Responses of Alpine Soil Nitrification and Denitrification Rates to Nitrogen Addition Gradient—The Role of Functional Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site and Experimental Design
2.2. Soil Sampling and Physico-Chemical Analysis
2.3. Quantitative PCR and High-Throughput Sequencing
2.4. Data Analysis
3. Results
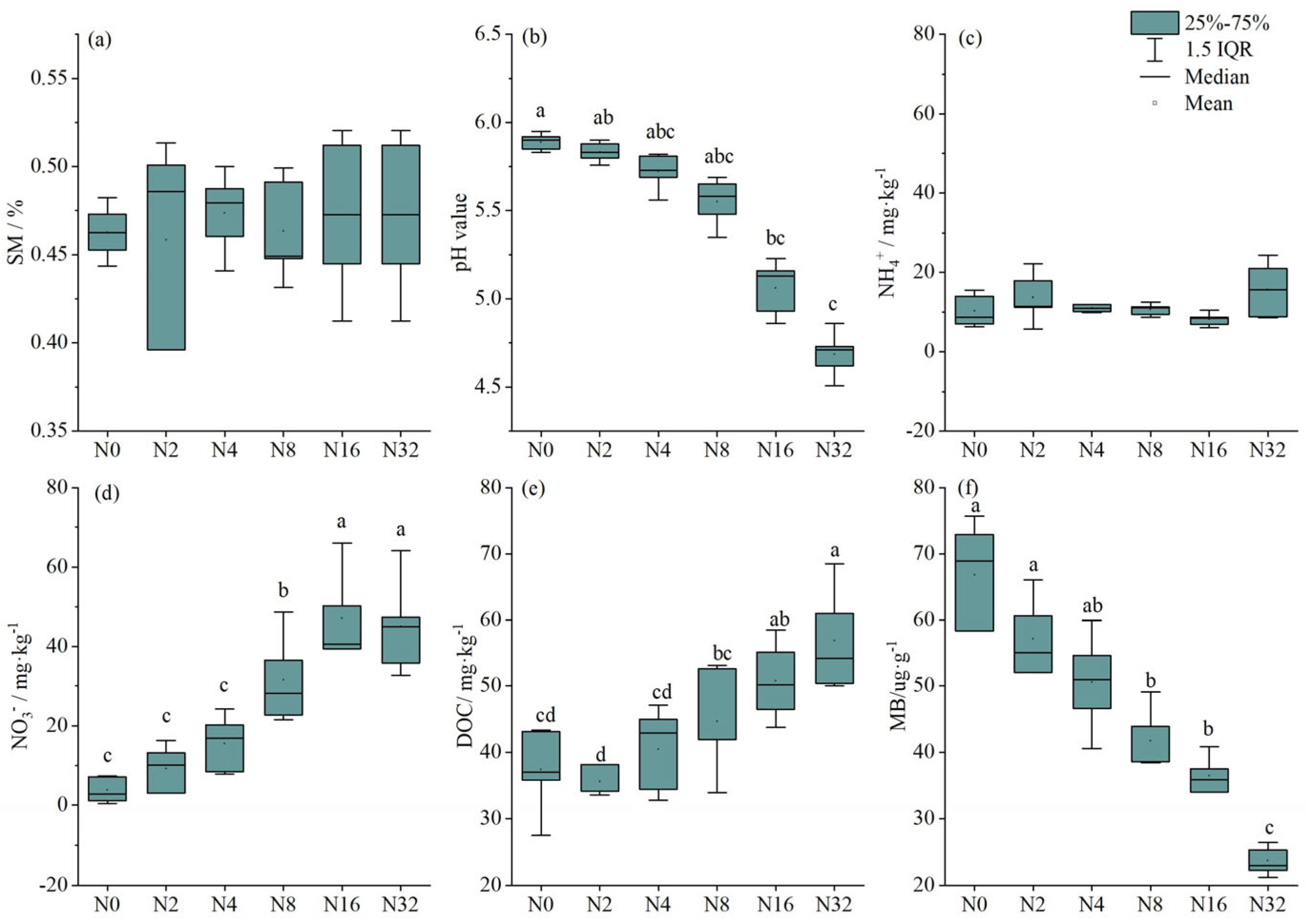
3.1. The Effects of N Application with Different Rates on Soil Physico-Chemical Properties
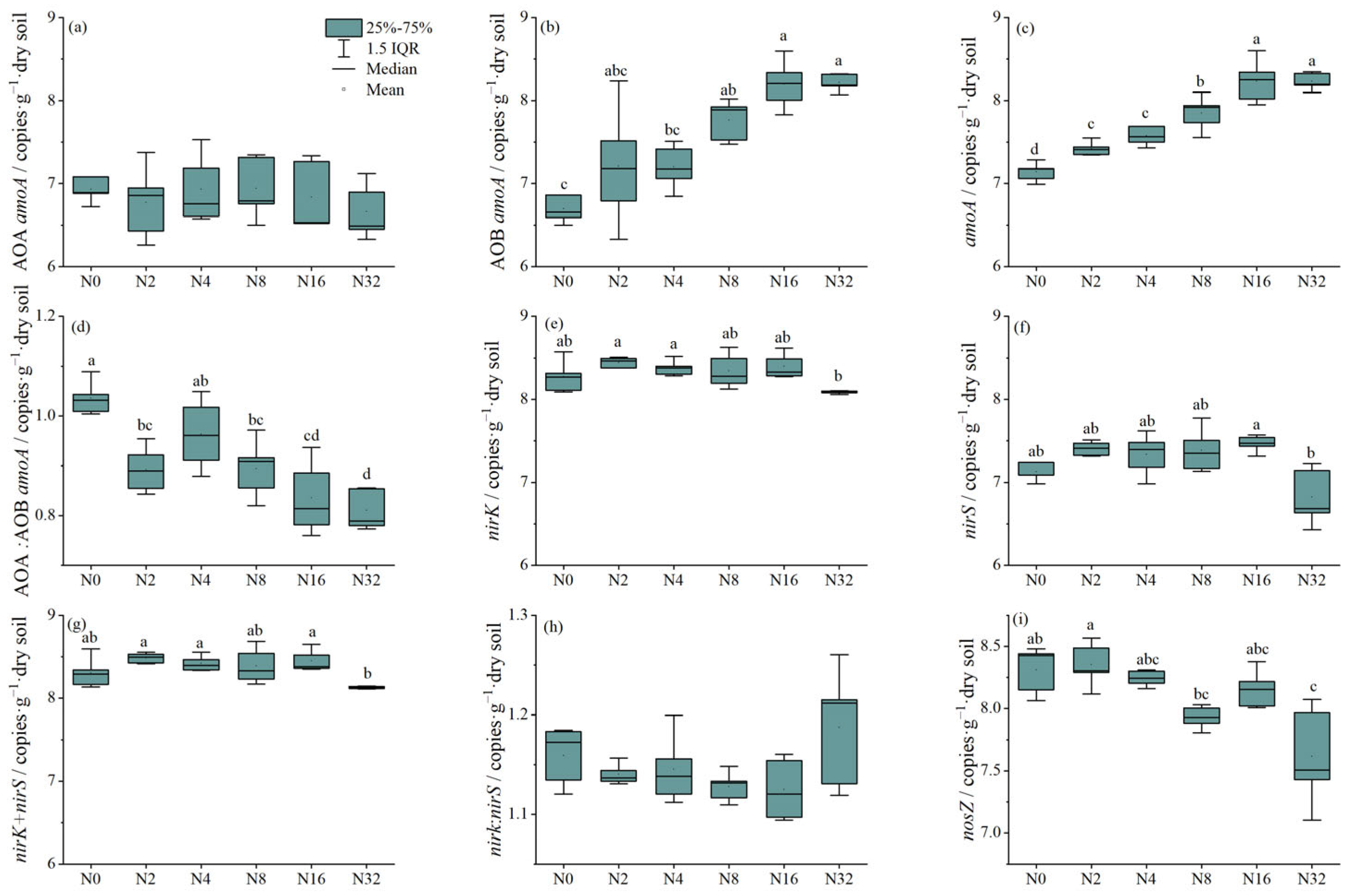
3.2. The Effect of Different N Application Rates on the Abundances of Nitrifiers and Denitrifiers
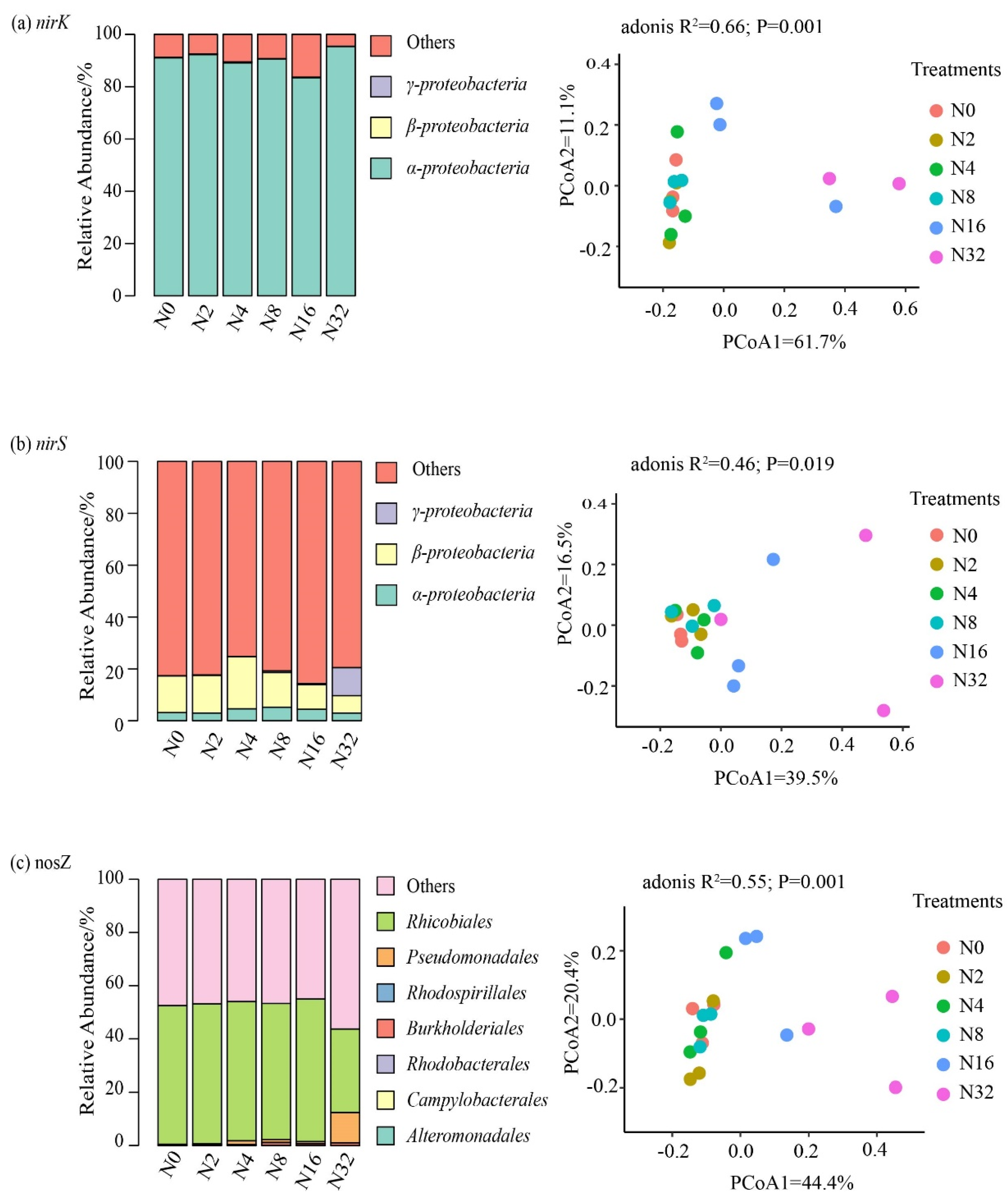
3.3. The Effects of the Different N Addition Rates on the Compositions of the Nitrifier and Denitrifier Communities
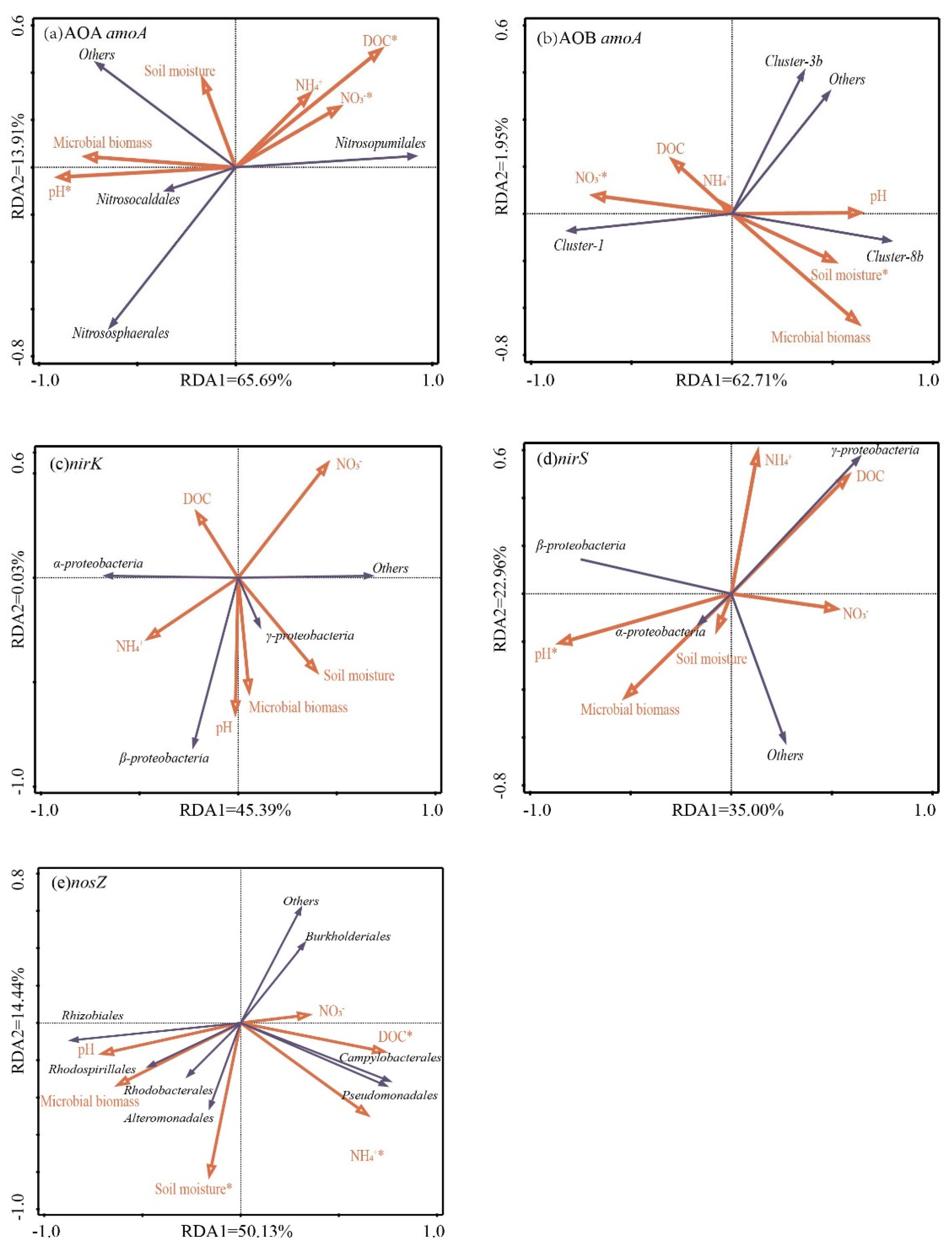
3.4. Relationships between the Soil Physical and Chemical Properties and the Abundances and Community Structure of Nitrifier and Denitrifier
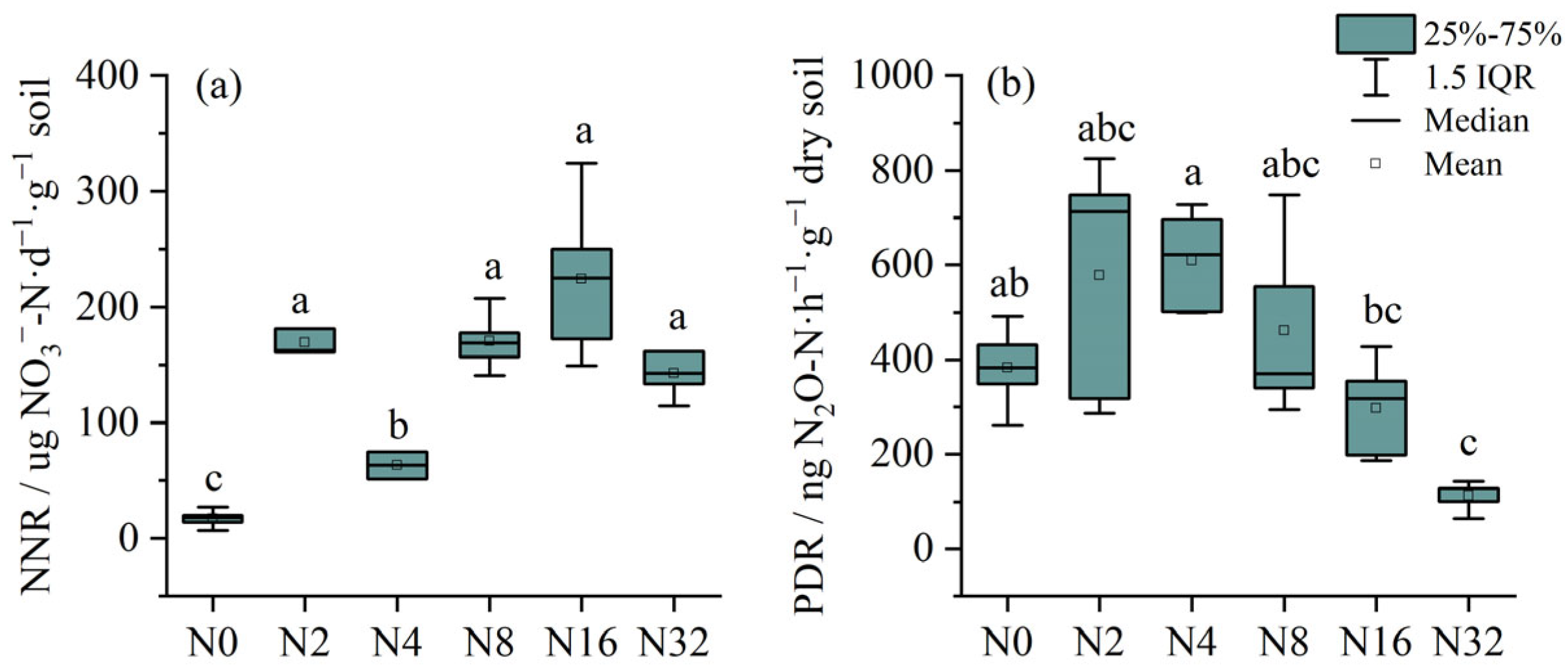
3.5. Soil Nitrification and Denitrification Rates in the Soils Treated with Different N Applications
4. Discussion
4.1. Impacts of N Applications on the Abundances of Soil Nitrifiers and Denitrifiers
4.2. Impacts of N Applications on the Nitrifier and Denitrifier Community Compositions
4.3. Impacts of N Applications on Nitrification and Denitrification Rates
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galloway, J.N.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.; Freney, J.R.; Martinelli, L.A.; Seitzinger, S.P.; Sutton, M.A. Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science 2008, 320, 889–892. