DDIT3 Governs Milk Production Traits by Targeting IL-6 to Induce Apoptosis in Dairy Cattle
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Phenotypic Data
2.3. DNA Extraction and SNP Identification
2.4. Genotyping and Haplotype Analysis
2.5. Association Analyses
2.6. Construction of Recombinant Plasmid and Luciferase Assay
2.7. siRNA Design, Synthesis, and Assessment
2.8. Construction of shRNA Vectors and Transfection in MAC-T cells
2.9. RNA Sequencing
2.10. Differential Expression Analysis and Pathway Enrichment Analysis
2.11. Validation of RNA-Seq Results by qPCR
2.12. Oil Red O Staining and TG Content Measurement
2.13. Calcein-AM/Propidium Iodide (PI) Staining
3. Results
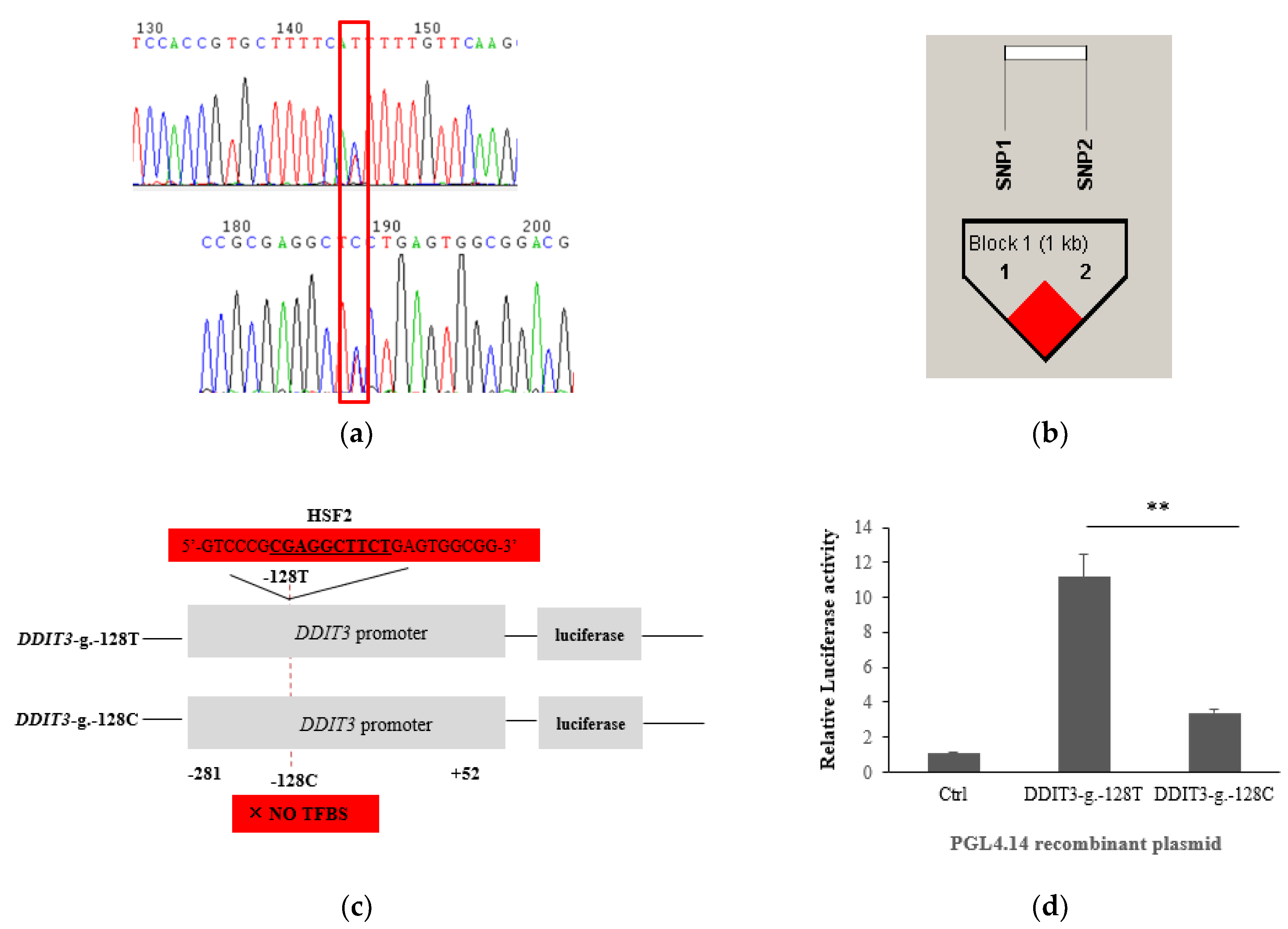
3.1. SNP Identification and Its Associations with the Five Milk Traits
3.2. Prediction of TFBSs and Its Effect on Transcriptional Activity
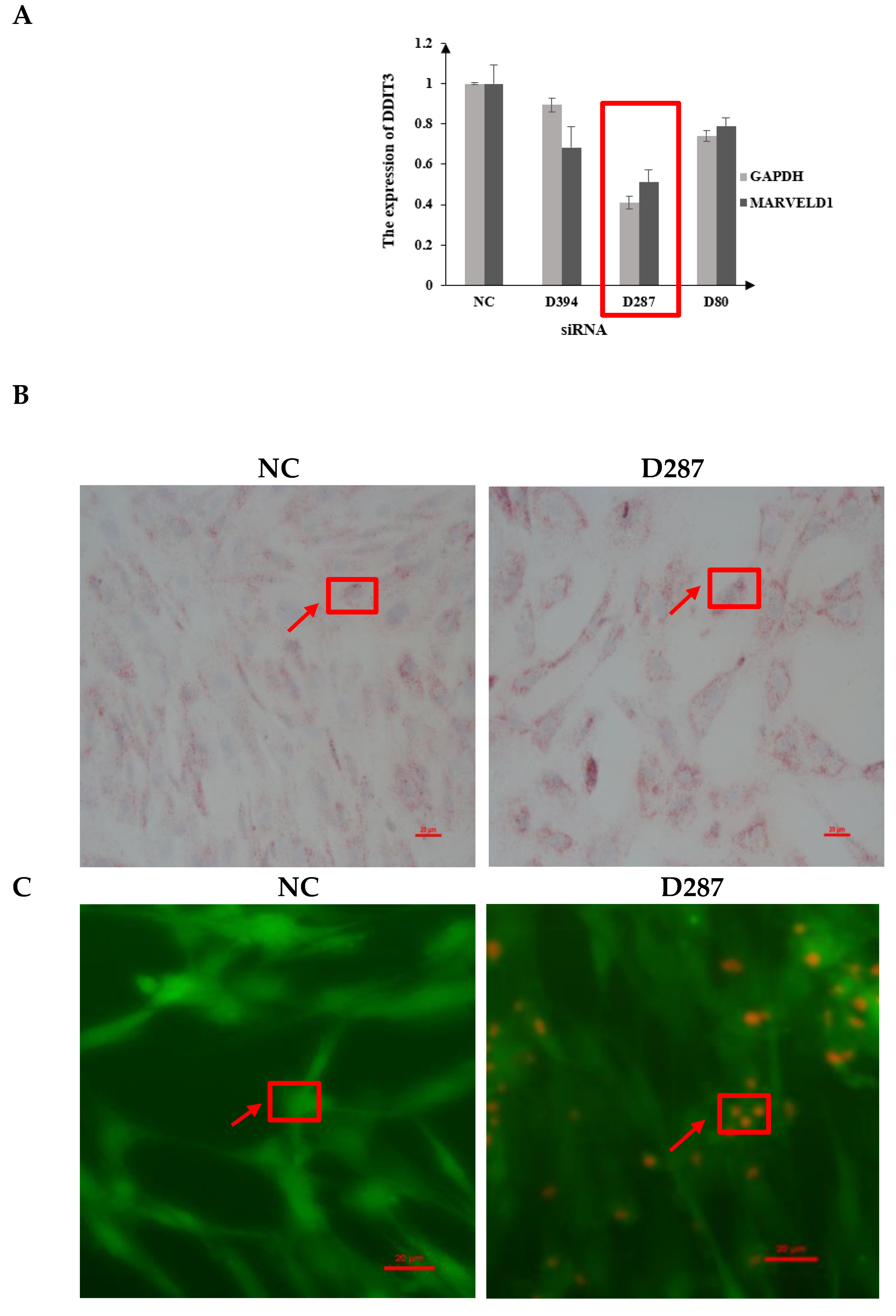
3.3. Interference Efficiency of Designed siRNAs in Bovine MAC-T Cells
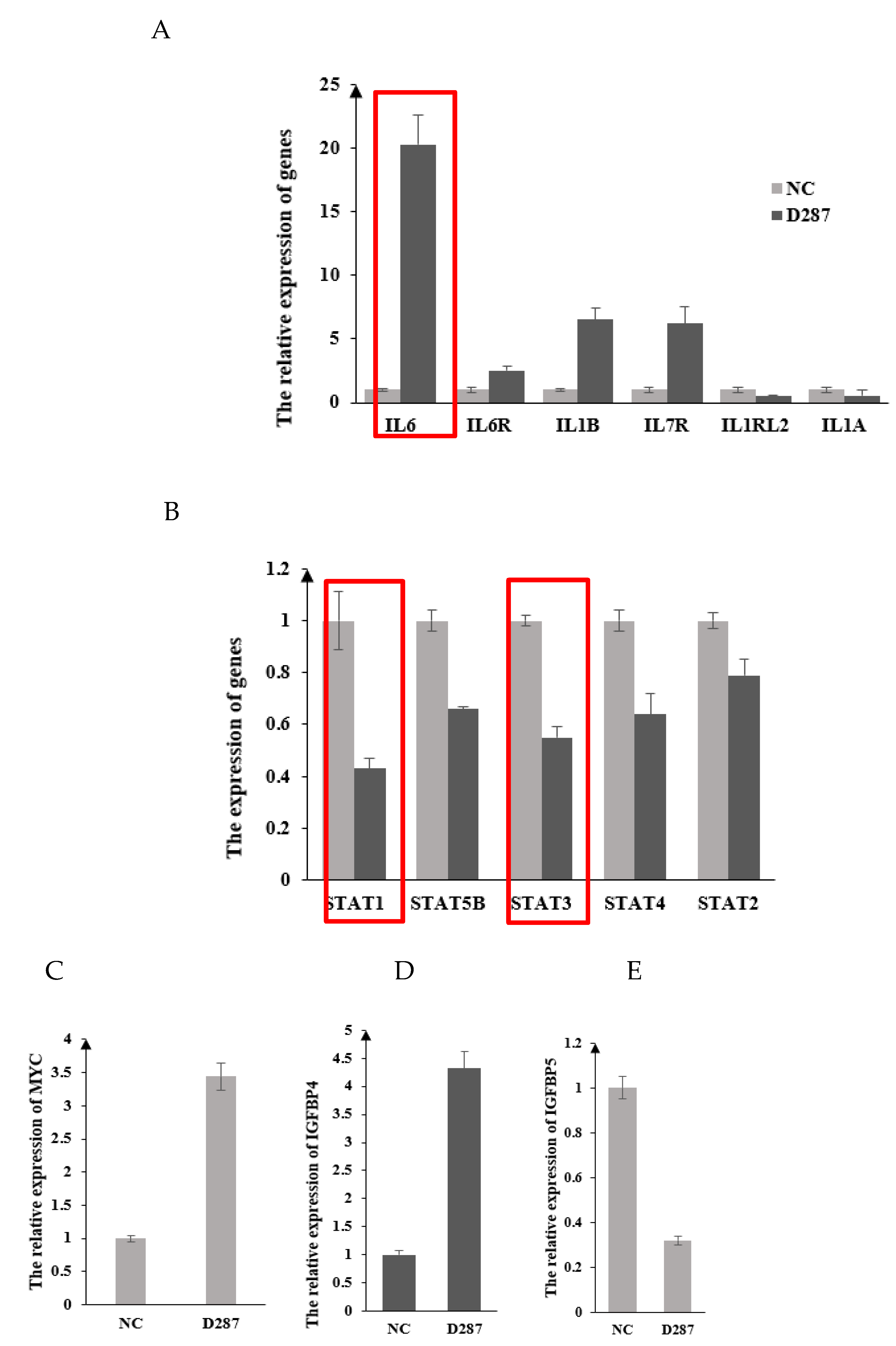
3.4. DEGs and Functional Annotation
3.5. DDIT3 Knocking down Caused Lipid Accumulation and Apoptosis in Bovine MAC-T Cells
3.6. The Expression Alteration of the Key Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Caroli, A.M.; Chessa, S.; Erhardt, G.J. Invited review: Milk protein polymorphisms in cattle: Effect on animal breeding and human nutrition. J. Dairy Sci. 2009, 92, 5335–5352. [Google Scholar] [CrossRef] [PubMed]
- Bovenhuis, H.; Van Arendonk, J.A.M.; Korver, S. Associations between milk protein polymorphisms and milk production traits. J. Dairy Sci. 1992, 75, 2549–2559. [Google Scholar] [CrossRef] [PubMed]
- McCabe, M.; Waters, S.; Morris, D.; Kenny, D.; Lynn, D.; Creevey, C. RNA-seq analysis of differential gene expression in liver from lactating dairy cows divergent in negative energy balance. BMC Genom. 2012, 13, 193. [Google Scholar] [CrossRef] [PubMed]
