Population Genetic Structure of Meloidogyne javanica Recovered from Different Regions of Iran
Abstract
:1. Introduction
2. Materials and Methods
2.1. Nematode Populations and Light Microscopy
2.2. DNA Extraction
2.3. PCR Using Species-Specific Primers
2.4. ISSR and RAPD PCR
2.5. ISSR and RAPD Data Analysis
3. Results
3.1. Characterization of the Recovered Populations
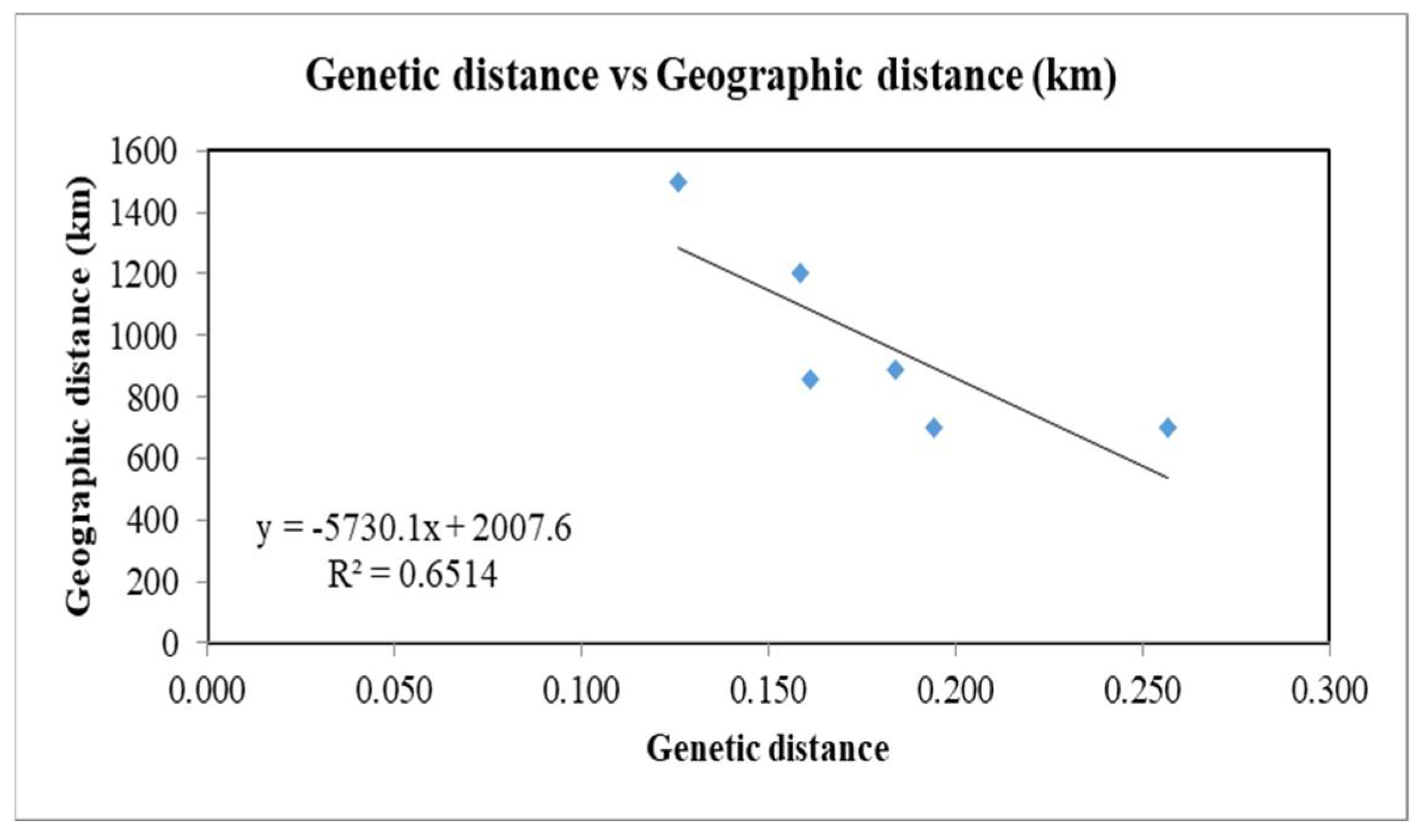
3.2. Genetic Diversity and Structure
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abad, P.B.; Favery, M.N.; Ross, P.; Serena, C. Root-knot nematode parasitism and host response: Molecular basis of a sophisticated interaction. Mol. Plant Pathol. 2003, 4, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Buenna, A.P.; Garcia-Alvarez, A.; Diez-Rojo, M.A.; Ros, C.; Fernandez, P.; Lacasa, A. Use of pepper crop residues for the control of root knot nematodes. Biores. Technol. 2007, 98, 2846–2851. [Google Scholar] [CrossRef]
- Ye, W.; Zeng, Y.; Kerns, J. Molecular characterisation and diagnosis of root-knot nematodes (Meloidogyne spp.) from turfgrasses in North Carolina, USA. PLoS ONE 2015, 10, e0143556. [Google Scholar] [CrossRef]
- Janssen, T.; Karssen, G.; Verhaeven, M.; Coyne, D.; Bert, W. Mitochondrial coding genome analysis of tropical root-knot nematodes (Meloidogyne) supports haplotype based diagnostics and reveals evidence of recent reticulate evolution. Sci. Rep. 2016, 6, 1–13. [Google Scholar]
- Cunha, T.G.D.; Visôtto, L.E.; Lopes, E.A.; Oliveira, C.M.G.; God, P.I.V.G. Diagnostic methods for identification of root-knot nematodes species from Brazil. Ciên. Rur. 2018, 48, 1–12. [Google Scholar] [CrossRef]
- Zijlstra, C. Identification of Meloidogyne chitwoodi, M. fallax and M. hapla based on SCAR-PCR: A powerful way of enabling reliable identification of populations or individuals that share common traits. Eur. J. Plant Pathol. 2000, 106, 283–290. [Google Scholar] [CrossRef]
- Zijlstra, C.; Donkers-Venne, D.T.H.M.; Fargette, M. Identification of Meloidogyne incognita, M. javanica and M. arenaria using sequence characterised amplified region (SCAR) based PCR assays. Nematology 2000, 2, 847–853. [Google Scholar]
- Askarian, H.; Sharifnabi, B.; Olia, M.; Moghadam, E.M.; Akhavan, A. Identification of Meloidogyne javanica using morphological and morphometrical characters and species-specific primers. J. Sci. Technol. Agric. Nat. Res. 2009, 13, 279–290. [Google Scholar]
- Mokaram Hesar, A.; Mehdikhani Moghadam, E.; Tanha Maafi, Z. Morphometrical and genetic diversity of Meloidogyne javanica isolates from the north east of Iran. J. Nematode Morph. System. 2011, 14, 1–11. [Google Scholar]
- Ghaderi, R.; Dehghan, A.A.; Hesar, A.M.; Karegar, A. Genetic intraspecific diversity of Meloidogyne javanica parasitizing vegetables in southern Iran. J. Nematol. 2020, 52, 1–13. [Google Scholar] [CrossRef]
- Adam, M.A.M.; Phillips, M.S.; Blok, V.C. Molecular diagnostic key for identification of single juveniles of seven common and economically important species of root-knot nematode (Meloidogyne spp.). Plant Pathol. 2007, 56, 190–197. [Google Scholar] [CrossRef]
- Baidoo, R.; Joseph, S.; Mengistu, T.M.; Brito, J.A.; McSorley, R.; Stamps, R.H.; Crow, W.T. Mitochondrial haplotype-based identification of root-knot nematodes (Meloidogyne spp.) on cut foliage crops in Florida. J. Nematol. 2016, 48, 193–202. [Google Scholar] [CrossRef]