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Jia, Y.; He, N.; Zhu, J.; Chen, Z.; Wang, Q.; Piao, S.; Liu, X.; He, H.; Guo, X.; et al. Stabilization of atmospheric nitrogen deposition in China over the past decade. Nat. Geosci. 2019, 12, 424–429. [Google Scholar] [CrossRef]
- Luo, Q.; Gong, J.; Yang, L.; Li, X.; Pan, Y.; Liu, M.; Zhai, Z.; Baoyin, T.-T. Impacts of nitrogen addition on the carbon balance in a temperate semiarid grassland ecosystem. Biol. Fertil. Soils 2017, 53, 911–927. [Google Scholar] [CrossRef]
- Xu, C.; Xu, X.; Ju, C.; Chen, H.Y.H.; Wilsey, B.J.; Luo, Y.; Fan, W. Long-term, amplified responses of soil organic carbon to nitrogen addition worldwide. Glob. Chang. Biol. 2021, 27, 1170–1180. [Google Scholar] [CrossRef]
- Zhang, J.; He, P.; Liu, Y.; Du, W.; Jing, H.; Nie, C. Soil properties and microbial abundance explain variations in N2O fluxes from temperate steppe soil treated with nitrogen and water in Inner Mongolia, China. Appl. Soil Ecol. 2021, 165, 103984. [Google Scholar] [CrossRef]
- Ma, L.N.; Lu, X.T.; Liu, Y.; Guo, J.X.; Zhang, N.Y.; Yang, J.Q.; Wang, R.Z. The effects of warming and nitrogen addition on soil nitrogen cycling in a temperate grassland, northeastern China. PLoS ONE 2011, 6, e27645. [Google Scholar] [CrossRef]
- Zhang, C.J.; Yang, Z.L.; Shen, J.P.; Sun, Y.F.; Wang, J.T.; Han, H.Y.; Wan, S.Q.; Zhang, L.M.; He, J.Z. Impacts of long-term nitrogen addition, watering and mowing on ammonia oxidizers, denitrifiers and plant communities in a temperate steppe. Appl. Soil Ecol. 2018, 130, 241–250. [Google Scholar] [CrossRef]
