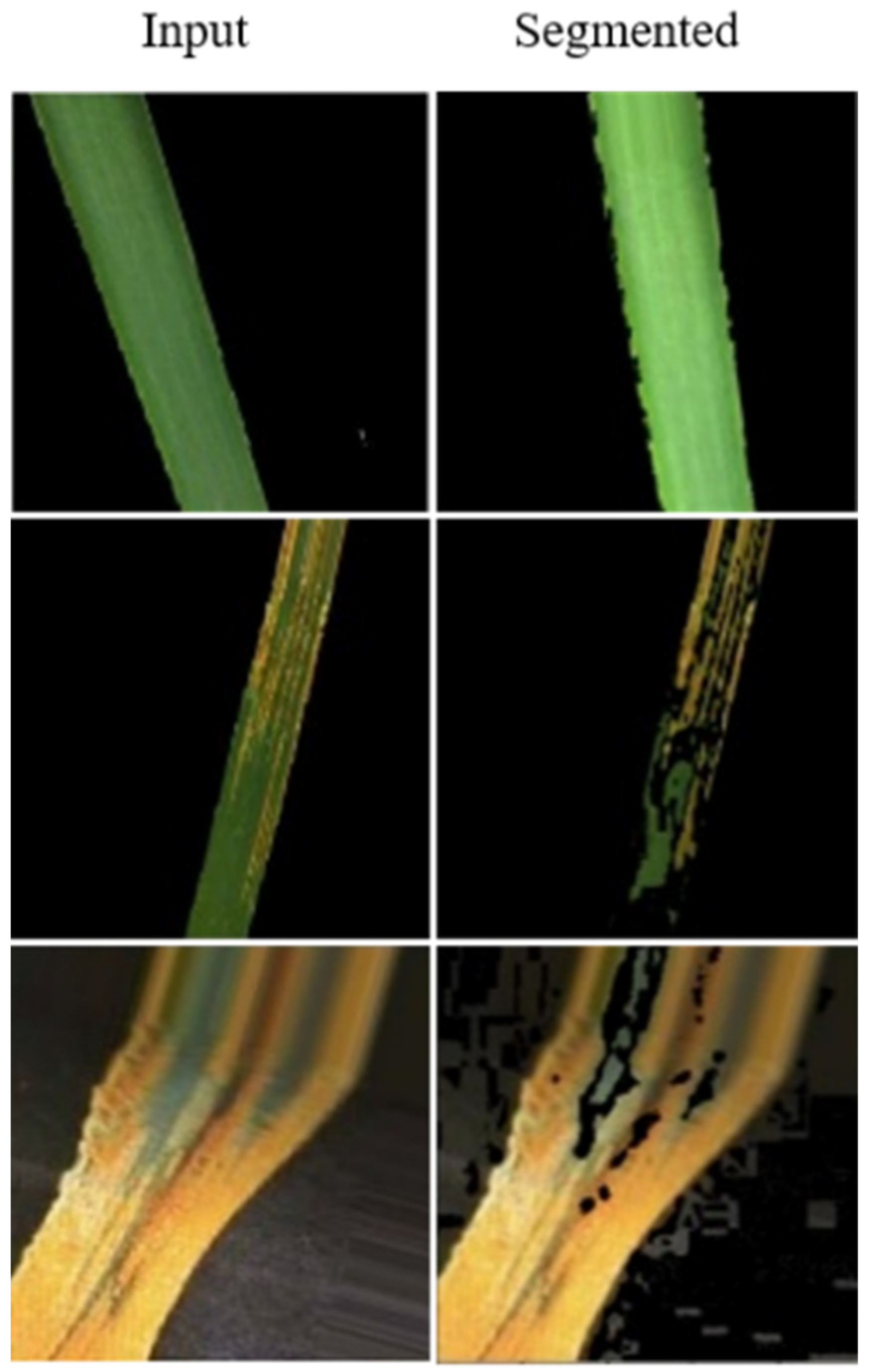
- We proposed a machine learning-based framework for the detection of salient cues regarding wheat diseases and accurately classify them into yellow and brown rust. Our model utilized a masked-based segmentation technique that automatically removes the background, noises, and identifies healthy, unhealthy wheat crops, and determines the affected and unaffected area of the crop. The proposed framework is lightweight and automatically identifies the wheat crop diseases with a high recognition rate.