- Wickramasinghe, S.; Rincon, G.; Islas-Trejo, A.; Medrano, J.F. Transcriptional profiling of bovine milk using RNA sequencing. BMC Genom. 2012, 13, 45. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Hou, Y.; Yang, S.; Xie, Y.; Zhang, S.; Zhang, Y.; Zhang, Q.; Lu, X.; Liu, G.E.; Sun, D. Transcriptional profiling of mammary gland in Holstein cows with extremely different milk protein and fat percentage using RNA sequencing. BMC Genom. 2014, 15, 226. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.B.; Wiggans, G.R.; Ma, L.; Sonstegard, T.S.; Lawlor, T.J., Jr.; Crooker, B.A.; Van Tassell, C.P.; Yang, J.; Wang, S.; Matukumalli, L.K.; et al. Genome-wide association analysis of thirty one production, health, reproduction and body conformation traits in contemporary U.S. Holstein cows. BMC Genom. 2011, 12, 408. [Google Scholar] [CrossRef]
- Bagnato, A.; Schiavini, F.; Rossoni, A.; Maltecca, C.; Dolezal, M.; Medugorac, I.; Sölkner, J.; Russo, V.; Fontanesi, L.; Friedmann, A.; et al. Quantitative trait loci affecting milk yield and protein percentage in a three-country Brown Swiss population. J. Dairy Sci. 2008, 91, 767–783. [Google Scholar] [CrossRef]
- Marola, O.J.; Syc-Mazurek, S.B.; Libby, R.T. DDIT3 (CHOP) contributes to retinal ganglion cell somal loss but not axonal degeneration in DBA/2J mice. Cell Death Discov. 2019, 5, 140. [Google Scholar] [CrossRef]
- Nashine, S.; Liu, Y.; Kim, B.J.; Clark, A.F.; Pang, I.H. Role of C/EBP homologous protein in retinal ganglion cell death after ischemia/reperfusion injury. Investig. Ophthalmol. Vis. Sci. 2014, 56, 221–231. [Google Scholar] [CrossRef]