- Rusinque, L.; Inácio, M.L.; Mota, M.; Nóbrega, F. Morphological, biochemical and molecular characterisation of Meloidogyne javanica, from North Portugal, in tomato. Rev. Ciên. Agrá. 2018, 41, 193–198. [Google Scholar] [CrossRef]
- Onkendi, E.M.; Kariuki, G.M.; Marais, M.; Moleleki, L.N. The threat of root-knot nematodes (Meloidogyne spp.) in Africa: A review. Plant Pathol. 2014, 63, 727–737. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.T.; Haegeman, A.; Danchin, E.G.; Gaur, H.S.; Helder, J.; Jones, M.G.; Perry, R.N. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 2013, 14, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Forghani, F.; Hajihassani, A. Recent advances in the development of environmentally benign treatments to control root-knot nematodes. Front. Plant Sci. 2020, 11, 1125. [Google Scholar] [CrossRef]
- Mahdikhani Moghadam, E. Nematodes of the family Meloidogynidae. In Plant-Parasitic Nematodes in Iran; Ghaderi, R., Kashi, L., Karegar, A., Eds.; Shiraz, Marjae elm & Iranian Society of Nematology: Shiraz, Iran, 2018; pp. 397–418. [Google Scholar]
- Lax, P.; Dueñas, J.R.; Gardenal, C.; Doucet, M. Assessment of genetic variability in populations of Nacobbus aberrans (Thorne, 1935) Thorne & Allen, 1944 (Nematoda: Pratylenchidae) from Argentina. Nematology 2007, 9, 261–270. [Google Scholar]
- Devran, Z.; Söğüt, M.A. Distribution and identification of root-knot nematodes from Turkey. J. Nematol. 2009, 41, 128–133. [Google Scholar]
- Hyman, B.C. Molecular systematics and population biology of phytonematodes: Some unifying principles. Fund. Appl. Nematol. 1996, 19, 309–313. [Google Scholar]
- Plantard, O.; Porte, C. Population genetic structure of the sugar beet cyst nematode Heterodera schachtii: A gonochorist and amphimictic species with highly inbred but weakly differentiated populations. Mol. Ecol. 2004, 13, 3341. [Google Scholar] [CrossRef]
- Rashidifard, M.; Fourie, H.; Véronneau, P.Y.; Marais, M.; Daneel, M.S.; Mimee, B. Genetic diversity and phylogeny of South African Meloidogyne populations using genotyping by sequencing. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Randig, O.; Bongiovanni, M.; Carneiro, R.M.D.G.; Castagnone-Sereno, P. Genetic diversity of root-knot nematodes from Brazil and development of SCAR markers specific for the coffee-damaging species. Genome 2002, 45, 862–870. [Google Scholar] [CrossRef] [PubMed]
- Cofcewicz, E.T.; Carneiro, R.M.D.G.; Castagnone-Sereno, P.; Quénéhervé, P. Enzyme phenotypes and genetic diversity of root-knot nematodes parasitising Musa in Brazil. Nematology 2004, 6, 85–95. [Google Scholar] [CrossRef]
- Medina, I.L.; Gomes, C.B.; Correa, V.R.; Mattos, V.S.; Castagnone-Sereno, P.; Carneiro, R.M.D.G. Genetic diversity of Meloidogyne spp. parasitising potato in Brazil and aggressiveness of M. javanica populations on susceptible cultivars. Nematology 2017, 19, 69–80. [Google Scholar] [CrossRef]
- de Grisse, A.T. Redescription ou modifications de quelques techniques utilisées dans l’étude des nématodes phytoparasitaires. Med. Van Den Rijks. Land. Gent. 1969, 34, 351–369. [Google Scholar]
- Tanha Maafi, Z.T.; Subbotin, S.A.; Moens, M. Molecular identification of cyst-forming nematodes (Heteroderidae) from Iran and a phylogeny based on ITS-rDNA sequences. Nematology 2003, 5, 99–111. [Google Scholar] [CrossRef]
- Rohlf, F.J. NTSYSpc: Numerical Taxonomy and Multivariate Analysis System, Version 2.02; Exeter Software: Setauket, NY, USA, 2000.
