Widely Targeted Metabolomics Analyses Clarify the Biosynthetic Pathways Responsible for Flavonoids in Sweet Potato (Ipomoea batatas (L.) Lam.) Storage Roots
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Sample Preparation and Extraction
2.3. Measurement of Flavonoid Levels in Sweet Potato Storage Roots
2.3.1. Rutin Standard Curve Preparation
2.3.2. Analysis of Flavonoid Content
2.4. UPLC Analyses
2.5. ESI-Q TRAP-MS/MS
2.6. Metabolite Quantification and Characterization
2.7. qRT-PCR
2.8. Statistical Analyses
3. Results
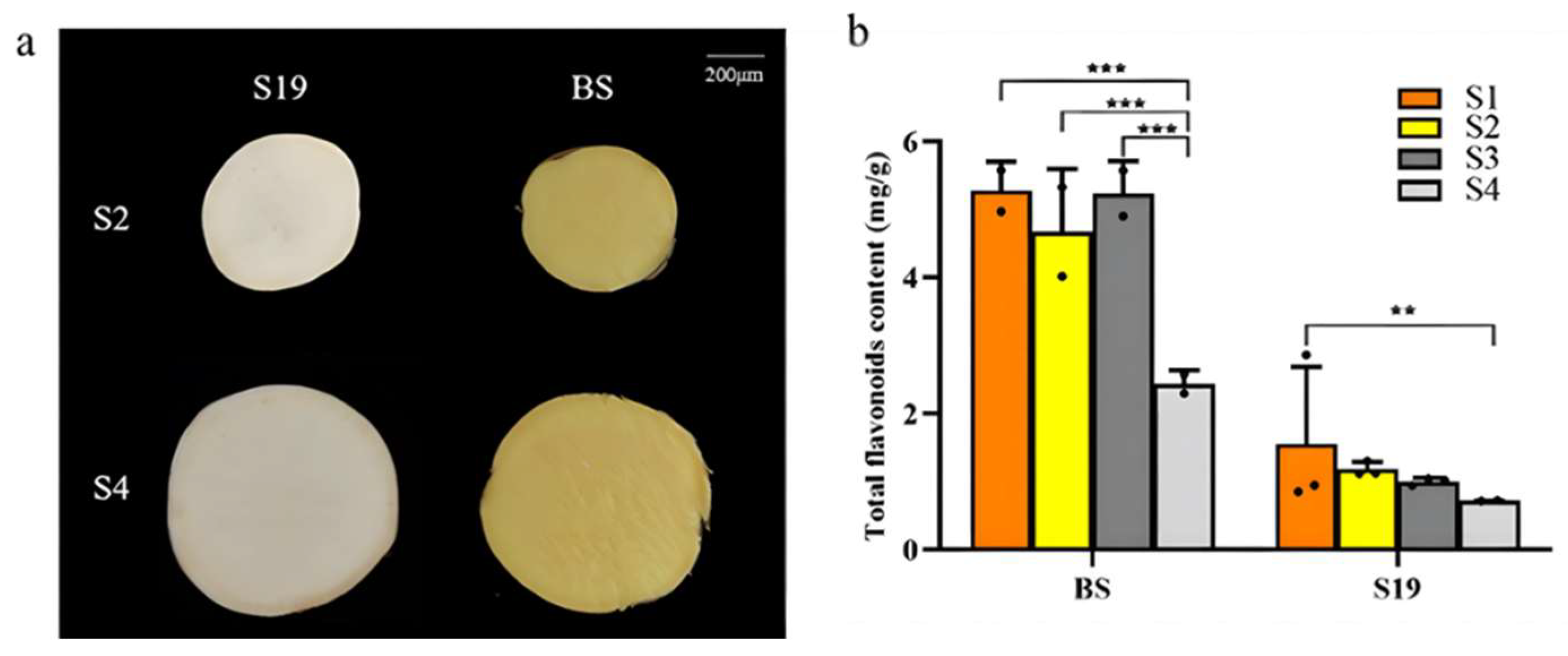
3.1. Analyses of Total Flavonoid Levels in Sweet Potato Storage Roots
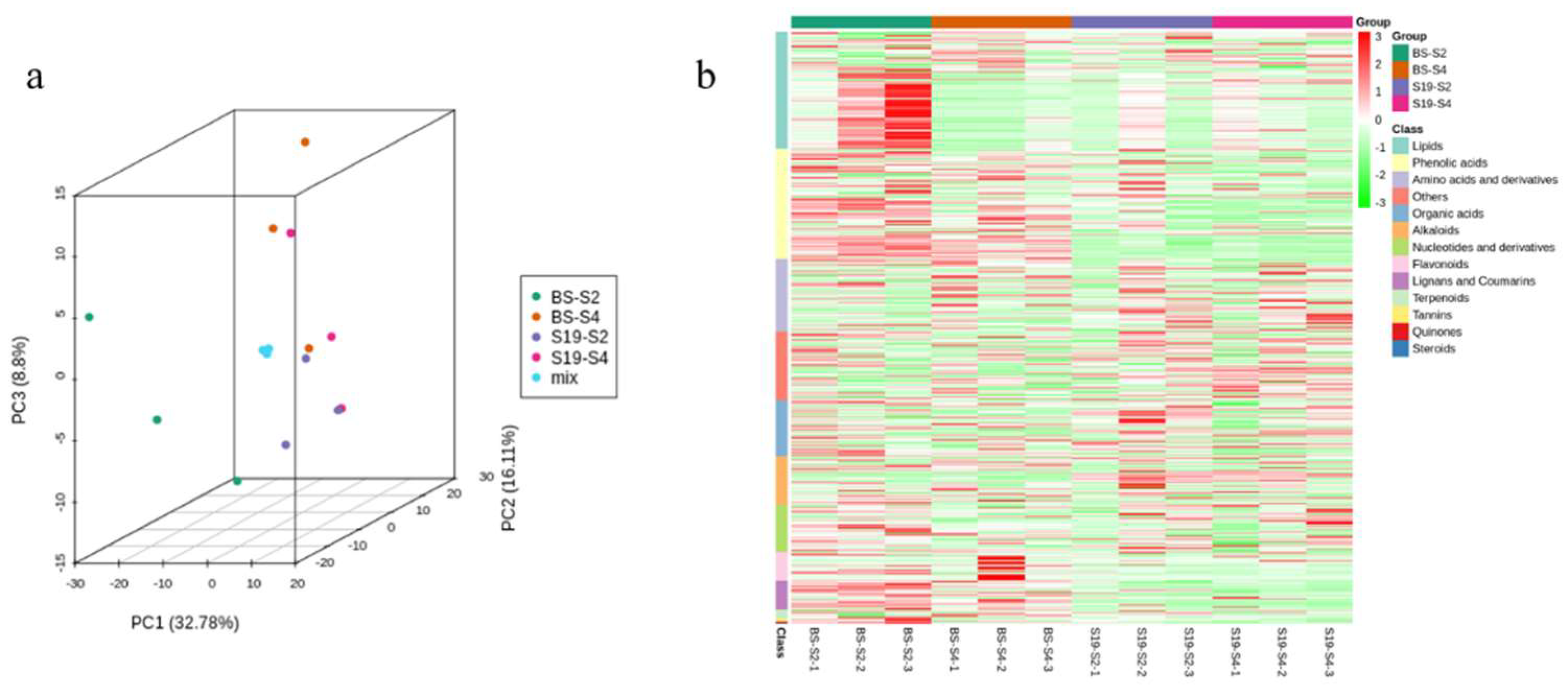
3.2. Widely Targeted Metabolomic Analysis of Differential Metabolites
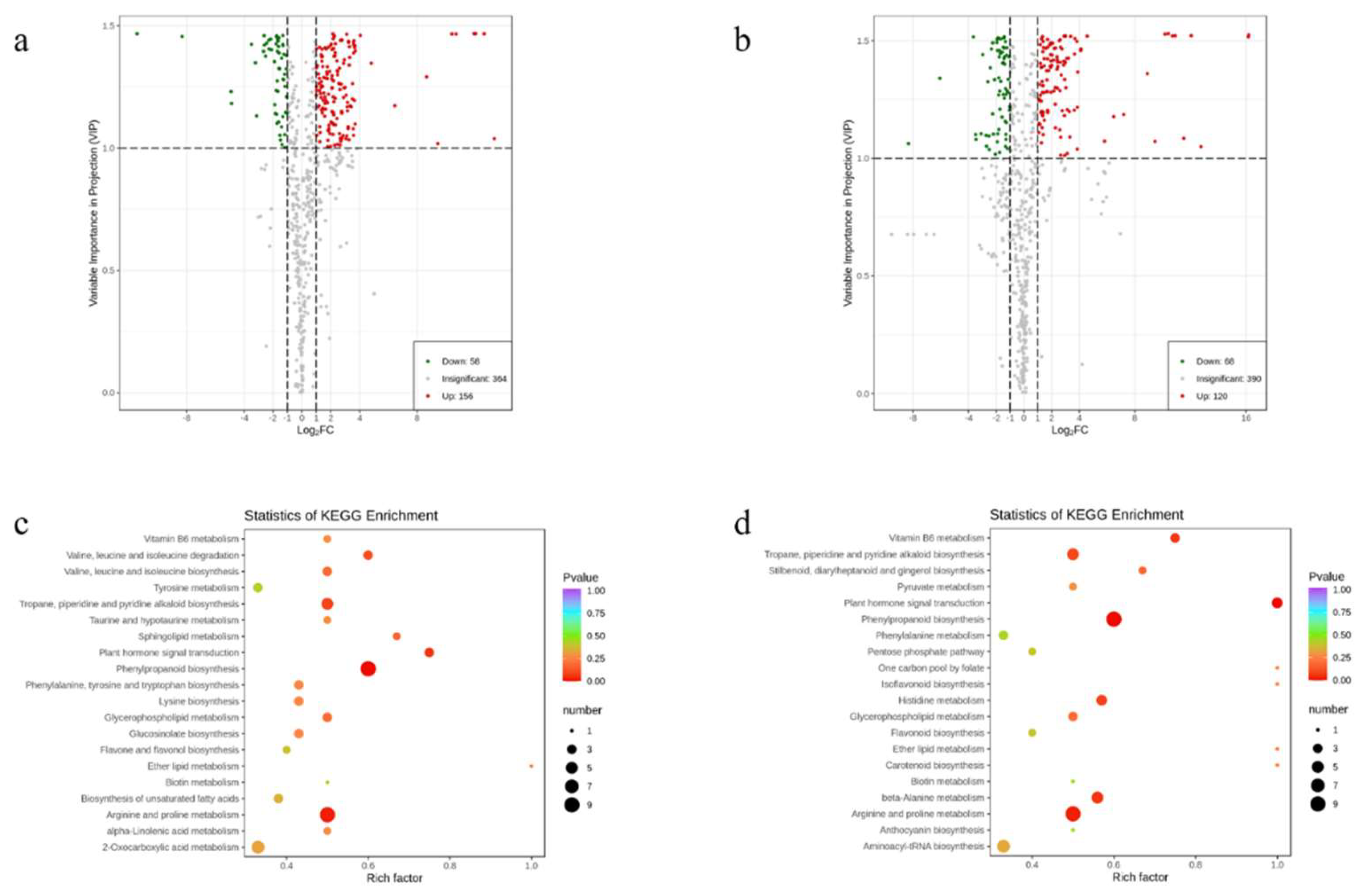
3.3. Identification of Differential Flavonoid Metabolites and Their Functional Annotation and Enrichment
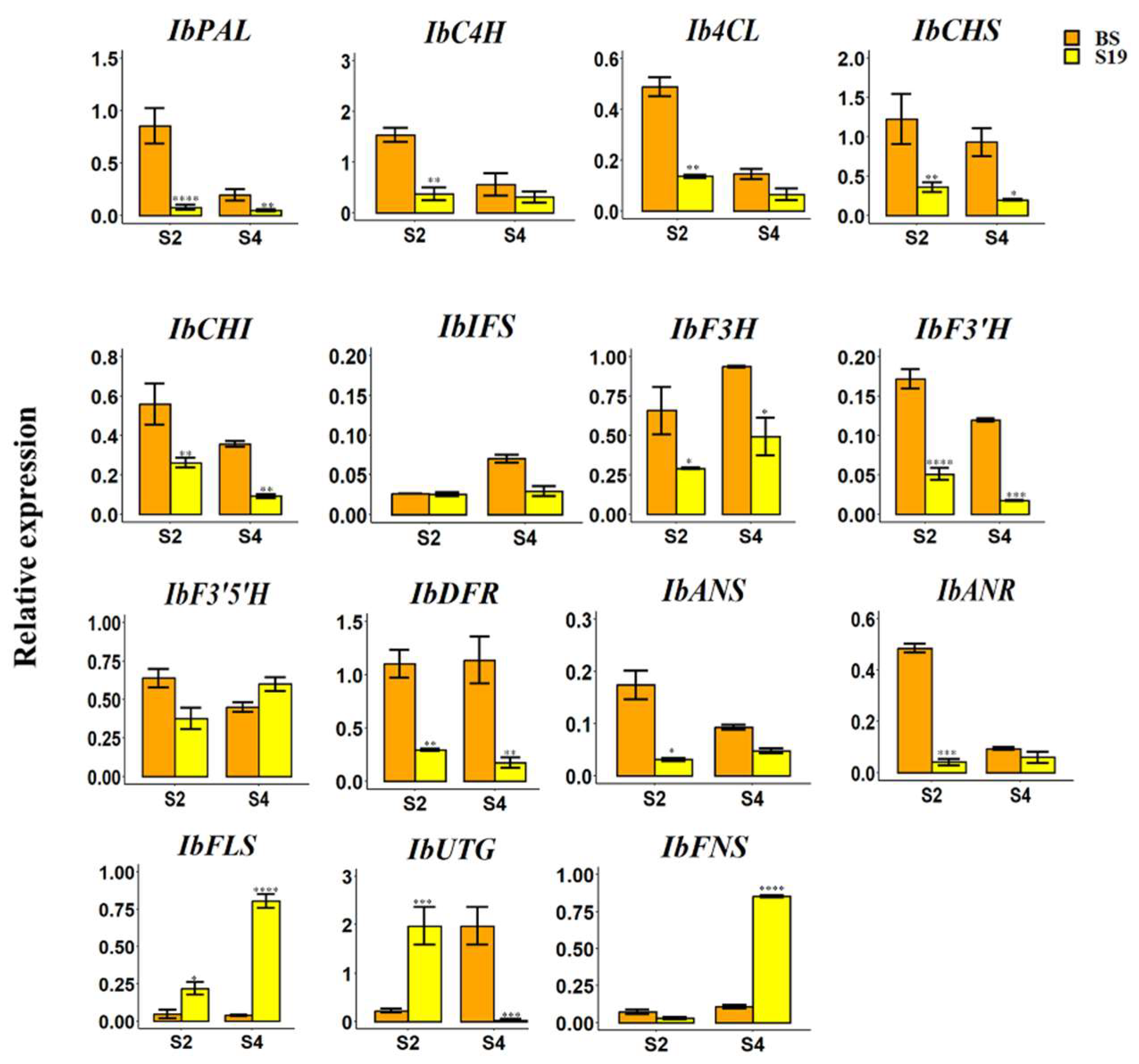
3.4. qRT-PCR
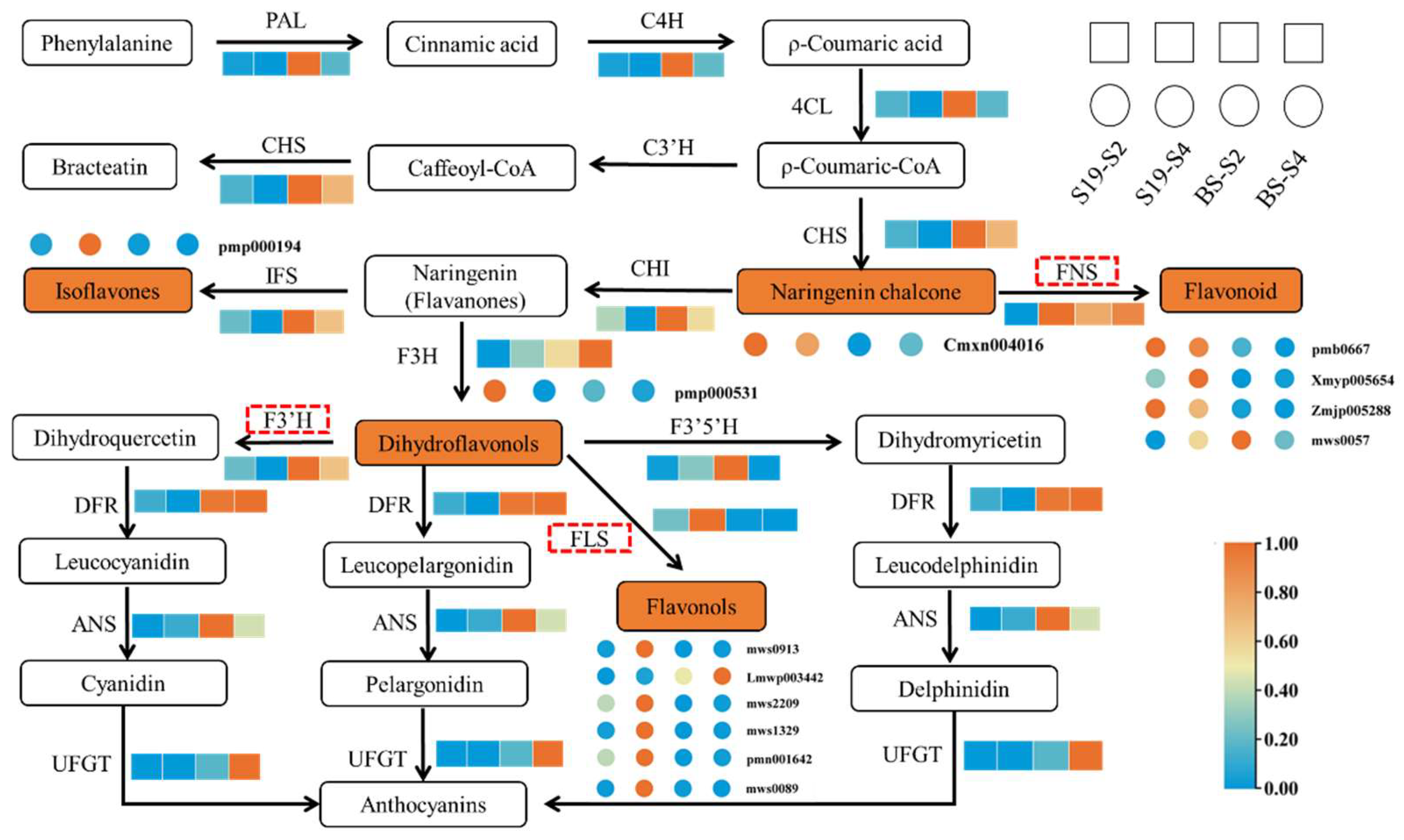
3.5. Flavonoid Biosynthesis Regulatory Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, L.; Gao, Y.; Deng, B.; Ru, W.; Tong, C.; Bao, J. Physicochemical, Nutritional, and Antioxidant Properties in seven sweet potato flours. Front. Nutr. 2022, 9, 923257. [Google Scholar] [CrossRef] [PubMed]
- Laveriano-Santos, E.P.; López-Yerena, A. Jaime-Rodríguez, C.; González-Coria, J.; Lamuela-Raventós, R.M.; Vallverdú-Queralt, A.; Romanyà, J.; Pérez, M. Sweet potato is not simply an abundant food crop: A Comprehensive Review of Its Phytochemical Constituents, Biological Activities, and the Effects of Processing. Antioxidants 2022, 11, 1648. [Google Scholar] [PubMed]