- Silva, R.M.; Ries, V.; Oo, T.F.; Yarygina, O.; Jackson-Lewis, V.; Ryu, E.J.; Lu, P.D.; Marciniak, S.J.; Ron, D.; Przedborski, S.; et al. CHOP/GADD153 is a mediator of apoptotic death in substantia nigra dopamine neurons in an in vivo neurotoxin model of parkinsonism. J. Neurochem. 2005, 95, 974–986. [Google Scholar] [CrossRef]
- Liao, Y.; Fung, T.S.; Huang, M.; Fang, S.G.; Zhong, Y.; Liu, D.X. Upregulation of CHOP/GADD153 during coronavirus infectious bronchitis virus infection modulates apoptosis by restricting activation of the extracellular signal-regulated kinase pathway. J. Virol. 2013, 87, 8124–8134. [Google Scholar] [CrossRef] [PubMed]
- Abreu, M.M.; Sealy, L. The C/EBPβ isoform, liver-inhibitory protein (LIP), induces autophagy in breast cancer cell lines. Exp. Cell Res. 2010, 316, 3227–3238. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, J.; Wang, L.; Ho, C.T.; Zhang, K.; Liu, Q.; Zhao, H. Garcinol from Garcinia indica Downregulates Cancer Stem-like Cell Biomarker ALDH1A1 in Nonsmall Cell Lung Cancer A549 Cells through DDIT3 Activation. J. Agric. Food Chem. 2017, 65, 3675–3683. [Google Scholar] [CrossRef] [PubMed]
- Barone, M.V.; Crozat, A.; Tabaee, A.; Philipson, L.; Ron, D. CHOP (GADD153) and its oncogenic variant, TLS-CHOP, have opposing effects on the induction of G1/S arrest. Genes Dev. 1994, 8, 453–464. [Google Scholar] [CrossRef]
- Wang, X.; Tomso, D.J.; Liu, X.; Bell, D.A. Single nucleotide polymorphism in transcriptional regulatory regions and expression of environmentally responsive genes. Toxicol. Appl. Pharmacol. 2005, 207, 84–90. [Google Scholar] [CrossRef]
- He, H.; Soncin, F.; Grammatikakis, N.; Li, Y.; Siganou, A.; Gong, J.; Brown, S.A.; Kingston, R.E.; Calderwood, S.K. Elevated expression of heat shock factor (HSF) 2A stimulates HSF1-induced transcription during stress. J. Biol. Chem. 2003, 278, 35465–35475. [Google Scholar] [CrossRef]