- Song, L.; Li, Z.; Niu, S. Global Soil Gross Nitrogen Transformation Under Increasing Nitrogen Deposition. Glob. Biogeochem. Cycle 2021, 35, e2020GB006711. [Google Scholar] [CrossRef]
- Lu, M.; Yang, Y.; Luo, Y.; Fang, C.; Zhou, X.; Chen, J.; Yang, X.; Li, B. Responses of ecosystem nitrogen cycle to nitrogen addition: A meta-analysis. New Phytol. 2011, 189, 1040–1050. [Google Scholar] [CrossRef]
- Wang, J.; Chadwick, D.R.; Cheng, Y.; Yan, X. Global analysis of agricultural soil denitrification in response to fertilizer nitrogen. Sci. Total Environ. 2018, 616, 908–917. [Google Scholar] [CrossRef]
- Song, L.; Niu, S. Increased soil microbial AOB amoA and narG abundances sustain long-term positive responses of nitrification and denitrification to N deposition. Soil Biol. Biochem. 2022, 166, 108539. [Google Scholar] [CrossRef]
- Thomson, A.J.; Giannopoulos, G.; Pretty, J.; Baggs, E.M.; Richardson, D.J. Biological sources and sinks of nitrous oxide and strategies to mitigate emissions. Philos. Trans. R. Soc. B Biol. Sci. 2012, 367, 1157–1168. [Google Scholar] [CrossRef]
- Canfield, D.E.; Glazer, A.N.; Falkowski, P.G. The Evolution and Future of Earth’s Nitrogen Cycle. Science 2010, 330, 192–196. [Google Scholar] [CrossRef]
- Prosser, J.I.; Nicol, G.W. Archaeal and bacterial ammonia-oxidisers in soil: The quest for niche specialisation and differentiation. Trends Microbiol. 2012, 20, 523–531. [Google Scholar] [CrossRef]
- Purkhold, U.; Pommereningröser, A.; Juretschko, S.; Schmid, M.C.; Koops, H.P.; Wagner, M. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: Implications for molecular diversity surveys. Appl. Environ. Microbiol. 2000, 66, 5368–5382. [Google Scholar] [CrossRef]