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Yap, I.V.; Nelson, R.J. WINBOOT: A Program for Performing Bootstrap Analysis for Binary Data to Determine the Confidence Limits of UPGMA-Based Dendrograms; Discussion Paper Series No. 14; International Rice Research Institute: Los Baños, Philippines, 1996. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Earl, D.A.; Vonholdt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Gen. Res. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Peakall, R.O.D.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes. 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Milgroom, M.G. Population Biology of Plant Pathogens: Genetics, Ecology, and Evolution; (No. 581.2 M644p); APS Press: St. Paul, MN, USA, 2015. [Google Scholar]
- Carneiro, R.M.D.G.; dos Santos, M.F.A.; Almeida, M.R.A.; Mota, F.C.; Gomes, A.C.M.M.; Tigano, M.S. Diversity of Meloidogyne arenaria using morphological, cytological and molecular approaches. Nematology 2008, 10, 819–834. [Google Scholar]
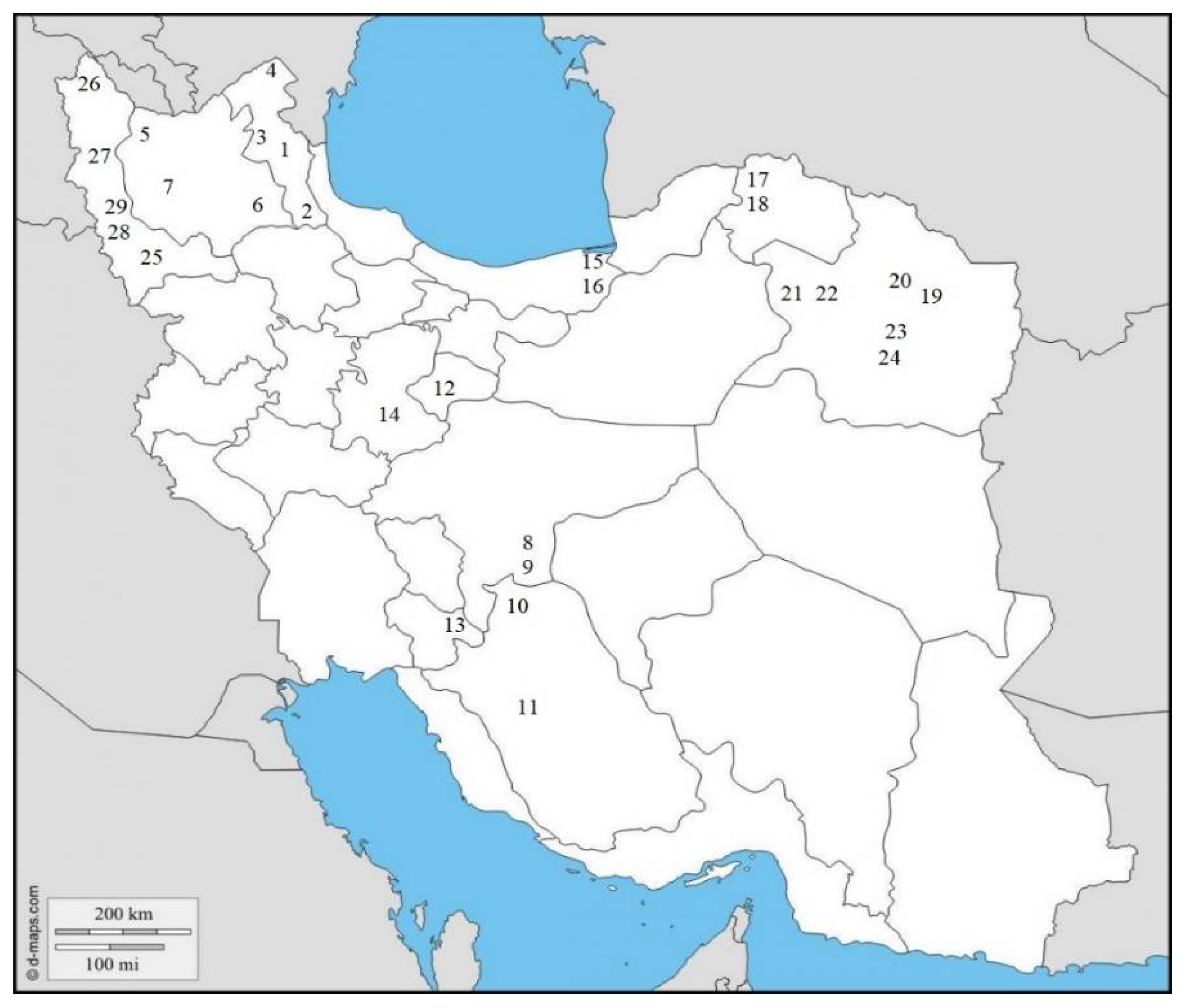
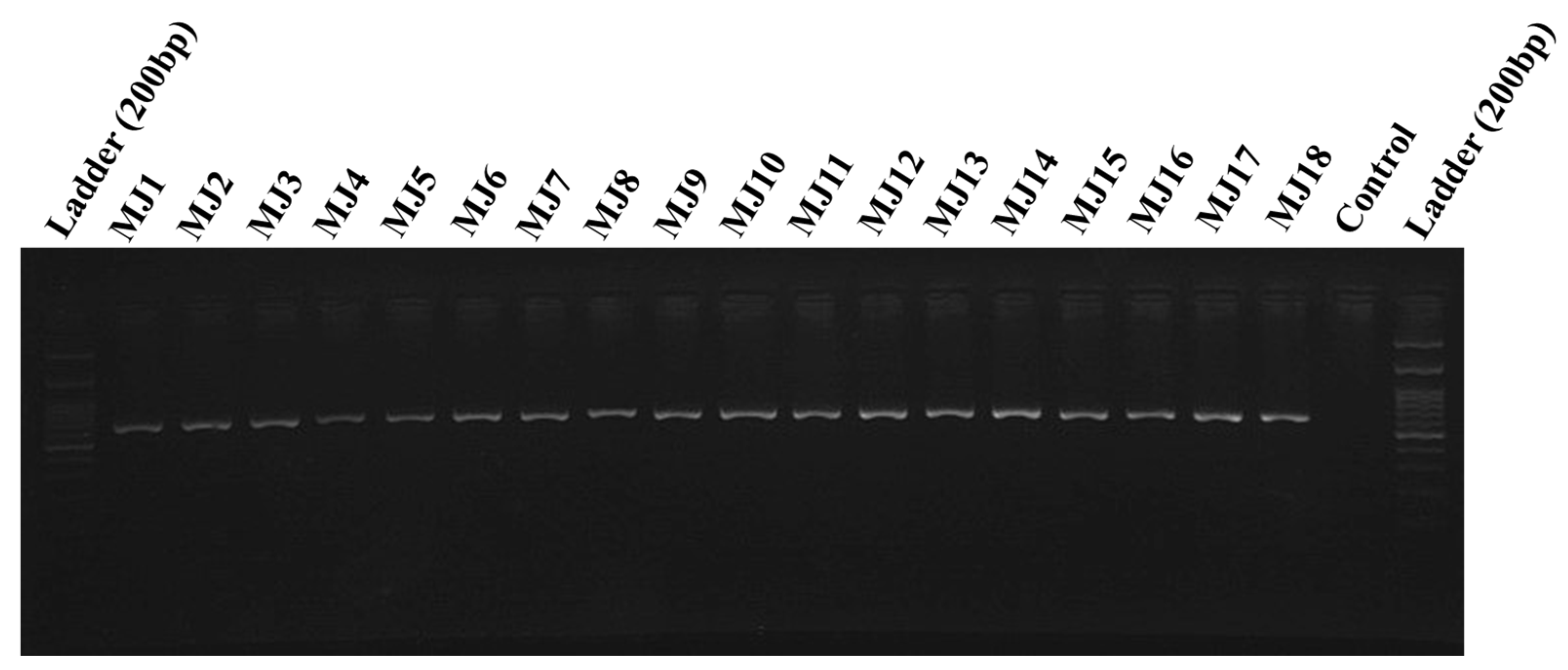

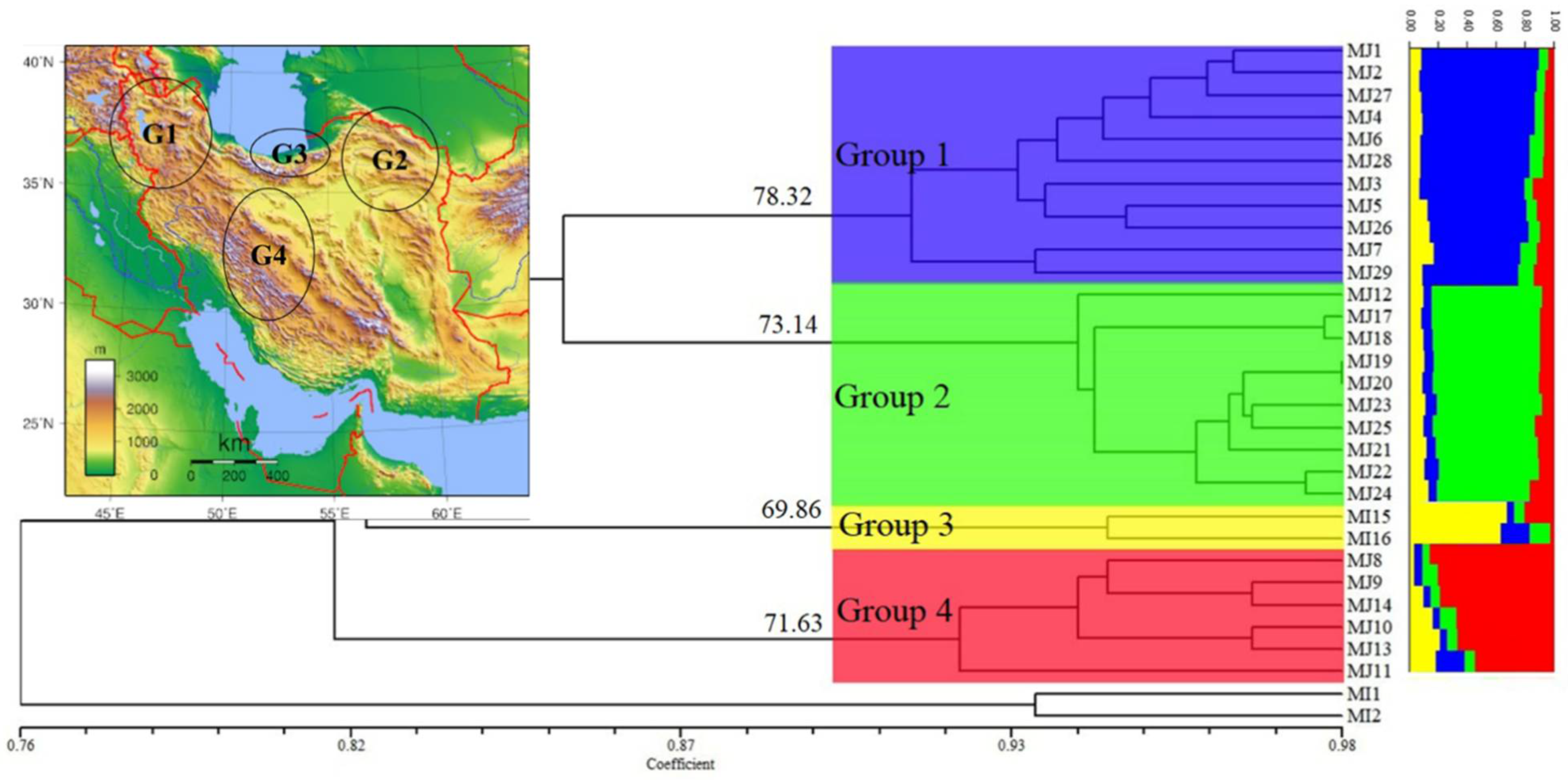
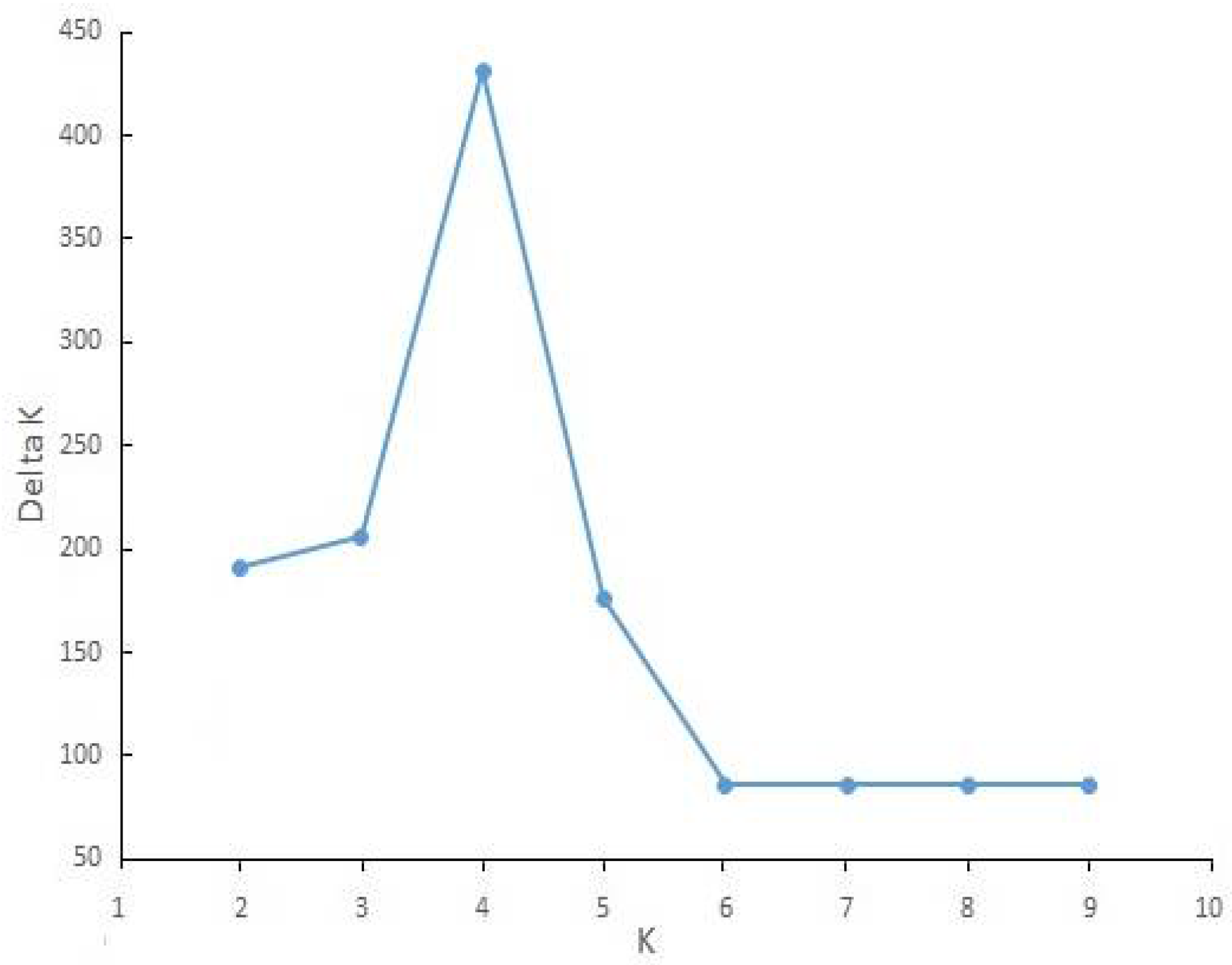

| Number | Sampling Area | N | E | H | Sample | Host Plant | Greenhouse/Field | Molecular Code |
|---|---|---|---|---|---|---|---|---|
| 1 | Ardabil province-Ardabil-Soumee | 38°17′22.92″ | 48°14′18.46″ | 1879 | Root | Cucumber | Greenhouse | MJ1 |
| 2 | Ardabil province-Khalkhal | 37°36′00.77″ | 48°32′40.52″ | 1778 | Soil & Root | Tomato | Field | MJ2 |
| 3 | Ardabil province-Meshkin Shahr-Parikhan | 38°25′14.88″ | 47°38′27.34″ | 1425 | Soil & Root | Tomato | Field | MJ3 |
| 4 | Ardabil province-Pars Abad-Ajirloo | 39°37′18.13″ | 47°55′42.54″ | 181 | Soil & Root | Tomato | Field | MJ4 |