- Ostling, P.; Björk, J.K.; Roos-Mattjus, P.; Mezger, V.; Sistonen, L. Heat shock factor 2 (HSF2) contributes to inducible expression of hsp genes through interplay with HSF1. J. Biol. Chem. 2007, 282, 7077–7086. [Google Scholar] [CrossRef]
- Sandqvist, A.; Björk, J.K.; Akerfelt, M.; Chitikova, Z.; Grichine, A.; Vourc’h, C.; Jolly, C.; Salminen, T.A.; Nymalm, Y.; Sistonen, L. Heterotrimerization of heat-shock factors 1 and 2 provides a transcriptional switch in response to distinct stimuli. Mol. Biol. Cell 2009, 20, 1340–1347. [Google Scholar] [CrossRef]
- Yang, L.N.; Ning, Z.Y.; Wang, L.; Yan, X.; Meng, Z.Q. HSF2 regulates aerobic glycolysis by suppression of FBP1 in hepatocellular carcinoma. Am. J. Cancer Res. 2019, 9, 1607–1621. [Google Scholar]
- Zi, C.; Wu, Z.; Wang, J.; Huo, Y.; Zhu, G.; Wu, S.; Bao, W. Transcriptional activity of the FUT1 gene promoter region in pigs. Int. J. Mol. Sci. 2013, 14, 24126–24134. [Google Scholar] [CrossRef]
- Posey, K.L.; Coustry, F.; Veerisetty, A.C.; Liu, P.; Alcorn, J.L.; Hecht, J.T. Chop (Ddit3) is essential for D469del-COMP retention and cell death in chondrocytes in an inducible transgenic mouse model of pseudoachondroplasia. Am. J. Pathol. 2012, 180, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sun, H.; Song, F.; Fu, D.; Wang, J. DDIT3 overexpression increases odontoblastic potential of human dental pulp cells. Cell Prolif. 2014, 47, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Hou, P.; Pan, W.; He, W.; He, D.C.; Wang, H.; He, H. DDIT3 Targets Innate Immunity via the DDIT3-OTUD1-MAVS Pathway To Promote Bovine Viral Diarrhea Virus Replication. J. Virol. 2021, 95, e02351-20. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Han, B.; Liu, L.; Zhao, F.; Liang, W.; Jiang, J.; Yang, Y.; Ma, Z.; Sun, D. Genetic association of DDIT3, RPL23A, SESN2 and NR4A1 genes with milk yield and composition in dairy cattle. Anim. Genet. 