- Parveen, A.; Choi, S.; Kang, J.H.; Oh, S.H.; Kim, S.Y. Trifostigmanoside I, an Active Compound from Sweet Potato, Restores the Activity of MUC2 and Protects the Tight Junctions through PKCα/β to Maintain Intestinal Barrier Function. Int. J. Mol. Sci. 2020, 22, 291. [Google Scholar] [CrossRef] [PubMed]
- Gou, S.; Zhang, X.; Han, W.; Li, Q.; Li, X. Research Progress on the Biosynthesis and Antistress Mechanisms of Plant Flavonoids. J. Hortic. Sci. 2023, 50, 209–224. [Google Scholar]
- Zhang, K.; Sun, Y.; Li, M.; Long, R. CrUGT87A1, a UDP-sugar glycosyltransferases (UGTs) gene from Carex rigescens, increases salt tolerance by accumulating flavonoids for antioxidation in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 159, 28–36. [Google Scholar]
- Lam, P.Y.; Wang, L.; Lui, A.C.W.; Liu, H.; Takeda-Kimura, Y.; Chen, M.X.; Zhu, F.Y.; Zhang, J.; Umezawa, T.; Tobimatsu, Y.; et al. Deficiency in flavonoid biosynthesis genes CHS, CHI, and CHIL alters rice flavonoid and lignin profiles. Plant Physiol. 2022, 188, 1993–2011. [Google Scholar]
- Peniche-Pavía, H.A.; Guzmán, T.J.; Magaña-Cerino, J.M.; Gurrola-Díaz, C.M.; Tiessen, A. Maize Flavonoid Biosynthesis, Regulation, and Human Health Relevance: A Review. Molecules 2022, 27, 5166. [Google Scholar] [CrossRef]
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 2020, 63, 180–209. [Google Scholar]