- A new dataset for wheat disease classification is introduced. The dataset is collected from different wheat fields in various regions of Peshawar, and Dir Pakistan. We focused on two categories of diseases, having a total of three classes, i.e., brown rust, severe yellow, and healthy leaves, respectively. The dataset will be publicly available to the research community.
- A comparative analysis has been conducted among ML techniques for wheat disease recognition. The proposed framework achieved 98.8% accuracy for the wheat diseases classification. Due to good generalization and a high recognition rate, the system can be employed in various real-time industrial applications.
2. Related Work
2.1. Statistical-Based Approaches
2.2. Machine Learning-Based Approaches
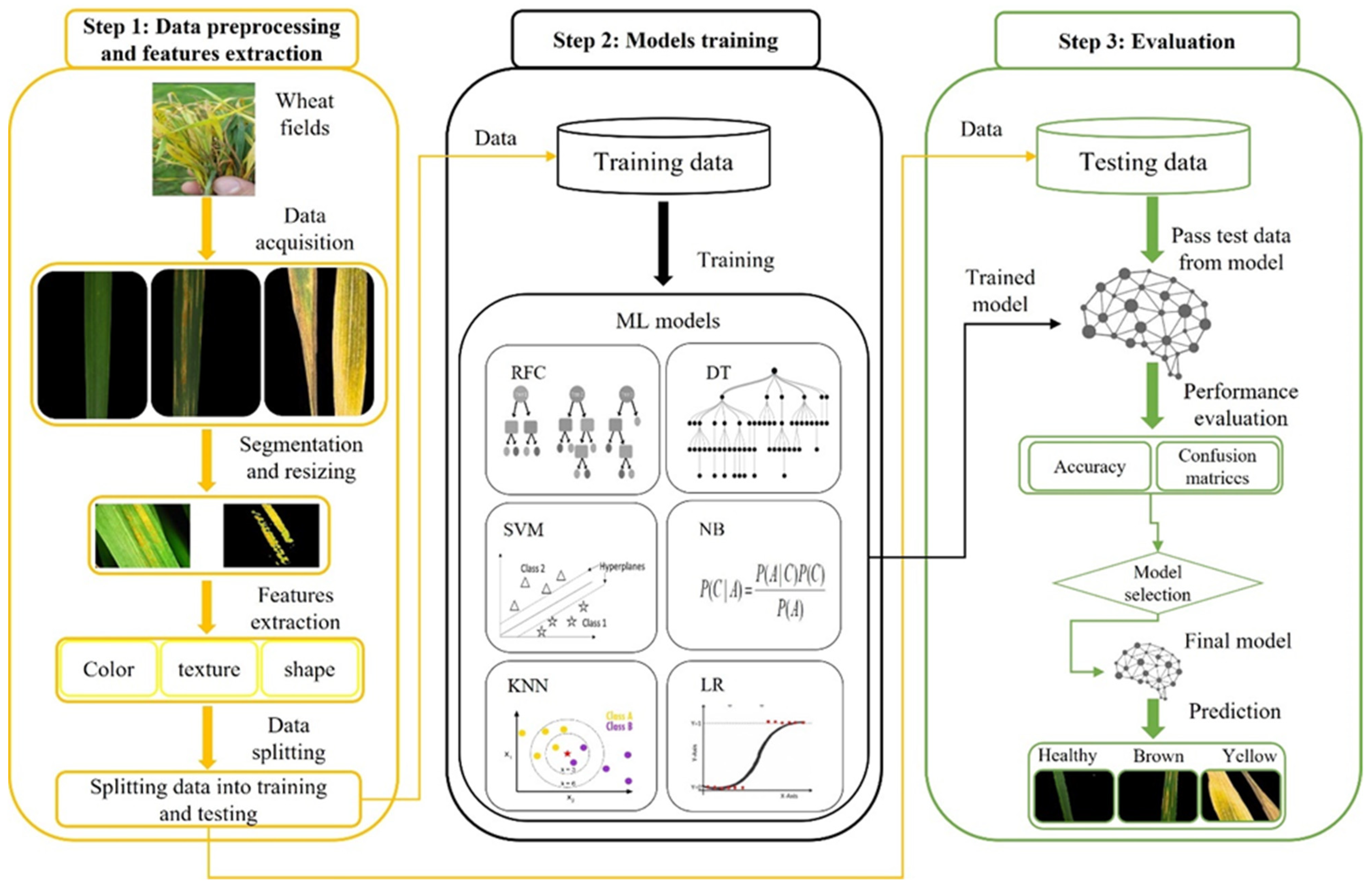
3. Materials and Methods
3.1. Real-Time Data Collection
3.2. Data Preprocessing and Features Extraction
3.2.1. Preprocessing
3.2.2. Feature Extraction
Hue Moments (HM)
Color Histogram (CH)
Haralick Texture (HT)
3.3. Proposed Fine-Tuned Framework
3.4. Comparative Analysis of Baseline Models
4. Experimental Results
4.1. Experimental Settings
4.2. Dataset
4.3. Evaluation Metrics
4.4. Results
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Aker, J.C. Dial “A” for agriculture: A review of information and communication technologies for agricultural extension in developing countries. Agric. Econ. 2011, 42, 631–647. [Google Scholar] [CrossRef]
- Barretto, R.; Buenavista, R.M.; Rivera, J.L.; Wang, S.; Prasad, P.V.; Siliveru, K. Teff (Eragrostis tef) processing, utilization and future opportunities: A review. Int. J. Food Sci. Technol. 2020, 56, 3125–3137. [Google Scholar] [CrossRef]
- Chakraborty, S.; Newton, A.C. Climate change, plant diseases and food security: An overview. Plant Pathol. 2011, 60, 2–14. [Google Scholar] [CrossRef]
- Nicholls, E.; Ely, A.; Birkin, L.; Basu, P.; Goulson, D. The contribution of small-scale food production in urban areas to the sustainable development goals: A review and case study. Sustain. Sci. 2020, 15, 1585–1599. [Google Scholar] [CrossRef]
- WHO. Available online: https://www.who.int/news-room/fact-sheets/detail/food-safety (accessed on 26 April 2022).
- Mottaleb, K.A.; Singh, P.K.; Sonder, K.; Kruseman, G.; Tiwari, T.P.; Barma, N.C.; Malaker, P.K.; Braun, H.-J.; Erenstein, O. Threat of wheat blast to South Asia’s food security: An ex-ante analysis. PLoS ONE 2018, 13, e0197555. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization of the United Nations (FAO). Supply and Deman Brief; Food and Agriculture Organization of the United Nations (FAO): Rome, Itlay, 2020. [Google Scholar]