| 5 | East Azerbaijan-Marand-Hejvan | 38°26′37.28″ | 45°50′20.31″ | 1334 | Soil & Root | Tomato | Field | MJ5 |
| 6 | East Azerbaijan-Miyaneh-Achachi | 37°24′11.93″ | 47°47′37.24″ | 1069 | Soil & Root | Cucumber | Greenhouse | MJ6 |
| 7 | East Azerbaijan-Tabriz-Sardroud | 38°00′13.71″ | 46°10′31.39″ | 1377 | Soil & Root | Tomato | Field | MJ7 |
| 8 | Esfahan province-Esfahan-Ghale shour | 32°28′41.16″ | 51°50′33.03″ | 1551 | Soil & Root | Tomato | Greenhouse | MJ8 |
| 9 | Esfahan province-Esfahan-Ghale shour | 32°28′41.15″ | 51°50′32.40″ | 1560 | Soil & Root | Tomato | Greenhouse | MJ9 |
| 10 | Fars province-Abadeh-Izad Khast | 31°32′18.77″ | 52°07′52.25″ | 2192 | Soil & Root | Tomato | Field | MJ10 |
| 11 | Fars province-Shiraz-Tange Khiareh | 29°55′41.12″ | 52°26′51.11″ | 1643 | Soil & Root | Tomato | Field | MJ11 |
| 12 | Ghom province-Ghom-Qanavat | 34°36′47.90″ | 51°02′11.32″ | 882 | Soil & Root | Tomato | Field | MJ12 |
| 13 | Kohgiluyeh and Boyer-Ahmad province-Yasuj | 30°39′00.57″ | 51°35′38.85″ | 1879 | Soil & Root | Tomato | Greenhouse | MJ13 |
| 14 | Markazi province-Arak-Mazijaran | 34°08′44.64″ | 49°39′43.87″ | 1750 | Soil & Root | Tomato | Greenhouse | MJ14 |
| 15 | Mazandaran province-Galugah-Lemrask | 36°43′09.71″ | 53°43′29.91″ | 40 | Root | Tobacco | Field | MJ15 |
| 16 | Mazandaran province-Galugah-Lemrask | 36°43′37.11″ | 53°43′22.40″ | 4 | Root | Tobacco | Field | MJ16 |
| 17 | North Khorasan-Ashkhaneh | 37°32′38.47″ | 56°54′56.11″ | 743 | Root | Tomato | Field | MJ17 |
| 18 | North Khorasan-Ashkhaneh | 37°34′17.53″ | 56°55′24.20″ | 752 | Root | Tomato | Field | MJ18 |
| 19 | Razavi Khorasan province-Mashhad-Tabadkan | 36°28′34.41″ | 59°54′14.58″ | 1218 | Root | Tomato | Field | MJ19 |