- Lin, S.; Singh, R.K.; Moehninsi, M.; Navarre, D.A. R2R3-MYB transcription factors, StmiR858 and sucrose mediate potato flavonol biosynthesis. Hortic. Res. 2021, 8, 25. [Google Scholar]
- Meng, X.; Li, Y.; Zhou, T.; Sun, W.; Shan, X.; Gao, X.; Wang, L. Functional Differentiation of Duplicated Flavonoid 3-O-Glycosyltransferases in the Flavonol and Anthocyanin Biosynthesis of Freesia hybrida. Front. Plant Sci. 2019, 10, 1330. [Google Scholar]
- LaFountain, A.M.; Yuan, Y.W. Repressorsof anthocyanin biosynthesis. New Phytol. 2021, 231, 933–949. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, X.J.; Zhao, S.N.; Yan, J.R.; Bu, X.; Zhang, Y.; Liu, Y.F.; Xu, T.; Qi, M.F.; Qi, H.Y.; et al. Light Regulation of Anthocyanin Biosynthesis in Horticultural Crops. Sci. Agric. Sin. 2020, 53, 4904–4917. [Google Scholar]
- Shen, N.; Wang, T.; Gan, Q.; Liu, S.; Wang, L.; Jin, B. Plant flavonoids: Classification, distribution, biosynthesis, and antioxidant activity. Food. Chem. 2022, 383, 132531. [Google Scholar] [PubMed]
- Chen, C.; Zhang, Y.; Fu, X.; Chen, C.; Wu, S.; Zhang, C.; Zhang, H.; Chang, Y.; Chen, S.; Zhao, J.; et al. Influential factors and transcriptome analyses of immature diploid embryo anthocyanin accumulation in maize. BMC. Plant. Biol. 2022, 22, 609. [Google Scholar]
- Lv, L.; Chen, X.; Li, H.; Huang, J.; Liu, Y.; Zhao, A. Different adaptive patterns of wheat with different drought tolerance under drought stresses and rehydration revealed by integrated metabolomic and transcriptomic analysis. Front. Plant. Sci. 2022, 13, 1008624. [Google Scholar]
