A High-Precision Detection Method of Apple Leaf Diseases Using Improved Faster R-CNN
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Data Analysis
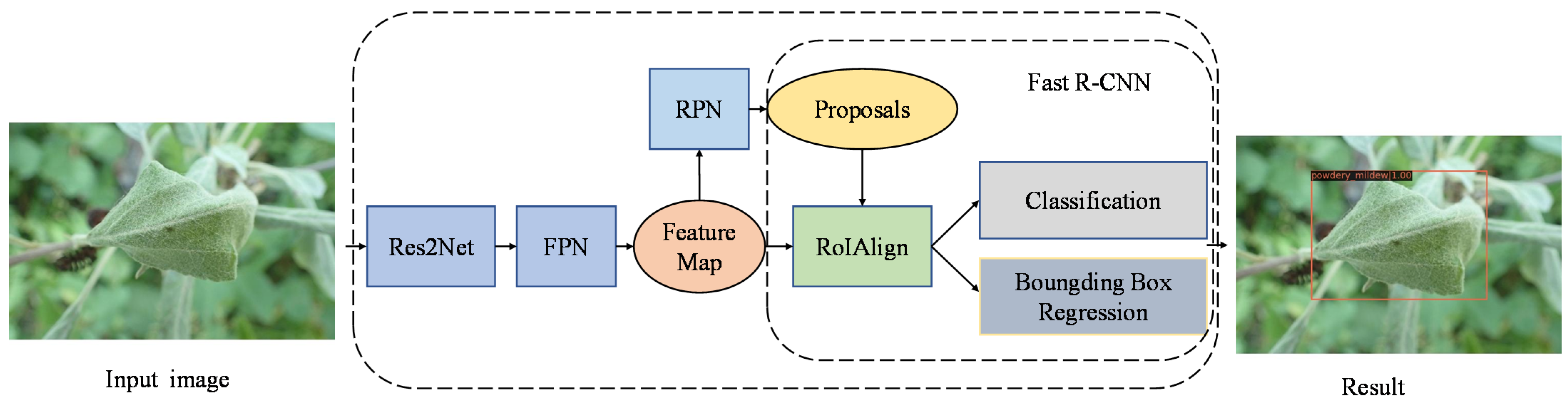
2.3. The Improved Faster R-CNN Model
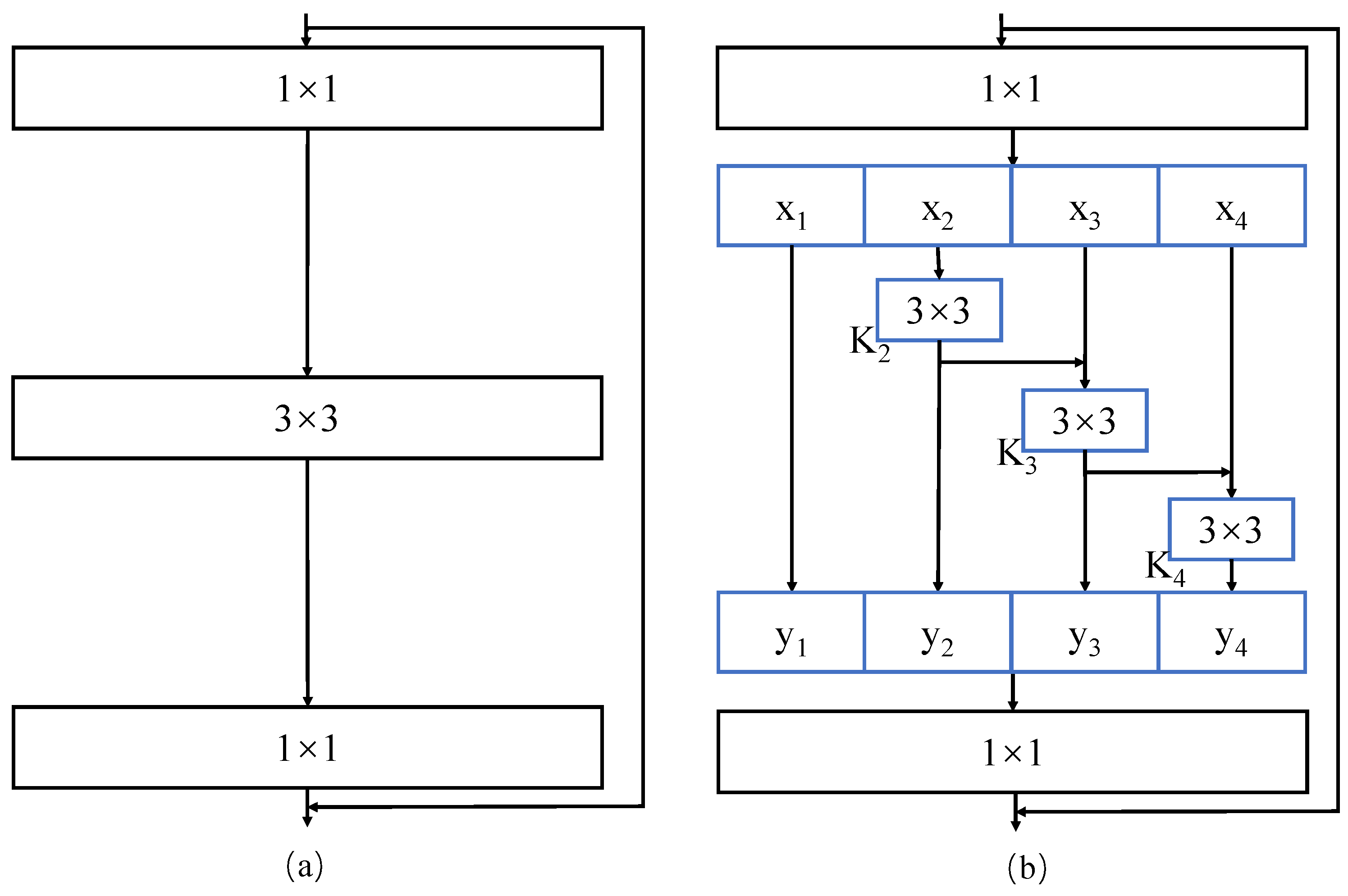
2.3.1. Res2Net Architecture
2.3.2. FPN
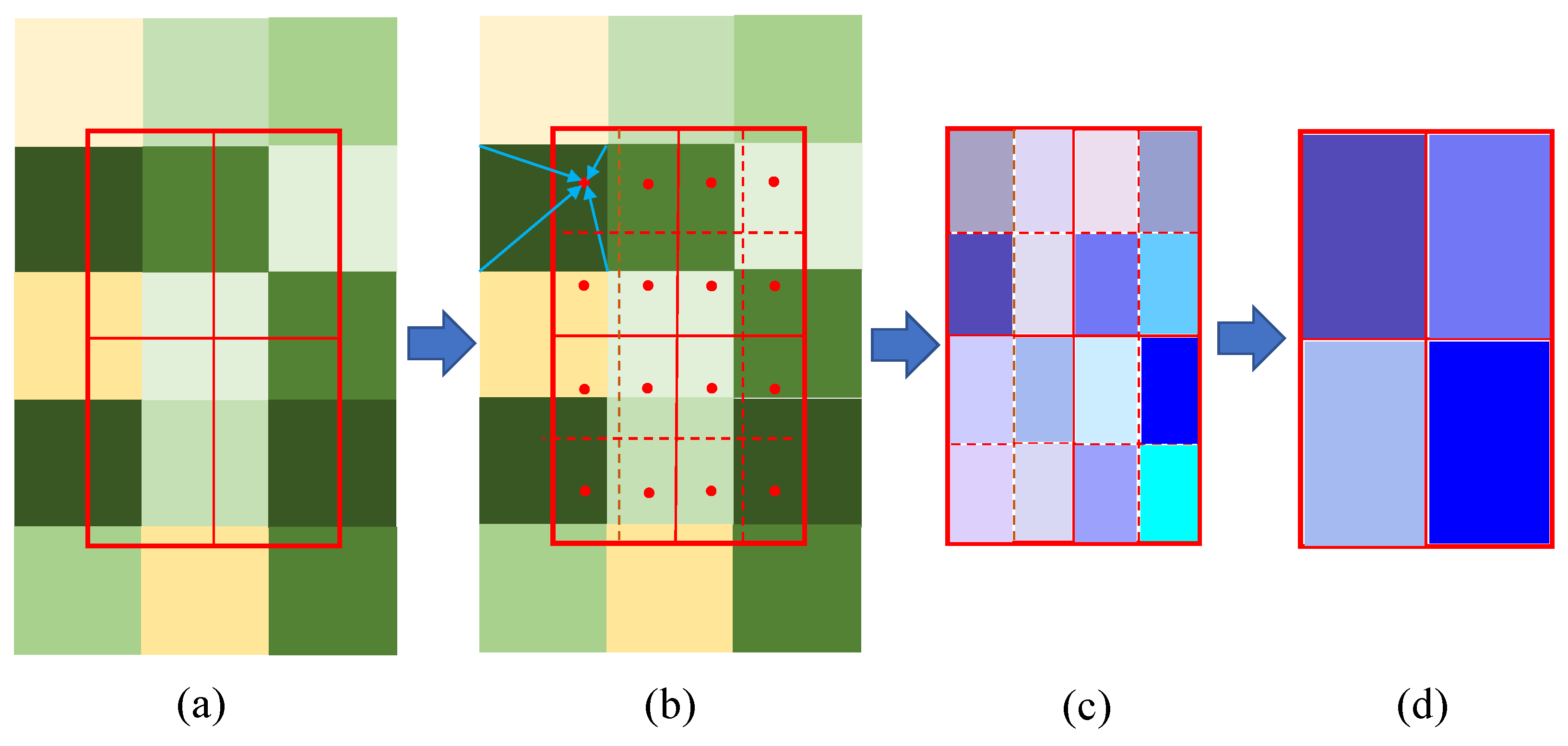
2.3.3. RoIAlign
2.3.4. Soft-NMS
2.4. Experimental Setup
2.4.1. Experiment Platform
2.4.2. Parameter Settings