- Figueroa, M.; Hammond-Kosack, K.E.; Solomon, P.S. A review of wheat diseases—A field perspective. Mol. Plant Pathol. 2018, 19, 1523–1536. [Google Scholar] [CrossRef]
- Huerta-Espino, J.; Singh, R.; German, S.; McCallum, B.; Park, R.; Chen, W.Q.; Bhardwaj, S.; Goyeau, H. Global status of wheat leaf rust caused by Puccinia triticina. Euphytica 2011, 179, 143–160. [Google Scholar] [CrossRef]
- Sankaran, S.; Mishra, A.; Ehsani, R.; Davis, C. A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 2010, 72, 1–13. [Google Scholar] [CrossRef]
- Jha, K.; Doshi, A.; Patel, P.; Shah, M. A comprehensive review on automation in agriculture using artificial intelligence. Artif. Intell. Agric. 2019, 2, 1–12. [Google Scholar] [CrossRef]
- Khan, N.; Muhammad, K.; Hussain, T.; Nasir, M.; Munsif, M.; Imran, A.S.; Sajjad, M. An adaptive game-based learning strategy for children road safety education and practice in virtual space. Sensors 2021, 21, 3661. [Google Scholar] [CrossRef] [PubMed]
- Haroon, U.; Ullah, A.; Hussain, T.; Ullah, W.; Sajjad, M.; Muhammad, K.; Lee, M.Y.; Baik, S.W. A Multi-Stream Sequence Learning Framework for Human Interaction Recognition. IEEE Trans. Hum.-Mach. Syst. 2022, 52, 435–444. [Google Scholar] [CrossRef]
- Khan, S.U.; Haq, I.U.; Khan, N.; Muhammad, K.; Hijji, M.; Baik, S.W. Learning to rank: An intelligent system for person reidentification. Int. J. Intell. Syst. 2022, 37, 5924–5948. [Google Scholar] [CrossRef]
- He, J.; Baxter, S.L.; Xu, J.; Xu, J.; Zhou, X.; Zhang, K. The practical implementation of artificial intelligence technologies in medicine. Nat. Med. 2019, 25, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.U.; Hussain, T.; Ullah, A.; Baik, S.W. Deep-ReID: Deep features and autoencoder assisted image patching strategy for person re-identification in smart cities surveillance. Multimed. Tools Appl. 2021, 1–22. [Google Scholar] [CrossRef]
- Ullah, W.; Ullah, A.; Hussain, T.; Muhammad, K.; Heidari, A.A.; Del Ser, J.; Baik, S.W.; De Albuquerque, V.H.C. Artificial Intelligence of Things-assisted two-stream neural network for anomaly detection in surveillance Big Video Data. Future Gener. Comput. Syst. 2022, 129, 286–297. [Google Scholar] [CrossRef]
- Ullah, W.; Ullah, A.; Haq, I.U.; Muhammad, K.; Sajjad, M.; Baik, S.W. CNN features with bi-directional LSTM for real-time anomaly detection in surveillance networks. Multimed. Tools Appl. 2021, 80, 16979–16995. [Google Scholar] [CrossRef]
- Yar, H.; Hussain, T.; Khan, Z.A.; Koundal, D.; Lee, M.Y.; Baik, S.W. Vision sensor-based real-time fire detection in resource-constrained IoT environments. Comput. Intell. Neurosci. 2021, 2021, 5195508. [Google Scholar] [CrossRef] [PubMed]