- Chen, X.; Yang, Y.; Yang, X.; Zhu, G.; Lu, X.; Jia, F.; Diao, B.; Yu, S.; Ali, A.; Zhang, H.; et al. Investigation of flavonoid components and their associated antioxidant capacity in different pigmented rice varieties. Food. Res. Int. 2022, 161, 111726. [Google Scholar] [CrossRef]
- Hall, R.D.; Brouwer, I.D.; Fitzgerald, M.A. Plant metabolomics and its potential application for human nutrition. Physiol. Plant 2008, 132, 162–175. [Google Scholar] [CrossRef]
- Zhou, J.; Hou, D.; Zou, W.; Wang, J.; Luo, R.; Wang, M.; Yu, H. Comparison of Widely Targeted Metabolomics and Untargeted Metabolomics of Wild Ophiocordyceps sinensis. Molecules 2022, 27, 3645. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, X.; Fu, D.; Zhao, Y. Integrated Analysis of Widely Targeted Metabolomics and Transcriptomics Reveals the Effects of Transcription Factor NOR-like1 on Alkaloids, Phenolic Acids, and Flavonoids in Tomato at Different Maturation Stages. Metabolites 2022, 12, 1296. [Google Scholar] [CrossRef]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant. 2013, 6, 1769–1780. [Google Scholar] [CrossRef]
- Wang, A.; Li, R.; Ren, L.; Gao, X.; Zhang, Y.; Ma, Z.; Ma, D.; Luo, Y. A comparative metabolomics study of flavonoids in sweet potato with different flesh colors (Ipomoea batatas (L.) Lam). Food Chem. 2018, 260, 124–134. [Google Scholar] [PubMed]