2.4.3. Evaluation Metrics
3. Results
3.1. Comparison of Different Feature Extraction Networks
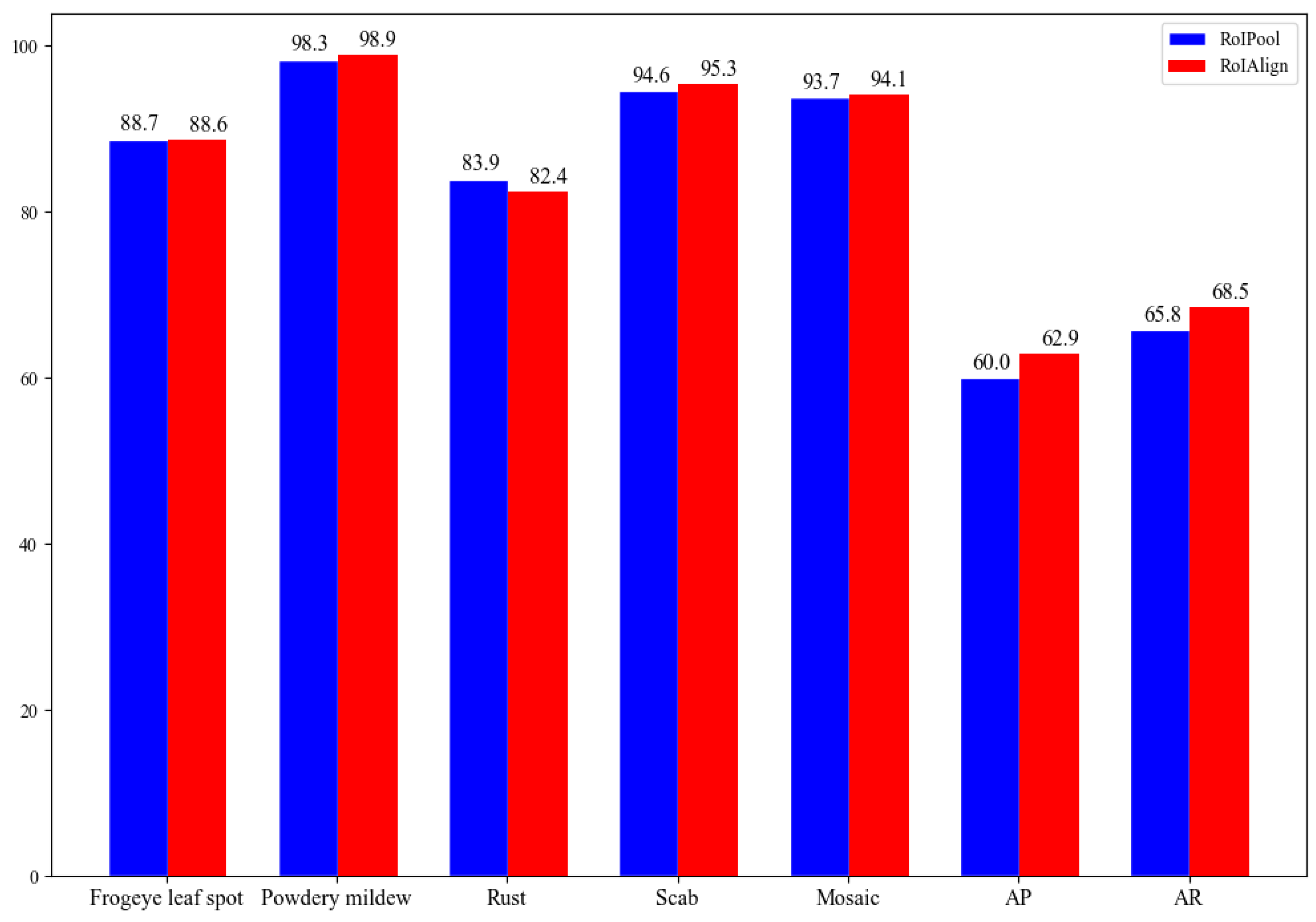
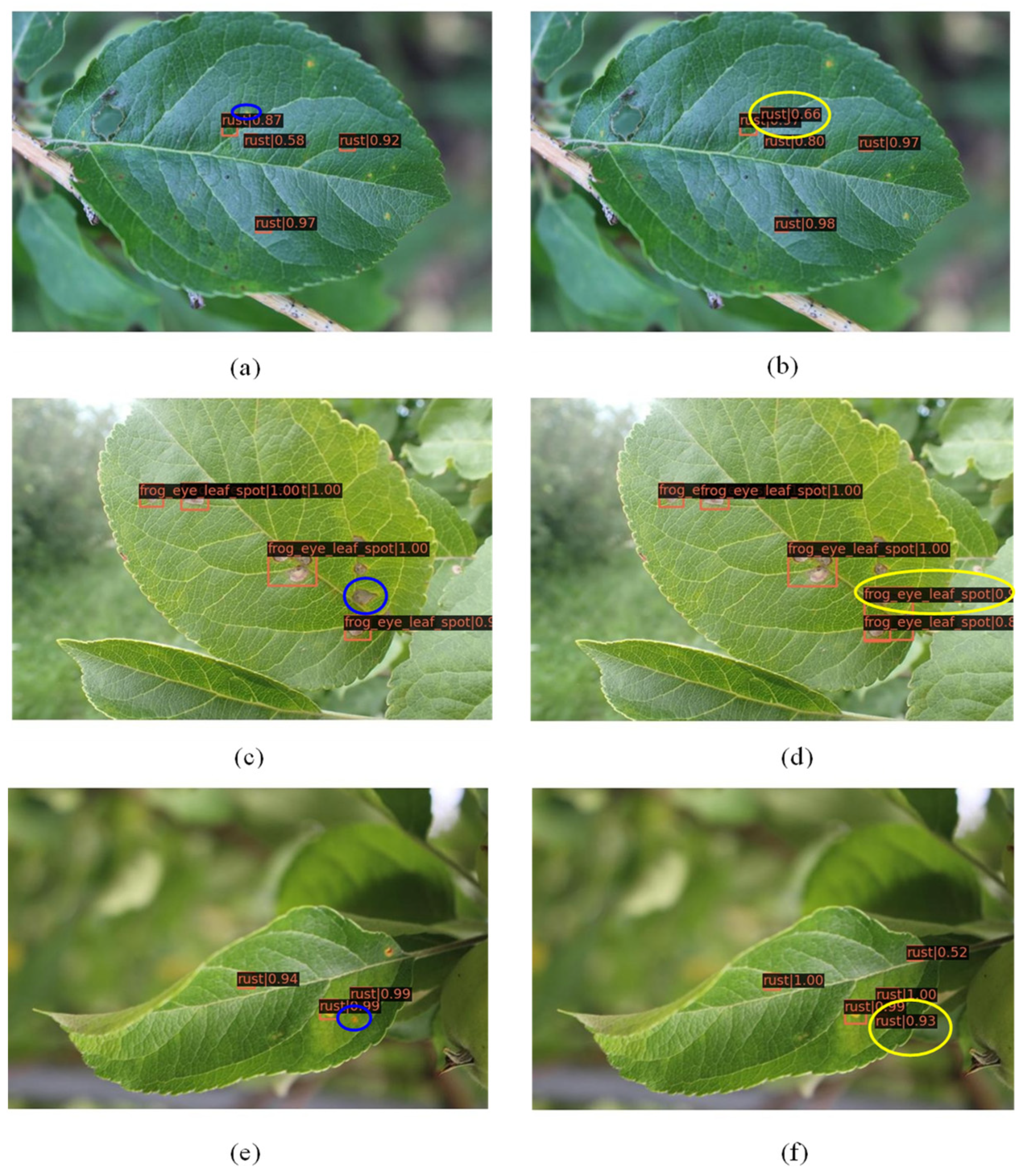
3.2. Detection Accuracy Comparison between RoIPool and RoIAlign
3.3. Comparison of Different Detection Techniques
3.4. Specific Class Performance Analysis
3.5. Result Comparison between NMS and Soft-NMS
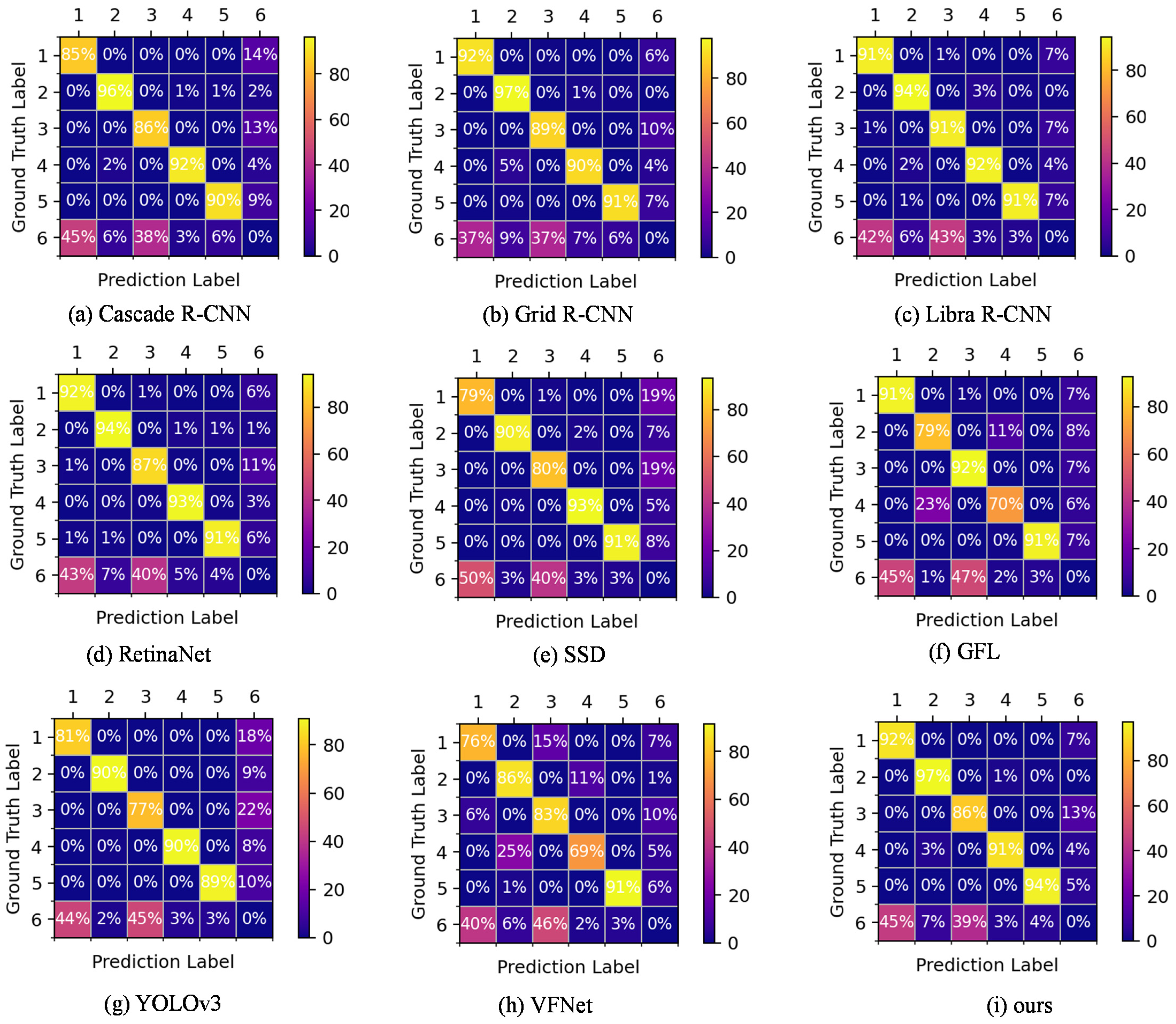
3.6. Comparison of Confusion Matrix
4. Discussion