- Patrício, D.I.; Rieder, R. Computer vision and artificial intelligence in precision agriculture for grain crops: A systematic review. Comput. Electron. Agric. 2018, 153, 69–81. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. A review on the main challenges in automatic plant disease identification based on visible range images. Biosyst. Eng. 2016, 144, 52–60. [Google Scholar] [CrossRef]
- Barbedo, J.G. Factors influencing the use of deep learning for plant disease recognition. Biosyst. Eng. 2018, 172, 84–91. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Automatic image-based detection and recognition of plant diseases—A critical view. In Proceedings of the XI Congresso Brasileiro de Agroinformática, Sao Paulo, Brazil, 2–6 October 2017. [Google Scholar]
- Hughes, D.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Barbedo, J.G.A.; Koenigkan, L.V.; Halfeld-Vieira, B.A.; Costa, R.V.; Nechet, K.L.; Godoy, C.V.; Junior, M.L.; Patricio, F.R.A.; Talamini, V.; Chitarra, L.G. Annotated plant pathology databases for image-based detection and recognition of diseases. IEEE Lat. Am. Trans. 2018, 16, 1749–1757. [Google Scholar] [CrossRef]
- Johannes, A.; Picon, A.; Alvarez-Gila, A.; Echazarra, J.; Rodriguez-Vaamonde, S.; Navajas, A.D.; Ortiz-Barredo, A. Automatic plant disease diagnosis using mobile capture devices, applied on a wheat use case. Comput. Electron. Agric. 2017, 138, 200–209. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. Plant disease identification from individual lesions and spots using deep learning. Biosyst. Eng. 2019, 180, 96–107. [Google Scholar] [CrossRef]
- Ngugi, L.C.; Abelwahab, M.; Abo-Zahhad, M. Recent advances in image processing techniques for automated leaf pest and disease recognition—A review. Inf. Process. Agric. 2021, 8, 27–51. [Google Scholar] [CrossRef]
- Wójtowicz, A.; Piekarczyk, J.; Czernecki, B.; Ratajkiewicz, H. A random forest model for the classification of wheat and rye leaf rust symptoms based on pure spectra at leaf scale. J. Photochem. Photobiol. B Biol. 2021, 223, 112278. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Zhao, J.; Hu, G.; Zhang, D.; Huang, L.; Liang, D. Identification of wheat leaf diseases and their severity based on elliptical-maximum margin criterion metric learning. Sustain. Comput. Inform. Syst. 2021, 30, 100526. [Google Scholar] [CrossRef]
- Paul, A.; Ghosh, S.; Das, A.K.; Goswami, S.; Choudhury, S.D.; Sen, S. A review on agricultural advancement based on computer vision and machine learning. In Emerging Technology in Modelling and Graphics; Springer: Cham, Switzerland, 2020; pp. 567–581. [Google Scholar]