- Shiina, Y.; Itakura, M.; Choi, H.; Saeki, Y.; Hayatsu, M.; Minamisawa, K. Relationship Between Soil Type and N2O Reductase Genotype (nosZ) of Indigenous Soybean Bradyrhizobia: nosZ-minus Populations are Dominant in Andosols. Microbes Environ. 2014, 29, 420–426. [Google Scholar] [CrossRef]
- Yin, C.; Fan, F.; Song, A.; Li, Z.; Yu, W.; Liang, Y. Different denitrification potential of aquic brown soil in Northeast China under inorganic and organic fertilization accompanied by distinct changes of nirS- and nirK-denitrifying bacterial community. Eur. J. Soil Biol. 2014, 65, 47–56. [Google Scholar] [CrossRef]
- Ullah, S.; Ai, C.; Huang, S.H.; Song, D.L.; Abbas, T.; Zhang, J.J.; Zhou, W.; He, P. Substituting ecological intensification of agriculture for conventional agricultural practices increased yield and decreased nitrogen losses in North China. Appl. Soil Ecol. 2020, 147, 103395. [Google Scholar] [CrossRef]
- Carey, C.J.; Dove, N.C.; Beman, J.M.; Hart, S.C.; Aronson, E.L. Meta-analysis reveals ammonia-oxidizing bacteria respond more strongly to nitrogen addition than ammonia-oxidizing archaea. Soil Biol. Biochem. 2016, 99, 158–166. [Google Scholar] [CrossRef]
- He, J.; Shen, J.; Zhang, L.; Zhu, Y.G.; Zheng, Y.; Xu, M.; Di, H.J. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ. Microbiol. 2007, 9, 2364–2374. [Google Scholar] [CrossRef]
- Isobe, K.; Ikutani, J.; Fang, Y.; Yoh, M.; Mo, J.; Suwa, Y.; Yoshida, M.; Senoo, K.; Otsuka, S.; Koba, K. Highly abundant acidophilic ammonia-oxidizing archaea causes high rates of nitrification and nitrate leaching in nitrogen-saturated forest soils. Soil Biol. Biochem. 2018, 122, 220–227. [Google Scholar] [CrossRef]