- Eriksson, E.J.L.; Kettaneh-Wold, N.; Trygg, J.; Wikström, C.; Wold, S. Multi- and Megavariate Data Analysis, Part I—Basic Principles and Applications, 2nd ed.; Umetrics Academy: Stockholm, Sweden, 2006. [Google Scholar]
- Chen, Y.; Zhang, R.; Song, Y.; He, J.; Sun, J.; Bai, J.; An, Z.; Dong, L.; Zhan, Q.; Abliz, Z. RRLC-MS/MS-based metabonomics combined with in-depth analysis of metabolic correlation network: Finding potential biomarkers for breast cancer. Analyst 2009, 134, 2003–2011. [Google Scholar] [CrossRef] [PubMed]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic. Acids. Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Xia, J. MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 2018, 34, 4313–4314. [Google Scholar] [CrossRef]
- Noguchi, A.; Nakamura, K.; Sakata, K.; Sato-Fukuda, N.; Ishigaki, T.; Mano, J.; Takabatake, R.; Kitta, K.; Teshima, R.; Kondo, K.; et al. Development and interlaboratory validation of a simple screening method for genetically modified maize using a∆∆C(q)-based multiplex real-time PCR assay. Anal. Chem. 2016, 88, 4285–4293. [Google Scholar] [CrossRef]
- Yang, C.; Luo, J. Establishment and research of method for determination of total flavonoid content of oily hemp blood vine in Guizhou. Guizhou Sci. 2022, 40, 11–14. [Google Scholar]
- Nguyen, H.C.; Chen, C.C.; Lin, K.H.; Chao, P.Y.; Lin, H.H.; Huang, M.Y. Bioactive Compounds, Antioxidants, and Health Benefits of Sweet Potato Leaves. Molecules 2021, 26, 1820. [Google Scholar] [CrossRef]
- Althwab, S.A.; Mousa, H.M.; El-Zaha, K.M.; Zaher, A.M.A. Protective effect of sweet potato peel against oxidative stress in hyperlipidemic albino rats. Food Nutr. Sci. 2019, 10, 503–516. [Google Scholar] [CrossRef]