| 20 | Razavi Khorasan province-Mashhad-Tous | 36°28′33.07″ | 59°30′35.78″ | 1001 | Root | Tomato | Field | MJ20 |
| 21 | Razavi Khorasan province-Sabzevar-Abaresh | 36°13′06.94″ | 57°36′47.93″ | 984 | Root | Tomato | Field | MJ21 |
| 22 | Razavi Khorasan province-Sabzevar-Kohne Aab | 36°13′08.84″ | 57°38′57.70″ | 993 | Soil & Root | Cucumber | Greenhouse | MJ22 |
| 23 | Razavi Khorasan province-Torbat Heydarieh | 35°14′30.16″ | 59°13′52.81″ | 1385 | Root | Tomato | Greenhouse | MJ23 |
| 24 | Razavi Khorasan province-Torbat Heydarieh | 35°17′47.97″ | 59°12′21.21″ | 1387 | Root | Cucumber | Greenhouse | MJ24 |
| 25 | West Azerbaijan-Mahabad-Eyrigash | 36°49′55.72″ | 45°45′37.69″ | 1472 | Soil & Root | Tomato | Field | MJ25 |
| 26 | West Azerbaijan-Maku-Ghale Jogh | 39°17′08.22″ | 44°27′49.46″ | 1305 | Soil & Root | Tomato | Greenhouse | MJ26 |
| 27 | West Azerbaijan-Salmas-Taze Shahr | 38°10′33.45″ | 44°42′26.37″ | 1442 | Root | Cucumber | Greenhouse | MJ27 |
| 28 | West Azerbaijan-Urmia-jarchlu | 37°35′34.69″ | 45°07′38.32″ | 1296 | Soil & Root | Tomato | Greenhouse | MJ28 |
| 29 | West Azerbaijan-Urmia-Kashtiban | 37°33′18.29″ | 45°15′09.30″ | 1280 | Soil & Root | Tomato | Field | MJ29 |
| ISSR | RAPD | ||
| (CCA)5 | 5′-CCACCACCACCACCA-3′ | OPA8 | 5′-GTGACGTAGG-3′ |
| (GTG)6 | 5′-GTGGTGGTGGTGGTGGTG-3′ | OPA10 | 5′-GTGATCGCAG-3′ |
| (ATG)6 | 5′-ATGATGATGATGATGATG-3′ | OPA13 | 5′-CAGCACCCAC-3′ |
| (GAG)4GC | 5′-GAGGAGGAGGAGGC-3′ | OPAD10 | 5′-AAGAGGCCAG-3′ |
| (GACA)4 | 5′-GACAGACAGACAGACA-3′ | OPE18 | 5′-GGACTGCAGA-3′ |
| (GA)8C | 5′-GAGAGAGAGAGAGAGAC-3′ | OPP17 | 5′-TGACCCGCCT-3′ |
| (AC)8T | 5′-ACACACACACACACACT-3′ | OPB11 | 5′-GTAGACCCGT-3′ |