- Sun, P.; Zhuge, Y.; Zhang, J.; Cai, Z. Soil pH was the main controlling factor of the denitrification rates and N-2/N2O emission ratios in forest and grassland soils along the Northeast China Transect (NECT). Soil Sci. Plant Nutr. 2012, 58, 517–525. [Google Scholar] [CrossRef]
- Cheng, Y.; Wang, J.; Wang, S.-Q.; Zhang, J.-B.; Cai, Z.-C. Effects of soil moisture on gross N transformations and N2O emission in acid subtropical forest soils. Biol. Fertil. Soils 2014, 50, 1099–1108. [Google Scholar] [CrossRef]
- Shrewsbury, L.H.; Smith, J.L.; Huggins, D.R.; Carpenter-Boggs, L.; Reardon, C.L. Denitrifier abundance has a greater influence on denitrification rates at larger landscape scales but is a lesser driver than environmental variables. Soil Biol. Biochem. 2016, 103, 221–231. [Google Scholar] [CrossRef]
- Tang, Y.G.; Yu, G.R.; Zhang, X.Y.; Wang, Q.F.; Tian, D.S.; Tian, J.; Niu, S.L.; Ge, J.P. Environmental variables better explain changes in potential nitrification and denitrification activities than microbial properties in fertilized forest soils. Sci. Total Environ. 2019, 647, 653–662. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Sun, J.; Zhou, Q.; Zong, N.; Li, L.; Niu, S. Initial shifts in nitrogen impact on ecosystem carbon fluxes in an alpine meadow: Patterns and causes. Biogeosciences 2017, 14, 3947–3956. [Google Scholar] [CrossRef]