- Kang, L.; Park, S.C.; Ji, C.Y.; Kim, H.S.; Lee, H.S.; Kwak, S.S. Metabolic engineering of carotenoids in transgenic sweetpotato. Breed. Sci. 2017, 67, 27–34. [Google Scholar]
- Zhou, W.; Gong, Y.; Lu, X.; Huang, C.; Gao, F. Molecular cloning and characterization of a flavonoid 3’-hydroxylase gene from purple-fleshed sweet potato (Ipomoea batatas). Mol. Biol. Rep. 2012, 39, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; Zhen, X.; Gao, H.; Liang, Y.; Li, H.; Zhao, J.; Yin, M.; Han, Y.; Zhang, B. Integrated Metabolomic and Transcriptomic Analyses Reveal the Basis for Carotenoid Biosynthesis in Sweet Potato (Ipomoea batatas (L.) Lam.) Storage Roots. Metabolites 2022, 12, 1010. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.R.; Zhang, H.; Li, X.; Bai, Y.W.; Peng, K.; Wang, Z.; Dai, Z.R.; Bian, X.F.; Zhang, Q.; Jia, L.C.; et al. The B-box transcription factor IbBBX29 regulates leaf development and flavonoid biosynthesis in sweet potato. Plant Physiol. 2023, 191, 496–514. [Google Scholar]
- Zhang, Y.; Bian, S.; Hu, J.; Liu, G.; Peng, S.; Chen, H.; Jiang, Z.; Wang, T.; Ye, Q.; Zhu, H. Natural Deep Eutectic Solvent-Based Microwave-Assisted Extraction of Total Flavonoid Compounds from Spent Sweet Potato (Ipomoea batatas L.) Leaves: Optimization and Antioxidant and Bacteriostatic Activity. Molecules 2022, 27, 5985. [Google Scholar] [CrossRef]