- Kumar, M.; Hazra, T.; Tripathy, S.S. Wheat leaf disease detection using image processing. Int. J. Latest Technol. Eng. Manag. Appl. Sci. (IJLTEMAS) 2017, 6, 73–76. [Google Scholar]
- Liakos, K.G.; Busato, P.; Moshou, D.; Pearson, S.; Bochtis, D. Machine learning in agriculture: A review. Sensors 2018, 18, 2674. [Google Scholar] [CrossRef] [PubMed]
- Dixit, A.; Nema, S. Wheat Leaf Disease Detection Using Machine Learning Method—A Review. Int. J. Comput. Sci. Mob. Comput. 2018, 7, 124–129. [Google Scholar]
- Xu, P.; Wu, G.; Guo, Y.; Yang, H.; Zhang, R. Automatic wheat leaf rust detection and grading diagnosis via embedded image processing system. Procedia Comput. Sci. 2017, 107, 836–841. [Google Scholar] [CrossRef]
- Islam, M.; Dinh, A.; Wahid, K.; Bhowmik, P. Detection of potato diseases using image segmentation and multiclass support vector machine. In Proceedings of the IEEE 30th Canadian Conference on Electrical and Computer Engineering (CCECE), Windsor, ON, Canada, 30 April–3 May 2017; pp. 1–4. [Google Scholar]
- Alehegn, E. Ethiopian maize diseases recognition and classification using support vector machine. Int. J. Comput. Vis. Robot. 2019, 9, 90–109. [Google Scholar] [CrossRef]
- Hossain, S.; Mou, R.M.; Hasan, M.M.; Chakraborty, S.; Razzak, M.A. Recognition and detection of tea leaf’s diseases using support vector machine. In Proceedings of the IEEE 14th International Colloquium on Signal Processing & Its Applications (CSPA), Penang, Malaysia, 9–10 March 2018; pp. 150–154. [Google Scholar]
- Ullah, W.; Muhammad, K.; Ul Haq, I.; Ullah, A.; Ullah Khattak, S.; Sajjad, M. Splicing sites prediction of human genome using machine learning techniques. Multimed. Tools Appl. 2021, 80, 30439–30460. [Google Scholar] [CrossRef]
- Ahmad, F.; Ikram, S.; Ahmad, J.; Ullah, W.; Hassan, F.; Khattak, S.U.; Rehman, I.U. GASPIDs Versus Non-GASPIDs-Differentiation Based on Machine Learning Approach. Curr. Bioinform. 2020, 15, 1056–1064. [Google Scholar] [CrossRef]
- Aurangzeb, K.; Akmal, F.; Khan, M.A.; Sharif, M.; Javed, M.Y. Advanced machine learning algorithm based system for crops leaf diseases recognition. In Proceedings of the IEEE 6th Conference on Data Science and Machine Learning Applications (CDMA), Riyadh, Saudi Arabia, 4–5 March 2020; pp. 146–151. [Google Scholar]
- Treboux, J.; Genoud, D. Improved machine learning methodology for high precision agriculture. In Proceedings of the IEEE Global Internet of Things Summit (GIoTS), Bilbao, Spain, 4–7 June 2018; pp. 1–6. [Google Scholar]