- Wen, Z.; Xu, W.; Li, Q.; Han, M.; Tang, A.; Zhang, Y.; Luo, X.; Shen, J.; Wang, W.; Li, K.; et al. Changes of nitrogen deposition in China from 1980 to 2018. Environ. Int. 2020, 144, 106022. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, J.; Li, Y.; He, P.; Dong, J. Do long-term N additions affect the soil organic carbon pool in temperate grasslands? Sci. Total Environ. 2021, 810, 152227. [Google Scholar] [CrossRef]
- Wang, C.; Wan, S.; Xing, X.; Zhang, L.; Han, X. Temperature and soil moisture interactively affected soil net N mineralization in temperate grassland in Northern China. Soil Biol. Biochem. 2006, 38, 1101–1110. [Google Scholar] [CrossRef]
- Smith, M.S.; Tiedje, J.M. Phases of denitrification following oxygen depletion in soil. Soil Biol. Biochem. 1979, 11, 261–267. [Google Scholar] [CrossRef]
- Treseder, K.K. Nitrogen additions and microbial biomass: A meta-analysis of ecosystem studies. Ecol. Lett. 2008, 11, 1111–1120. [Google Scholar] [CrossRef]
- Xiang, X.; He, D.; He, J.-S.; Myrold, D.D.; Chu, H. Ammonia-oxidizing bacteria rather than archaea respond to short-term urea amendment in an alpine grassland. Soil Biol. Biochem. 2017, 107, 218–225. [Google Scholar] [CrossRef]
- Offre, P.; Kerou, M.; Spang, A.; Schleper, C. Variability of the transporter gene complement in ammonia-oxidizing archaea. Trends Microbiol. 2014, 22, 665–675. [Google Scholar] [CrossRef]
- Dong, J.Y.; Zhang, J.Q.; Liu, Y.H.; Jing, H.C. How climate and soil properties affect the abundances of nitrogen-cycling genes in nitrogen-treated ecosystems: A meta-analysis. Plant Soil 2022, 477, 389–404. [Google Scholar] [CrossRef]