- Li, G.; Lin, Z.; Zhang, H.; Liu, Z.; Xu, Y.; Xu, G.; Li, H.; Ji, R.; Luo, W.; Qiu, Y.; et al. Anthocyanin Accumulation in the Leaves of the Purple Sweet Potato (Ipomoea batatas L.) Cultivars. Molecules 2019, 24, 3743. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.G.; Wu, S.Q.; Li, P.; Yang, H. Advancement in the chemical analysis and quality control of flavonoid in Ginkgo biloba. J. Pharm. Biomed. Anal. 2015, 113, 212–225. [Google Scholar]
- Tang, Y.; Cai, W.; Xu, B. Profiles of phenolics, carotenoids and antioxidative capacities of thermal processed white, yellow, orange and purple sweet potatoes grown in Guilin, China. Food Sci. Hum. Wellness 2015, 4, 123–132. [Google Scholar] [CrossRef]
- Lou, Q.; Liu, Y.L.; Qi, Y.Y.; Jiao, S.Z.; Tian, F.F.; Jiang, L.; Wang, Y.J. Transcriptome sequencing and metabolite analysis reveals the role of delphinidin metabolism in flower color in grape hyacinth. J. Exp. Bot. 2014, 65, 3157–3164. [Google Scholar] [CrossRef]
- Takahashi, R.; Githiri, S.M.; Hatayama, K.; Dubouzet, E.G.; Shimada, N.; Aoki, T.; Ayabe, S.; Iwashina, T.; Toda, K.; Matsumura, H. A single-base deletion in soybean flavonol synthase gene is associated with magenta flower color. Plant. Mol. Biol. 2007, 63, 125–135. [Google Scholar] [CrossRef]
- Nielsen, K.; Deroles, S.C.; Markham, K.R.; Bradley, M.J.; Podivinsky, E.; Manson, D. Antisense flavonol synthase alters copigmentation and flower color in lisianthus. Mol. Breed. 2002, 9, 217–229. [Google Scholar] [CrossRef]