5. Conclusions
- The AP and AR of the improved Faster R-CNN model with Res2Net-50-FPN are 62.9% and 68.5%, it is highest among other different backbones including ResNet-50-FPN, ResNet-101-FPN, ResNeXt-101-FPN, ResNeSt-50-FPN, and Reg-Net-FPN.
- The AP50 of the improved Faster R-CNN with RoIAlign is 0.6%, 0.7% and 0.4% higher than that of Faster R-CNN with RoIPool in detecting powdery mildew, scab and mosaic, respectively. The AP and AR of the Faster R-CNN with RoIAlign also has an improvement of 2.9% and 2.7% compared with Faster R-CNN with RoIPool.
- To compare the recognition results of specific apple leaf diseases, eight different detection methods are used. The AP50 of our improved Faster R-CNN in detecting frogeye leaf spot, powdery mildew, rust, scab and mosaic was 88.6%, 98.9%, 82.4%, 95.3% and 94.1%, respectively. Moreover, the AP50 of powdery mildew and mosaic has the best performance. This indicates that our method has a wide application in practice.
- The improved Faster R-CNN using soft-NMS can achieve a 63.1% AP and 71.4 AR, outperforming the Faster R-CNN using NMS. There is also an advantage in detecting dense disease objects compared with the original Faster R-CNN.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- National Bureau of Statistics of China, Agricultural Data. Available online: https://data.stats.gov.cn/easyquery.htm?cn=C01 (accessed on 1 January 2023).