2019, 50, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Bobbo, V.C.; Engel, D.F.; Jara, C.P.; Mendes, N.F.; Haddad-Tovolli, R.; Prado, T.P.; Sidarta-Oliveira, D.; Morari, J.; Velloso, L.A.; Araujo, E.P. Interleukin-6 actions in the hypothalamus protects against obesity and is involved in the regulation of neurogenesis. J. Neuroinflam. 2021, 18, 192. [Google Scholar] [CrossRef]
- Jo, H.A.; Kim, J.Y.; Yang, S.H.; Han, S.S.; Joo, K.W.; Kim, Y.S.; Kim, D.K. The role of local IL6/JAK2/STAT3 signaling in high glucose-induced podocyte hypertrophy. Kidney Res. Clin. Pract. 2016, 35, 212–218. [Google Scholar] [CrossRef]
- Serrano, A.L.; Baeza-Raja, B.; Perdiguero, E.; Jardí, M.; Muñoz-Cánoves, P. Interleukin-6 is an essential regulator of satellite cell-mediated skeletal muscle hypertrophy. Cell Metab. 2008, 7, 33–44. [Google Scholar] [CrossRef]
- Begue, G.; Douillard, A.; Galbes, O.; Rossano, B.; Vernus, B.; Candau, R.; Py, G. Early activation of rat skeletal muscle IL-6/STAT1/STAT3 dependent gene expression in resistance exercise linked to hypertrophy. PloS ONE 2013, 8, e57141. [Google Scholar] [CrossRef]
- Mir, S.A.; Chatterjee, A.; Mitra, A.; Pathak, K.; Mahata, S.K.; Sarkar, S. Inhibition of signal transducer and activator of transcription 3 (STAT3) attenuates interleukin-6 (IL-6)-induced collagen synthesis and resultant hypertrophy in rat heart. J. Biol. Chem. 2012, 287, 2666–2677. [Google Scholar] [CrossRef]
- Ropelle, E.R.; Flores, M.B.; Cintra, D.E.; Rocha, G.Z.; Pauli, J.R.; Morari, J.; de Souza, C.T.; Moraes, J.C.; Prada, P.O.; Guadagnini, D.; et al. IL-6 and IL-10 anti-inflammatory activity links exercise to hypothalamic insulin and leptin sensitivity through IKKβ and ER stress inhibition. PLoS Biol. 2010, 8, e1000465. [Google Scholar] [CrossRef]