| (CA)8G | 5′-CACACACACACACACAG-3′ | OPB17 | 5′-AGGGAACGAG-3′ |
| (CTC)4GC | 5′-CTCCTCCTCCTCGC-3′ | OPD8 | 5′-GTGTGCCCCA-3′ |
| (GAGA)4GG | 5′-GAGAGAGAGAGAGAGAGG-3′ | OPC8 | 5′-TGGACCGGTG-3′ |
| Species-specific primers | |||
| M. javanica | Fjav (5′-GGTGCGCGATTGAACTGAGC-3′) | M. incognita | Finc (5′-CTCTGCCCAATGAGCTGTCC-3′) |
| Rjav (5′-CAGGCCCTTCAGTGGAACTATAC-3′) | Rinc (5′-CTCTGCCCTCACATTAAG-3′) | ||
| M. arenaria | Far (5′-TCGGCGATAGAGGTAAATGAC-3′) | M. hapla | F (5′-TGACGGCGGTGAGTGCGA-3′) |
| Rar (5′-TCGGCGATAGACACTACAAACT-3′) | R (5′-TGACGGCGGTACCTCATAG-3′) | ||
| Diagnostic Characters | Females | Males | J2s | |||
|---|---|---|---|---|---|---|
| 54 | CV | 17 | CV | 61 | CV | |
| L (including neck) | 693 ± 102 (521–986) | 16 | 1327 ± 133 (1078–1563) | 15 | 452 ± 14 (414–504) | 11 |
| Body width (W) | 418 ± 79 (296–587) | 20 | 35 ± 3.6 (27–40) | 14 | 16 ± 2.1 (10–21) | 12 |
| Neck length | 231 ± 70 (141–352) | 26 | - | - | - | - |
| Stylet | 16 ± 2 (13–19) | 15 | 17 ± 1.7 (15–21) | 9 | 11 ± 0.4 (9–13) | 8 |
| DGO | 3.4 ± 0.8 (2–5) | 23 | 4.2 ± 0.4 (3.3–5.1) | 11 | 3.1 ± 0.4 (1.5–4.2) | 26 |
| Excretory pore | 175 ± 21 (133–261) | 16 | - | - | - | - |
| Vulval slit length | 22 ± 4 (14–29) | 16 | - | - | - | - |
| Vulva-anus distance | 14 ± 5 (10–31) | 29 | - | - | - | - |
| Inter-phasmidial distance | 81 ± 12 (63–97) | 16 | - | - | - | - |
| Tail length | - | - | 16 ± 3.2 (11–20) | 18 | 49 ± 7 (38–61) | 13 |
| Hyaline length | - | - | - | - | 15 ± 3 (10–22) | 23 |
| Pharynx (glands end) | - | - | 491 ± 62 (374–587) | 16 | 172 ± 29 (112–197) | 14 |
| Body length to width (a) | 1.8 ± 0.3 (1.2–2.5) | 25 | 42 ± 3.4 (36–48) | 13 | 29 ± 2 (24–36) | 12 |
| b | - | - | 5.9 ± 1.4 (4.6–7.4) | 22 | 6.1 ± 1.3 (4.5–9.4) | 21 |