- Rumpf, T.; Mahlein, A.-K.; Steiner, U.; Oerke, E.-C.; Dehne, H.-W.; Plümer, L. Early detection and classification of plant diseases with support vector machines based on hyperspectral reflectance. Comput. Electron. Agric. 2010, 74, 91–99. [Google Scholar] [CrossRef]
- Ramesh, S.; Hebbar, R.; Niveditha, M.; Pooja, R.; Shashank, N.; Vinod, P. Plant disease detection using machine learning. In Proceedings of the IEEE International Conference on Design Innovations for 3Cs Compute Communicate Control (ICDI3C), Bangalore, India, 25–28 April 2018; pp. 41–45. [Google Scholar]
- Phadikar, S.; Sil, J.; Das, A.K. Classification of rice leaf diseases based on morphological changes. Int. J. Inf. Electron. Eng. 2012, 2, 460–463. [Google Scholar]
- Prajapati, H.B.; Shah, J.P.; Dabhi, V.K. Detection and classification of rice plant diseases. Intell. Decis. Technol. 2017, 11, 357–373. [Google Scholar] [CrossRef]
- Ahmed, K.; Shahidi, T.R.; Alam, S.M.I.; Momen, S. Rice leaf disease detection using machine learning techniques. In Proceedings of the IEEE International Conference on Sustainable Technologies for Industry 4.0 (STI), Dhaka, Bangladesh, 24–25 December 2019; pp. 1–5. [Google Scholar]
- Panigrahi, K.P.; Das, H.; Sahoo, A.K.; Moharana, S.C. Maize leaf disease detection and classification using machine learning algorithms. In Progress in Computing, Analytics and Networking; Springer: Cham, Switzerland, 2020; pp. 659–669. [Google Scholar]
- Waghmare, H.; Kokare, R.; Dandawate, Y. Detection and classification of diseases of grape plant using opposite colour local binary pattern feature and machine learning for automated decision support system. In Proceedings of the IEEE 3rd International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 11–12 February 2016; pp. 513–518. [Google Scholar]
- Zhao, J.; Fang, Y.; Chu, G.; Yan, H.; Hu, L.; Huang, L. Identification of leaf-scale wheat powdery mildew (Blumeria graminis f. sp. Tritici) combining hyperspectral imaging and an SVM classifier. Plants 2020, 9, 936. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Ma, Z.; Wang, H. Image recognition of wheat stripe rust and wheat leaf rust based on support vector machine. J. China Agric. Univ. 2012, 17, 72–79. [Google Scholar]
- Azadbakht, M.; Ashourloo, D.; Aghighi, H.; Radiom, S.; Alimohammadi, A. Wheat leaf rust detection at canopy scale under different LAI levels using machine learning techniques. Comput. Electron. Agric. 2019, 156, 119–128. [Google Scholar] [CrossRef]
- Tursunov, A.; Choeh, J.Y.; Kwon, S. Age and gender recognition using a convolutional neural network with a specially designed multi-attention module through speech spectrograms. Sensors 2021, 21, 5892. [Google Scholar] [CrossRef] [PubMed]