| SNPs | Genotype (No. Individuals) | MY | FY | FP | PY | PP |
|---|---|---|---|---|---|---|
| g.-1194 C>T | CC(313) | 426.07 ± 63.03 a | 9.29 ± 2.81 A | −0.04 ± 0.029 | 11.34 ± 1.97 a | −0.014 ± 0.009 |
| TC(321) | 302.21 ± 62.33 b | 2.90 ± 2.78 B | −0.06 ± 0.029 | 9.16 ± 1.95 ab | 0.001 ± 0.009 | |
| TT(83) | 232.53 ± 82.51 b | −0.96 ± 3.05 B | −0.07 ± 0.036 | 6.20 ± 2.45 b | −0.004 ± 0.012 | |
| p value | 0.0063 ** | 0.0001 ** | 0.5409 | 0.0216 * | 0.1536 | |
| g.-128 C>T | CC(313) | 426.07 ± 63.03 a | 9.29 ± 2.81 A | −0.04 ± 0.029 | 11.34 ± 1.97 a | −0.014 ± 0.009 |
| TC(321) | 302.21 ± 62.33 b | 2.90 ± 2.78 B | −0.06 ± 0.029 | 9.16 ± 1.95 ab | 0.001 ± 0.009 | |
| TT(83) | 232.53 ± 82.51 b | −0.96 ± 3.05 B | −0.07 ± 0.036 | 6.20 ± 2.45 b | −0.004 ± 0.012 | |
| p value | 0.0063 ** | 0.0001 ** | 0.5409 | 0.0216 * | 0.1536 |
| siRNA Name | Sense (5′-3′) | Target Sites in DDIT3 | Target Sites in DDIT3 Gene |
|---|---|---|---|
| D394 | GCTGGCTGAAGAGAATGAACG | 394–415 bp | Exon 4 |
| D287 | GACCAAGGAAGAACCAGAAAA | 287–308 bp | Exon 4 |
| D80 | GTGCTGTCCTCAGATGAAAAT | 80–101 bp | Exon 3 |
| NC | GATCCGTTCTCCGAACGTGTCACGT |
| Symbol | Gene Name | Regulation | Log2 Fold Change | padj | Gene Function |
|---|---|---|---|---|---|
| IL6 | interleukin 6 | up | 4.22 | 3.49 × 10−112 | plays important roles in fatty acid biosynthesis |
| CXCL8 | C-X-C motif chemokine ligand 8 | up | 3.01 | 2.04 × 10−90 | catalyzes the first step in the hydrolysis of triglycerides |
| GRO1 | chemokine (C-X-C motif) ligand 1 | up | 2.68 | 1.21 × 10−87 | involved in the biosynthesis of unsaturated fatty acids |
| TNFAIP3 | TNF alpha-induced protein 3 | up | 2.08 | 6.41 × 10−80 | linoleic acid metabolism, n-3, and n-6 fatty acid metabolism |