- Zhong, Y.; Zhao, M. Research on deep learning in apple leaf disease recognition. Comput. Electron. Agric. 2020, 168, 105146. [Google Scholar] [CrossRef]
- Bansal, P.; Kumar, R.; Kumar, S. Disease detection in apple leaves using deep convolutional neural network. Agriculture 2021, 11, 617. [Google Scholar] [CrossRef]
- Hlaing, C.S.; Maung Zaw, S.M. Tomato plant diseases classification using statistical texture feature and color feature. In Proceedings of the 2018 IEEE/ACIS 17th International Conference on Computer and Information Science (ICIS), Singapore, 6–8 June 2018. [Google Scholar]
- Shrivastava, V.K.; Pradhan, M.K. Rice plant disease classification using color features: A machine learning paradigm. J. Plant Pathol. 2021, 103, 17–26. [Google Scholar] [CrossRef]
- Yang, N.; Qian, Y.; El-Mesery, H.S.; Zhang, R.; Wang, A.; Tang, J. Rapid detection of rice disease using microscopy image identification based on the synergistic judgment of texture and shape features and decision tree-confusion matrix method. J. Sci. Food Agric. 2019, 99, 6589–6600. [Google Scholar] [CrossRef]
- Mahmud, M.S.; Zaman, Q.U.; Esau, T.J.; Price, G.W. Development of an artificial cloud lighting condition system using machine vision for strawberry powdery mildew disease detection. Comput. Electron. Agric. 2019, 158, 219–225. [Google Scholar] [CrossRef]
- Chaudhary, P.; Chaudhari, A.K.; Godara, S. Color transform based approach for disease spot detection on plant leaf. Int. J. Comput. Sci. Telecommun. 2012, 3, 65–70. [Google Scholar]
- Pupitasari, T.D.; Basori, A.H.; Riskiawan, H.Y.; Setyohadi, D.P.S.; Kurniasari, A.A.; Firgiyanto, R.; Mansur, A.B.F.; Yunianta, A. Intelligent detection of rice leaf diseases based on histogram color and closing morphological. Emir. J. Food Agric. 2022, 34, 404–410. [Google Scholar] [CrossRef]
- Arivazhagan, S.; Shebiah, R.N.; Ananthi, S.; Varthini, S.V. Detection of unhealthy region of plant leaves and classification of plant leaf diseases using texture features. Agric. Eng. Int. CIGR J. 2013, 15, 211–217. [Google Scholar]
- Li, Z.; Guo, R.; Li, M.; Chen, Y.; Li, G. A review of computer vision technologies for plant phenotyping. Comput. Electron. Agric. 2020, 176, 105672. [Google Scholar] [CrossRef]
- Chen, J.; Yin, H.; Zhang, D. A self-adaptive classification method for plant disease detection using GMDH-logistic model. Sustain. Comput. Inform. 2020, 28, 100415. [Google Scholar] [CrossRef]
- Shuaibu, M.; Lee, W.S.; Schueller, J.; Gaderc, P.; Hong, Y.K.; Kim, S. Unsupervised hyperspectral band selection for apple Marssonina blotch detection. Comput. Electron. Agric. 2018, 148, 45–53. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, S.; Yang, J.; Shi, Y.; Chen, J. Apple leaf disease identification using genetic algorithm and correlation based feature selection method. Int. J. Agric. Biol. Eng. 2017, 10, 74–83. [Google Scholar]
- Omrani, E.; Khoshnevisan, B.; Shamshirband, S.; Saboohi, H.; Anuar, N.B.; Nasir, M. Potential of radial basis function-based support vector regression for apple disease detection. Measurement 2014, 55, 512–519. [Google Scholar] [CrossRef]
- Tian, Y.; Yang, G.; Wang, Z.; Li, E.; Liang, Z. Detection of apple lesions in orchards based on deep learning methods of CycleGAN and YOLOV3-Dense. J. Sens. 2019, 2019, 7630926. [Google Scholar] [CrossRef]