- Mustaqeem; Ishaq, M.; Kwon, S. A CNN-Assisted deep echo state network using multiple Time-Scale dynamic learning reservoirs for generating Short-Term solar energy forecasting. Sustain. Energy Technol. Assess. 2022, 52, 102275. [Google Scholar] [CrossRef]
- Maji, B.; Swain, M.; Mustaqeem. Advanced Fusion-Based Speech Emotion Recognition System Using a Dual-Attention Mechanism with Conv-Caps and Bi-GRU Features. Electronics 2022, 11, 1328. [Google Scholar] [CrossRef]


| Article | Crop | Preprocessing | Features | Algorithms/ Models | Accuracy |
|---|---|---|---|---|---|
| Xu et al., 2017 [35] | Wheat | Conversion images to G single gray RGB model, background removal | Binary features point set | Flood filling algorithm | 92.3% |
| Islam et al., 2017 [36] | Potato | Color based segmentation | Statistical features | SVM | 95% |
| Alehegen et al., 2019 [37] | Maize | Segmentation | Texture and morphological | SVM | 95.63% |
| Hossain et al., 2018 [38] | Tea | Image resizing and cropping | Statistical Features | SVM | 93% |
| Aurangzeb et al., 2020 [41] | Corn and Potato | Image resizing | LTP, HOG, SFTA | MSVM | 92.8% and 98.7% |
| Treboux et al., 2018 [42] | Vineyards | Morphological operation (Opening and closing) | First order statistic, Tamura, Haralick | DTE | 94.275% |
| Rumpf et al., 2010 [43] | Sugar beet | Image resizing, Clustering | Physiological parameters | SVM | 97% |
| Ramesh et al., 2018 [44] | Papaya | Image resizing and Normalization | HOG | RFC | 70% |
| Phadikar et al., 2012 [45] | Rice | Enhancement via mean filters and segmentation | Colors descriptors | SVM, NB | 68.1% and 79.5% |
| Prajapati et al., 2017 [46] | Rice | Back removal, segmentation | Texture, Color, and shape | SVM | 93.33% |
| Ahmed et al., 2019 [47] | Rice | Augmentation | Pure statistical features | DT | 97.91% |
| Panigrahi et al.,2020 [48] | Maize | Resizing, denoising, segmentation | Grayscale pixel values | NB, KNN, DT, SVM and RFC | 79.23% (highest with RFC) |
| Waghmare et al., 2016 [49] | Graphs | Back removal | Texture | SVM | 96.6% |
| Zhao et al., 2020 [50] | Wheat | Image smoothing via S-G filter and derivative function | Disease level of severity, and affected leaf spots | SVM, PNN, and RFC | 93.33% |
| Li et al., 2012 [51] | Wheat | Cropping, denoising | Colored and texture | SVM with RBF | 96.67% |
| Azadbakht et al., 2019 [52] | Wheat | Noise reduction | Disease severity level, leaf area index, and pixel values | V-SVR, and RFR | 99% and 79% |
| Proposed framework | Wheat | Resizing, Masked based segmentation | Haralick texture, color histogram, and hue moments | Fine-tuned RFC | 99.8% |