| b′ | - | - | 2.9 ± 0.4 (2.2–3.3) | 15 | 3.1 ± 0.7 (2.1–4.2) | 24 |
| c | - | - | 122 ± 19 (91–151) | 27 | 9.1 ± 0.6 (7.1–10.2) | 13 |
| c′ | - | - | 0.6 ± 0.2 (0.3–1.2) | 22 | 5.2 ± 0.4 (3.9–6.4) | 15 |
| Spicules | - | - | 25 ± 6.3 (14–31) | 22 | - | - |
| Gubernaculum | - | - | 12 ± 2.7 (9–18) | 17 | - | - |
| RAPD | Total Band | Polymorph | Monomorph | ISSR | Total Band | Polymorph | Monomorph |
|---|---|---|---|---|---|---|---|
| OPA8 | 18 | 15 | 3 | (CCA)5 | 17 | 12 | 5 |
| OPA10 | 16 | 15 | 1 | (GTG)6 | 15 | 16 | 1 |
| OPA13 | 18 | 13 | 5 | (ATG)6 | 18 | 13 | 5 |
| OPAD10 | 21 | 17 | 4 | (GAG)4GC | 20 | 14 | 6 |
| OPE18 | 20 | 16 | 4 | (GACA)4 | 17 | 14 | 3 |
| OPP17 | 16 | 11 | 5 | (GA)8C | 16 | 12 | 4 |
| OPB11 | 16 | 13 | 3 | (AC)8T | 16 | 13 | 3 |
| OPB17 | 18 | 15 | 3 | (CA)8G | 18 | 16 | 2 |
| OPD8 | 18 | 16 | 2 | (CTC)4GC | 18 | 17 | 1 |
| OPC8 | 14 | 11 | 3 | (GAGA)4GG | 13 | 11 | 2 |
| Total | 175 | 142 | 33 | Total | 168 | 138 | 32 |
| Mean | 17.5 | 14.2 | 3.3 | Mean | 16.8 | 13.8 | 3.2 |
| Percentage | - | 81.14 | 18.85 | Percentage | - | 82.14 | 19.04 |
| Population | ISSR-RAPD | |||
|---|---|---|---|---|
| %P † | I | H | uHe | |
| Group 1 | 27.43% | 0.167 | 0.115 | 0.121 |
| Group 2 | 18.58% | 0.112 | 0.078 | 0.082 |
| Group 3 | 5.90% | 0.036 | 0.024 | 0.033 |
| Group 4 | 16.52% | 0.109 | 0.078 | 0.085 |
| Mean | 17.11% | 0.106 | 0.074 | 0.08 |
| Group 1 | Group 2 | Group 3 | Group 4 | |
|---|---|---|---|---|
| *** | 1500 | 860 | 890 | Group 1 |
| 0.126 | *** | 700 | 1200 | Group 2 |
| 0.161 | 0.194 | *** | 700 | Group 3 |
| 0.184 | 0.159 | 0.257 | *** | Group 4 |
| Source of Variation | Degree of Freedom | Sum of Squares | Variance Components | Percentage of Variation | Fixation Index (FST) | Number of Migrants (Nm) |