- Khan, A.I.; Quadri, S.M.K.; Banday, S.; Shah, J.L. Deep diagnosis: A real-time apple leaf disease detection system based on deep learning. Comput. Electron. Agric. 2022, 198, 1070931. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, Y.; He, D.; Li, Y. Identification of apple leaf diseases based on deep convolutional neural networks. Symmetry 2018, 10, 11. [Google Scholar] [CrossRef]
- Mahato, D.K.; Pundir, A.; Saxena, G.J. An improved deep convolutional neural network for image-based apple plant leaf disease detection and identification. J. Inst. Eng. India Ser. A 2022, 103, 975–987. [Google Scholar] [CrossRef]
- Atila, Ü.; Uçar, M.; Akyol, K.; Uçar, E. Plant leaf disease classification using EfficientNet deep learning model. Ecol. Inform. 2021, 61, 101182. [Google Scholar] [CrossRef]
- Sun, H.; Xu, H.; Liu, B.; He, D.; He, J.; Zhang, H.; Geng, N. MEAN-SSD: A novel real-time detector for apple leaf diseases using improved light-weight convolutional neural networks. Comput. Electron. Agric. 2021, 189, 106379. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Zhao, J. MGA-YOLO: A lightweight one-stage network for apple leaf disease detection. Front. Plant Sci. 2022, 13, 927424. [Google Scholar] [CrossRef]
- Rehman, Z.u.; Khan, M.A.; Ahmed, F.; Damaševicius, R.; Naqvi, S.R.; Nisar, W.; Javed, K. Recognizing apple leaf diseases using a novel parallel real-time processing framework based on mask rcnn and transfer learning: An application for smart agriculture. IET Image Process. 2021, 15, 2157–2168. [Google Scholar] [CrossRef]
- Jiang, P.; Chen, Y.; Liu, B.; He, D.; Liang, C. Real-Time Detection of Apple Leaf Diseases Using Deep Learning Approach Based on Improved Convolutional Neural Networks. IEEE Access 2019, 7, 59069–59080. [Google Scholar] [CrossRef]
- Lin, T.Y.; Dollar, P.; Girshick, R.; He, K.; Hariharan, B.; Belongie, S. Feature pyramid networks for object detection. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017. [Google Scholar]
- He, K.; Georgia, G.; Piotr, D.; Ross, G. Mask R-CNN. In Proceedings of the 2017 IEEE Transactions on Pattern Analysis & Machine Intelligence (ICCV), Venice, Italy, 22–29 October 2017. [Google Scholar]
- Bodla, N.; Singh, B.; Chellappa, R.; Davis, L.S. Improving object detection with one line of code. In Proceedings of the 2017 IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.B.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. Adv. Neural Inf. Process. Syst. 2015, 39, 1137–1149. [Google Scholar] [CrossRef] [PubMed]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Columbus, OH, USA, 24–27 June 2014. [Google Scholar]
- Girshick, R. Fast R-CNN. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015. [Google Scholar]
- Gao, S.H.; Cheng, M.M.; Zhao, K.; Zhang, X.Y.; Yang, M.H.; Torr, P. Res2net: A new multi-scale backbone architecture. IEEE Trans. Pattern Anal. Mach. Intel. 2021, 43, 652–662. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar]
- Kai, C.; Jiaqi, W.; Jiangmiao, P.; Yuhang, C.; Yu, X.; Xiaoxiao, L.; Shuyang, S.; Wansen, F.; Ziwei, L.; Jiarui, X.; et al. MMDetection: OpenMMLab detection toolbox and benchmark. arXiv 2019, arXiv:1906.07155. [Google Scholar]
- Alruwaili, M.; Siddiqi, M.H.; Khan, A.; Azad, M.; Khan, A.; Alanazi, S. RTF-RCNN: An architecture for real-time tomato plant leaf diseases detection in video streaming using Faster-RCNN. Bioengineering 2022, 9, 565. [Google Scholar] [CrossRef]