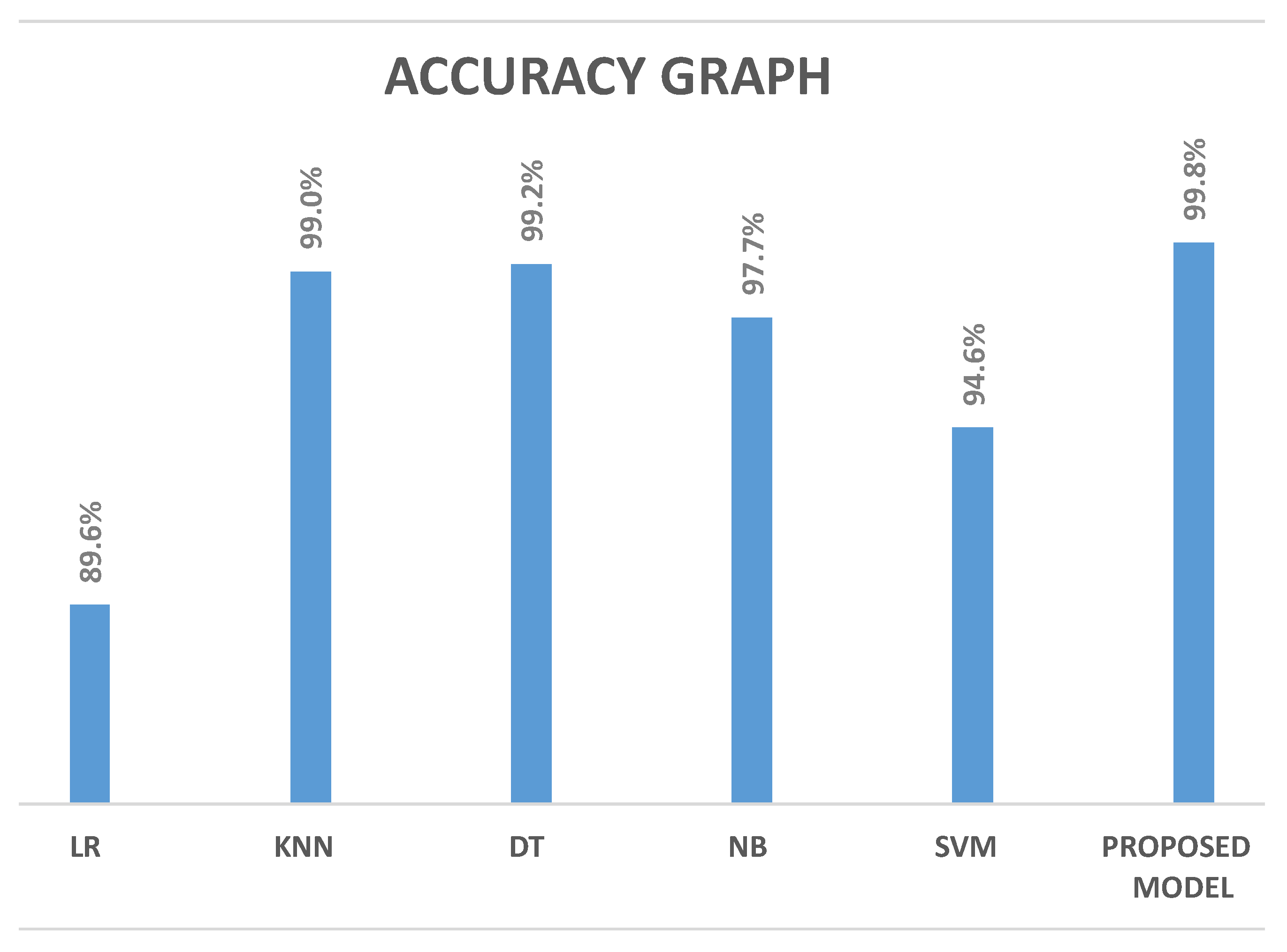
| Model | LR | SVM | NB | KNN | DT | Proposed Framework |
|---|---|---|---|---|---|---|
| Accuracy (%) | 89.6 | 94.4 | 97.7 | 99.0 | 99.2 | 99.8 |
| The Proposed Framework | ||||
|---|---|---|---|---|
| Predicted Classes ↓ | Healthy | Rusted | Yellow rusted | |
| Actual Classes → | ||||
| Healthy | 215 | 1 | 0 | |
| Rusted | 0 | 201 | 0 | |
| Yellow Rusted | 0 | 0 | 212 | |
| Overall Accuracy (%) | 99.8 | |||
| DT | ||||
| Predicted Classes ↓ | Healthy | Rusted | Yellow rusted | |
| Actual Classes → | ||||
| Healthy | 213 | 1 | 1 | |
| Rusted | 0 | 205 | 1 | |
| Yellow Rusted | 1 | 1 | 207 | |
| Overall Accuracy (%) | 99.2 | |||
| LR | ||||
| Predicted Classes ↓ | Healthy | Rusted | Yellow rusted | |
| Actual Classes → | ||||
| Healthy | 167 | 48 | 0 | |
| Rusted | 2 | 204 | 0 | |
| Yellow Rusted | 13 | 2 | 194 | |
| Overall Accuracy (%) | 89.6 | |||
| Class | Total Testing Images | Accurate Prediction |
|---|---|---|
| Healthy | 16 | 13 |
| Yellow | 18 | 12 |
| Rusted | 16 | 11 |
| Authors | Year | Features | Classifiers | Accuracy (%) |
|---|---|---|---|---|
| Azadbakht et al. [52] | 2019 | Texture | SVR, RFR, GRR, BRT | 99 |
| Zhao et al. [50] | 2020 | Diseased area/total area of leaf | SVM, PNN, RFC | 93.33 |
| Bao et al. [30] | 2021 | Color, texture, and combination of these two | Elliptical-Maximum Margin Criterion (E-MMC) metric learning | 94.16 |
| Proposed method | 2022 | Haralick-texture, Color-histogram, Hue-moment, LBP, HOG | Fine-Tuned RFC | 99.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, H.; Haq, I.U.; Munsif, M.; Mustaqeem; Khan, S.U.; Lee, M.Y. Automated Wheat Diseases Classification Framework Using Advanced Machine Learning Technique. Agriculture 2022, 12, 1226. https://doi.org/10.3390/agriculture12081226
Khan H, Haq IU, Munsif M, Mustaqeem, Khan SU, Lee MY. Automated Wheat Diseases Classification Framework Using Advanced Machine Learning Technique. Agriculture. 2022; 12(8):1226. https://doi.org/10.3390/agriculture12081226
Chicago/Turabian StyleKhan, Habib, Ijaz Ul Haq, Muhammad Munsif, Mustaqeem, Shafi Ullah Khan, and Mi Young Lee. 2022. "Automated Wheat Diseases Classification Framework Using Advanced Machine Learning Technique" Agriculture 12, no. 8: 1226. https://doi.org/10.3390/agriculture12081226
APA StyleKhan, H., Haq, I. U., Munsif, M., Mustaqeem, Khan, S. U., & Lee, M. Y. (2022). Automated Wheat Diseases Classification Framework Using Advanced Machine Learning Technique. Agriculture, 12(8), 1226. https://doi.org/10.3390/agriculture12081226