|---|---|---|---|---|---|---|
| Between populations | 3 | 395.908 | 18.18 * | 63 | ||
| Within populations | 25 | 269.333 | 10.77 * | 37 | 0.628 | 0.148 |
| Total | 28 | 665.241 | 28.95 |
| Group 1 | Group 2 | Group 3 | Group 4 | |
|---|---|---|---|---|
| *** | 0.184 | 0.176 | 0.150 | Group 1 |
| 0.575 | *** | 0.095 | 0.123 | Group 2 |
| 0.586 | 0.724 | *** | 0.094 | Group 3 |
| 0.624 | 0.670 | 0.726 | *** | Group 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hesar, A.M.; Rostami, M.; Ghaderi, R.; Danesh, Y.R.; Jalal, A.; da Silva Oliveira, C.E.; Teixeira Filho, M.C.M. Population Genetic Structure of Meloidogyne javanica Recovered from Different Regions of Iran. Agriculture 2022, 12, 1374. https://doi.org/10.3390/agriculture12091374
Hesar AM, Rostami M, Ghaderi R, Danesh YR, Jalal A, da Silva Oliveira CE, Teixeira Filho MCM. Population Genetic Structure of Meloidogyne javanica Recovered from Different Regions of Iran. Agriculture. 2022; 12(9):1374. https://doi.org/10.3390/agriculture12091374
Chicago/Turabian StyleHesar, Abbas Mokaram, Mahsa Rostami, Reza Ghaderi, Younes Rezaee Danesh, Arshad Jalal, Carlos Eduardo da Silva Oliveira, and Marcelo Carvalho Minhoto Teixeira Filho. 2022. "Population Genetic Structure of Meloidogyne javanica Recovered from Different Regions of Iran" Agriculture 12, no. 9: 1374. https://doi.org/10.3390/agriculture12091374
APA StyleHesar, A. M., Rostami, M., Ghaderi, R., Danesh, Y. R., Jalal, A., da Silva Oliveira, C. E., & Teixeira Filho, M. C. M. (2022). Population Genetic Structure of Meloidogyne javanica Recovered from Different Regions of Iran. Agriculture, 12(9), 1374. https://doi.org/10.3390/agriculture12091374