| EGR1 | early growth response 1 | up | 3.03 | 7.80 × 10−78 | involved in sterol biosynthesis |
| PHLDA1 | pleckstrin homology-like domain family A1 | up | 1.72 | 7.19 × 10−53 | involved in adipocyte differentiation and glucose homeostasis |
| SERPINE1 | serpin family E member 1 | up | 1.48 | 7.74 × 10−49 | plays a key role in lipid biosynthesis and fatty acid degradation |
| CYR61 | cysteine rich angiogenic inducer 61 | up | 1.66 | 8.90 × 10−49 | catalyzes the carboxylation of acetyl-CoA to malonyl-CoA |
| CCL20 | C-C motif chemokine ligand 20 | up | 4.27 | 2.00 × 10−46 | involved in lipid synthesis and energy generation |
| IFI27 | putative ISG12(a) protein | down | −3.01 | 1.06 × 10−46 | plays critical roles in lipid metabolism |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, X.; Li, C.; Wei, Z.; Meng, H.; Zhang, F.; Liu, Y.; Wu, C.; Yang, S. DDIT3 Governs Milk Production Traits by Targeting IL-6 to Induce Apoptosis in Dairy Cattle. Agriculture 2023, 13, 117. https://doi.org/10.3390/agriculture13010117
Cui X, Li C, Wei Z, Meng H, Zhang F, Liu Y, Wu C, Yang S. DDIT3 Governs Milk Production Traits by Targeting IL-6 to Induce Apoptosis in Dairy Cattle. Agriculture. 2023; 13(1):117. https://doi.org/10.3390/agriculture13010117
Chicago/Turabian StyleCui, Xiaogang, Changqing Li, Zhangqi Wei, Hangting Meng, Fengfeng Zhang, Yue Liu, Changxin Wu, and Shaohua Yang. 2023. "DDIT3 Governs Milk Production Traits by Targeting IL-6 to Induce Apoptosis in Dairy Cattle" Agriculture 13, no. 1: 117. https://doi.org/10.3390/agriculture13010117
APA StyleCui, X., Li, C., Wei, Z., Meng, H., Zhang, F., Liu, Y., Wu, C., & Yang, S. (2023). DDIT3 Governs Milk Production Traits by Targeting IL-6 to Induce Apoptosis in Dairy Cattle. Agriculture, 13(1), 117. https://doi.org/10.3390/agriculture13010117