- Lin, Q.F.; Liu, T.T.; Liu, J.R.; Cai, M.; Cheng, T.R.; Wang, J.; Zhang, Q.X.; Pan, H.T. Flavonoids composition and content in petals of Lagerstroemia and Heimia species and cultivars. Acta Hortic. Sin. 2021, 48, 1956–1968. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, H.; Zhang, Y.; Duan, Q.; Ren, Q.; Deng, L.; Huo, Y.; Zhang, B.; Zhen, X. Widely Targeted Metabolomics Analyses Clarify the Biosynthetic Pathways Responsible for Flavonoids in Sweet Potato (Ipomoea batatas (L.) Lam.) Storage Roots. Agriculture 2023, 13, 1955. https://doi.org/10.3390/agriculture13101955
Gao H, Zhang Y, Duan Q, Ren Q, Deng L, Huo Y, Zhang B, Zhen X. Widely Targeted Metabolomics Analyses Clarify the Biosynthetic Pathways Responsible for Flavonoids in Sweet Potato (Ipomoea batatas (L.) Lam.) Storage Roots. Agriculture. 2023; 13(10):1955. https://doi.org/10.3390/agriculture13101955
Chicago/Turabian StyleGao, Huiyu, Yuyang Zhang, Qian Duan, Qingming Ren, Lin Deng, Yiqiong Huo, Bin Zhang, and Xiaoxi Zhen. 2023. "Widely Targeted Metabolomics Analyses Clarify the Biosynthetic Pathways Responsible for Flavonoids in Sweet Potato (Ipomoea batatas (L.) Lam.) Storage Roots" Agriculture 13, no. 10: 1955. https://doi.org/10.3390/agriculture13101955
APA StyleGao, H., Zhang, Y., Duan, Q., Ren, Q., Deng, L., Huo, Y., Zhang, B., & Zhen, X. (2023). Widely Targeted Metabolomics Analyses Clarify the Biosynthetic Pathways Responsible for Flavonoids in Sweet Potato (Ipomoea batatas (L.) Lam.) Storage Roots. Agriculture, 13(10), 1955. https://doi.org/10.3390/agriculture13101955