- Chen, Z.; Guo, Y.; Du, Z.; Wu, W.; Meng, F. Change in the abundance and community composition of ammonia-oxidizing bacteria and archaea at soil aggregate level as native pasture converted to cropland in a semiarid alpine steppe of central Asia. J. Soils Sediments 2015, 16, 243–254. [Google Scholar] [CrossRef]
- Nicol, G.W.; Leininger, S.; Schleper, C.; Prosser, J.I. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 2008, 10, 2966–2978. [Google Scholar] [CrossRef]
- Yang, Y.D.; Hu, Y.G.; Wang, Z.M.; Zeng, Z.H. Variations of the nirS-, nirK-, and nosZ-denitrifying bacterial communities in a northern Chinese soil as affected by different long-term irrigation regimes. Environ. Sci. Pollut. Res. Int. 2018, 25, 14057–14067. [Google Scholar] [CrossRef]
- Yang, Y.; Nie, J.; Wang, S.; Shi, L.; Li, Z.; Zeng, Z.; Zang, H. Differentiated responses of nirS- and nirK-type denitrifiers to 30 years of combined inorganic and organic fertilization in a paddy soil. Arch. Agron. Soil Sci. 2020, 67, 79–92. [Google Scholar] [CrossRef]
- vanGestel, M.; Merckx, R.; Vlassak, K. Spatial distribution of microbial biomass in microaggregates of a silty-loam soil and the relation with the resistance of microorganisms to soil drying. Soil Biol. Biochem. 1996, 28, 503–510. [Google Scholar] [CrossRef]
- Zhaolei, L.; Tang, Z.; Song, Z.; Chen, W.; Tian, D.; Tang, S.; Wang, X.; Wang, J.; Liu, W.; Wang, Y.; et al. Variations and controlling factors of soil denitrification rate. Glob. Chang. Biol. 2021, 28, 2133–2145. [Google Scholar] [CrossRef]

| AOA | AOB | amoA | nirK | nirS | nir | nosZ | MB | |
|---|---|---|---|---|---|---|---|---|
| N | −0.185 | 0.819 ** | 0.941 ** | −0.333 | −0.05 | −0.316 | −0.671 ** | −0.940 ** |
| SM | −0.006 | 0.217 | −0.035 | 0.433 * | 0.530 ** | 0.467 ** | 0.368 * | 0.328 |