- Zhang, Y.; Song, C.; Zhang, D. Deep Learning-Based Object Detection Improvement for Tomato Disease. IEEE Access 2020, 8, 56607–56614. [Google Scholar] [CrossRef]
- Sardogan, M.; Ozen, Y.; Tuncer, A. Detection of apple leaf diseases using Faster R-CNN. Düzce Univ. J. Sci. Tech. 2020, 8, 1110–1117. [Google Scholar]
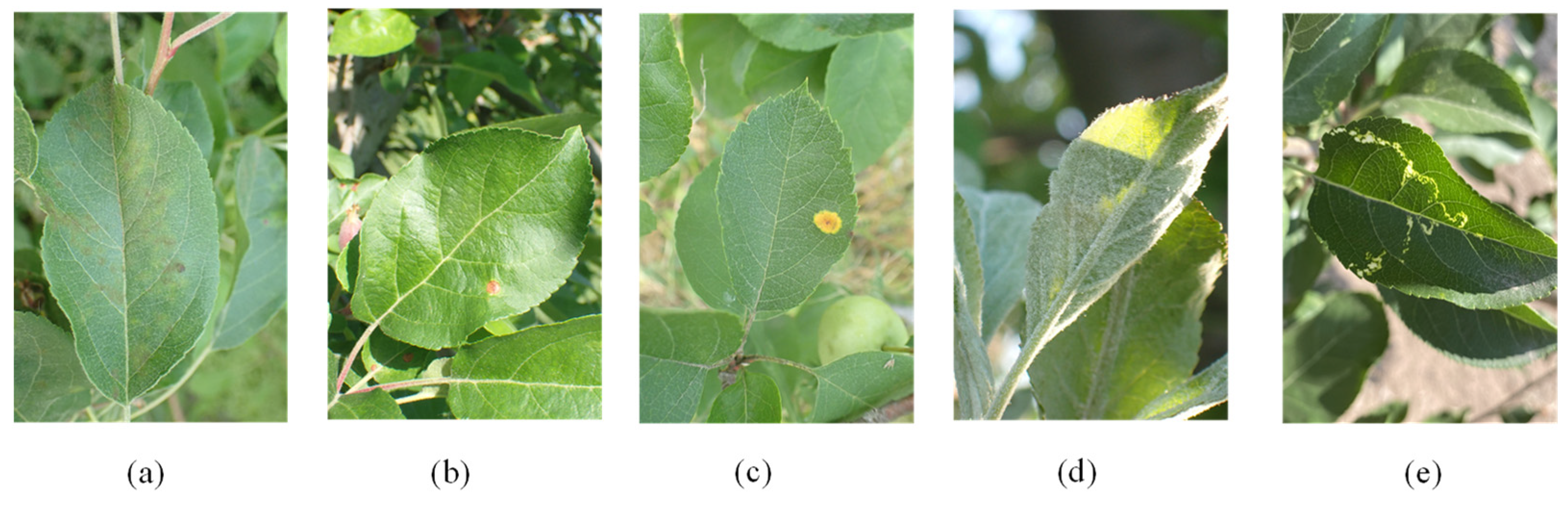

| Disease | Number of Images | Training Set | Enhanced Training Set | Validation Set | Test Set |
|---|---|---|---|---|---|
| Scab | 915 | 733 | 733 | 91 | 91 |
| Frogeye leaf spot | 952 | 762 | 762 | 95 | 95 |
| Rust | 928 | 743 | 743 | 93 | 92 |
| Powdery mildew | 903 | 723 | 723 | 90 | 90 |
| Mosaic | 378 | 190 | 760 | 94 | 94 |
| Scab and frogeye leaf spot | 54 | 33 | 66 | 11 | 10 |
| Rust and frogeye leaf spot | 52 | 32 | 64 | 10 | 10 |
| Total | 4182 | 3216 | 3851 | 484 | 482 |
| Framework | Backbone | AP | AP50 | AP75 | AR |
|---|---|---|---|---|---|
| Faster R-CNN | Res2Net-50-FPN | 62.9 | 91.3 | 69.8 | 68.5 |
| ResNet-50-FPN | 61.9 | 92.2 | 70.4 | 67.6 | |
| ResNet-101-FPN | 62.5 | 90.9 | 70.6 | 68.1 | |
| ResNeXt-101-FPN | 61.8 | 91.0 | 69.6 | 67.7 | |
| ResNeSt-50-FPN | 61.0 | 91.3 | 68.6 | 67.0 | |
| RegNet-FPN | 62.1 | 89.3 | 70.0 | 67.9 |
| Method | Backbone | AP | AP50 | AP75 | AR | FPS |
|---|---|---|---|---|---|---|
| Cascade R-CNN | ResNet-50-FPN | 62.6 | 90.1 | 68.2 | 68.4 | 12.0 |
| Grid R-CNN | ResNet-50-FPN | 61.5 | 91.1 | 65.9 | 68.3 | 12.5 |
| Libra R-CNN | ResNet-50-FPN | 60.3 | 91.3 | 66.7 | 67.4 | 13.3 |
| RetinaNet | ResNet-101-FPN | 61.2 | 91.4 | 68.2 | 67.7 | 14.6 |
| SSD | SSDVGG | 59.8 | 88.9 | 64.8 | 68.5 | 64.4 |
| GFL | ResNet-50-FPN | 61.2 | 89.7 | 68.0 | 71.2 | 14.5 |
| YOLOv3 | DarkNet-53 | 59.3 | 88.0 | 63.9 | 68.0 | 76.6 |
| VFNet | ResNet-50-FPN | 59.6 | 88.7 | 64.7 | 69.9 | 13.6 |
| Ours | 62.9 | 91.3 | 69.8 | 68.5 | 12.2 |
| Method | Backbone | Frogeye Leaf Spot | Powdery Mildew | Rust | Scab | Mosaic |
|---|---|---|---|---|---|---|
| Cascade R-CNN | ResNet-50-FPN | 83.7 | 98.5 | 83.0 | 94.8 | 90.2 |
| Grid R-CNN | ResNet-50-FPN | 89.6 | 98.0 | 84.1 | 94.9 | 89.1 |
| Libra R-CNN | ResNet-50-FPN | 87.8 | 98.0 | 85.0 | 94.8 | 90.8 |
| RetinaNet | ResNet-101-FPN | 87.8 | 96.3 | 84.0 | 96.3 | 92.7 |
| SSD | SSDVGG | 83.1 | 96.4 | 80.1 | 93.8 | 91.1 |
| GFL | ResNet-50-FPN | 88.5 | 91.2 | 86.0 | 90.0 | 92.9 |
| YOLOv3 | DarkNet-53 | 80.0 | 94.3 | 78.0 | 93.9 | 93.6 |
| VFNet | ResNet-50-FPN | 83.0 | 93.5 | 82.4 | 92.8 | 91.7 |
| Ours | 88.6 | 98.9 | 82.4 | 95.3 | 94.1 |
| Method | Backbone | AP | AR |
|---|---|---|---|
| Faster R-CNN using NMS | Res2Net-50-FPN | 62.9 | 68.5 |
| Faster R-CNN using soft-NMS | Res2Net-50-FPN | 63.1 | 71.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, X.; Zhang, S. A High-Precision Detection Method of Apple Leaf Diseases Using Improved Faster R-CNN. Agriculture 2023, 13, 240. https://doi.org/10.3390/agriculture13020240
Gong X, Zhang S. A High-Precision Detection Method of Apple Leaf Diseases Using Improved Faster R-CNN. Agriculture. 2023; 13(2):240. https://doi.org/10.3390/agriculture13020240
Chicago/Turabian StyleGong, Xulu, and Shujuan Zhang. 2023. "A High-Precision Detection Method of Apple Leaf Diseases Using Improved Faster R-CNN" Agriculture 13, no. 2: 240. https://doi.org/10.3390/agriculture13020240
APA StyleGong, X., & Zhang, S. (2023). A High-Precision Detection Method of Apple Leaf Diseases Using Improved Faster R-CNN. Agriculture, 13(2), 240. https://doi.org/10.3390/agriculture13020240