| pH | 0.161 | −0.846 ** | −0.922 ** | 0.304 | 0.034 | 0.295 | 0.582 ** | 0.876 ** |
| NH4+ | 0.214 | 0.07 | 0.015 | 0.013 | −0.085 | −0.037 | −0.15 | 0.113 |
| NO3− | −0.1 | 0.788 ** | 0.902 ** | −0.126 | 0.205 | −0.098 | −0.555 ** | −0.789 ** |
| DOC | −0.136 | 0.696 ** | 0.729 ** | −0.236 | −0.175 | −0.221 | −0.486 ** | −0.684 ** |
| Factor (n = 30) | Standardized Beta | p | ss | |
|---|---|---|---|---|
| NNR | amoA abundance | 0.694 | <0.001 | 0.463 |
| Total explained | 0.463 | |||
| PDR | SM | 0.555 | <0.001 | 0.44 |
| pH | 0.535 | <0.001 | 0.271 | |
| Total explained | 0.711 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Liu, Y.; Dong, J. Responses of Alpine Soil Nitrification and Denitrification Rates to Nitrogen Addition Gradient—The Role of Functional Genes. Agriculture 2023, 13, 245. https://doi.org/10.3390/agriculture13020245
Zhang J, Liu Y, Dong J. Responses of Alpine Soil Nitrification and Denitrification Rates to Nitrogen Addition Gradient—The Role of Functional Genes. Agriculture. 2023; 13(2):245. https://doi.org/10.3390/agriculture13020245
Chicago/Turabian StyleZhang, Jiaqi, Yinghui Liu, and Jingyi Dong. 2023. "Responses of Alpine Soil Nitrification and Denitrification Rates to Nitrogen Addition Gradient—The Role of Functional Genes" Agriculture 13, no. 2: 245. https://doi.org/10.3390/agriculture13020245
APA StyleZhang, J., Liu, Y., & Dong, J. (2023). Responses of Alpine Soil Nitrification and Denitrification Rates to Nitrogen Addition Gradient—The Role of Functional Genes. Agriculture, 13(2), 245. https://doi.org/10.3390/agriculture13020245






