Integrated Metabolome and Transcriptome Analysis Unveils the Underlying Molecular Response of Panax ginseng Plants to the Phytophthora cactorum Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Methods
2.2. Plant Materials
2.3. Sample Preparation for Non-Targeted Metabolomics Analysis
2.4. UPLC-MS Based Non-Targeted Metabolomics Analysis
2.5. Sample Preparation for Quantitative Analysis of Plant Hormones
2.6. UPLC-MS/MS Conditions for Phytohormone Analysis
2.7. RNA Isolation and Transcriptomics Analysis
2.8. Quantitative PCR Assays
2.9. Integrated Analysis of Metabolome and Transcriptome
2.10. Statistical Analysis
3. Results
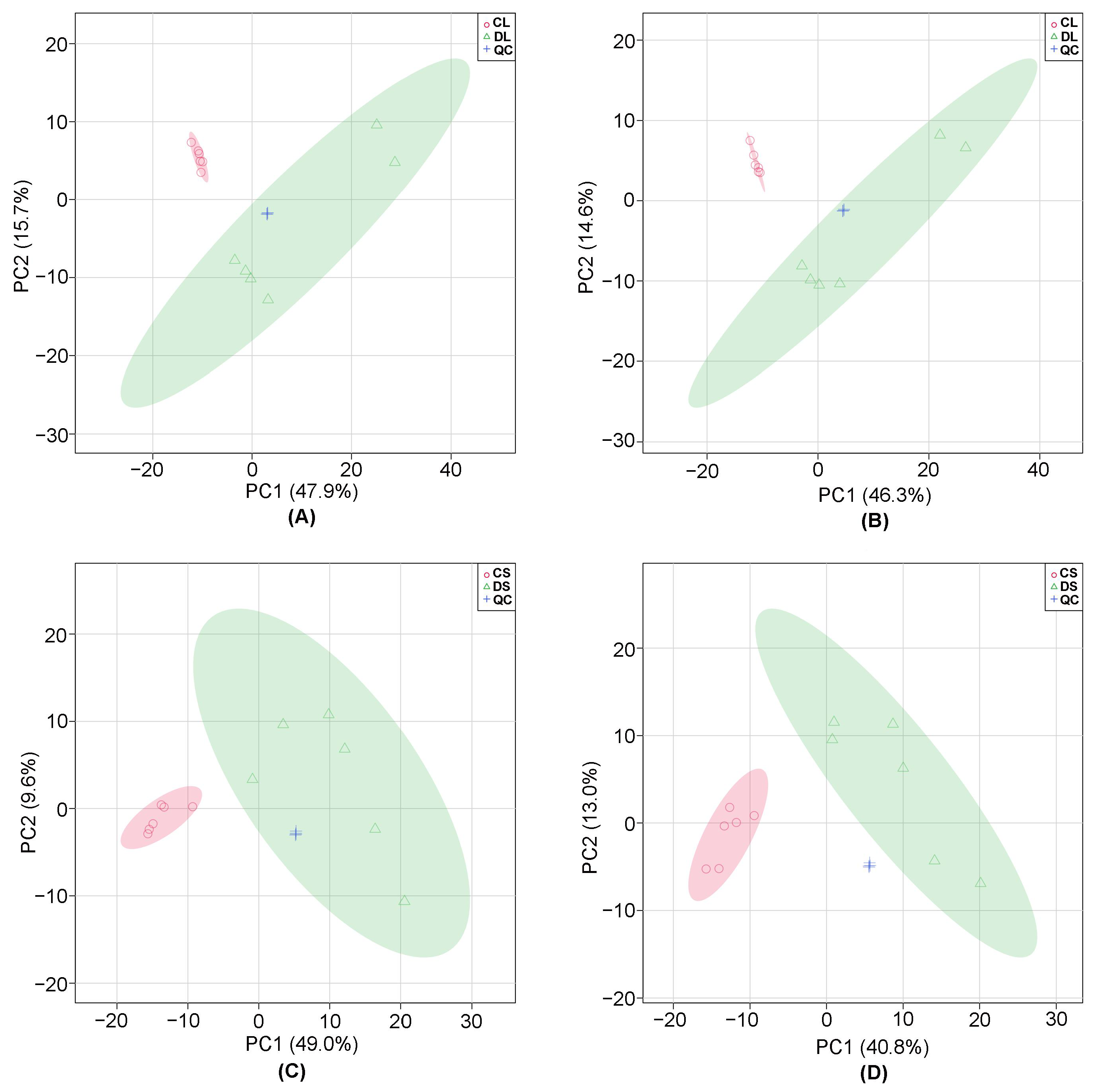
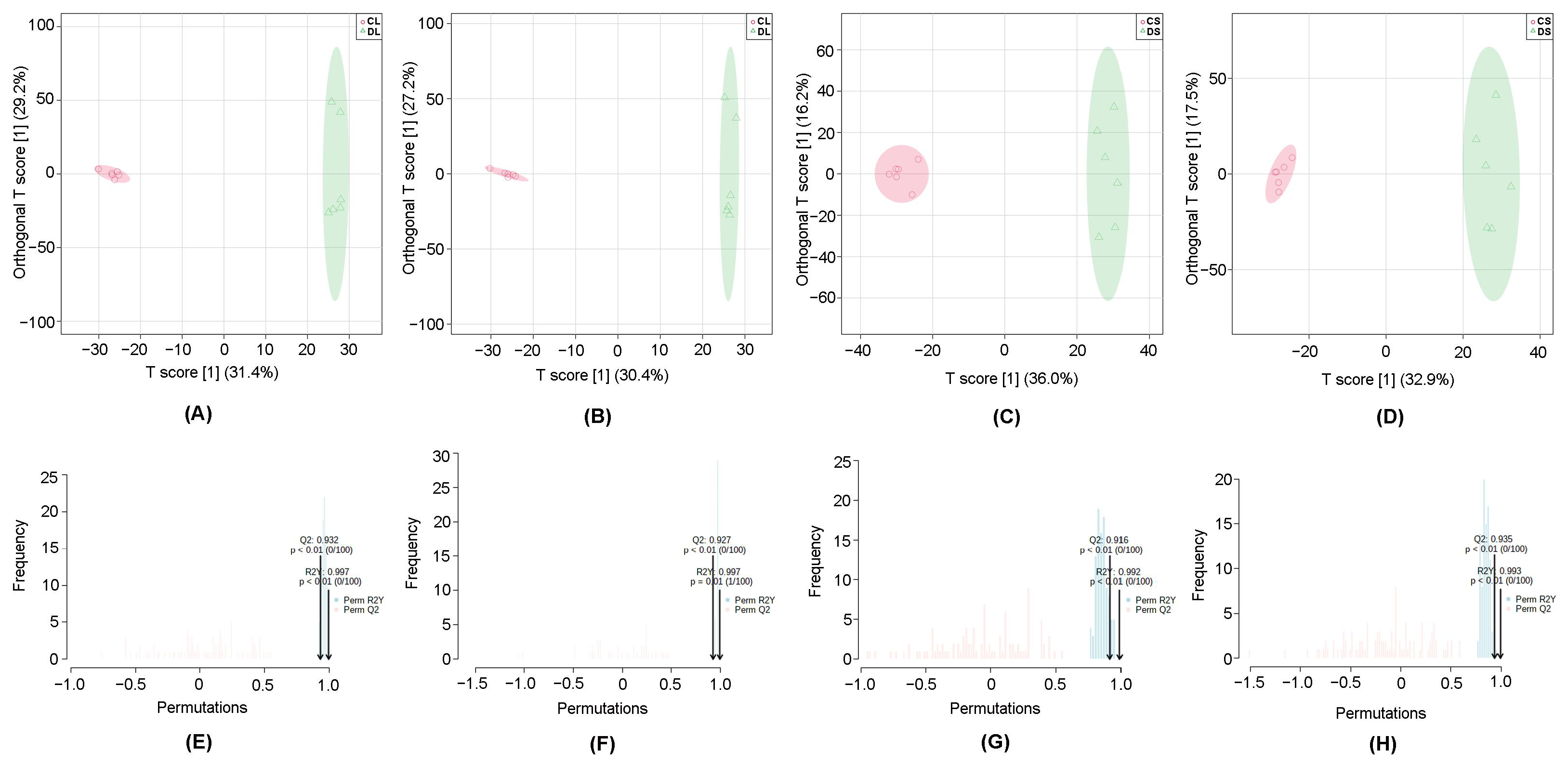
3.1. Multivariate Statistical Analysis
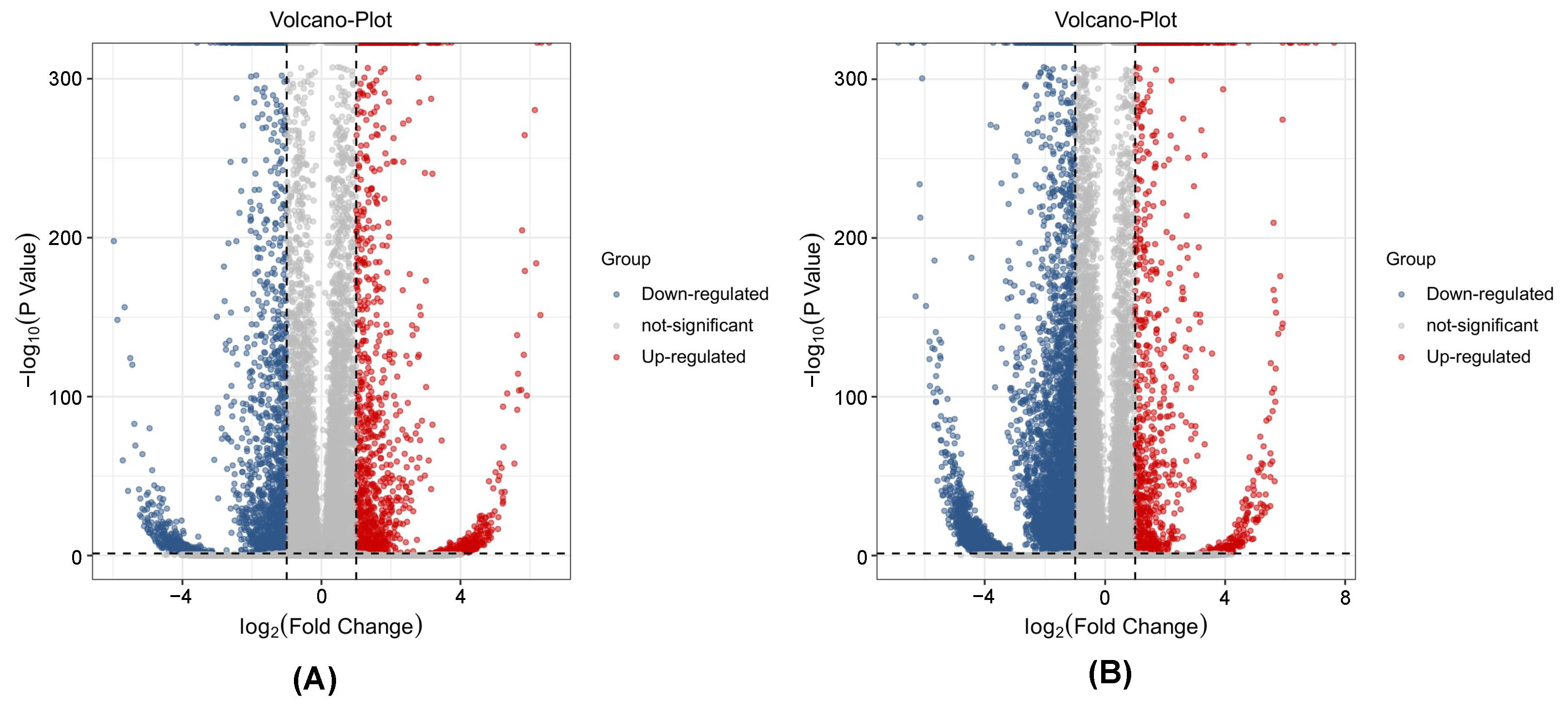
3.2. Significant Differences in Metabolites
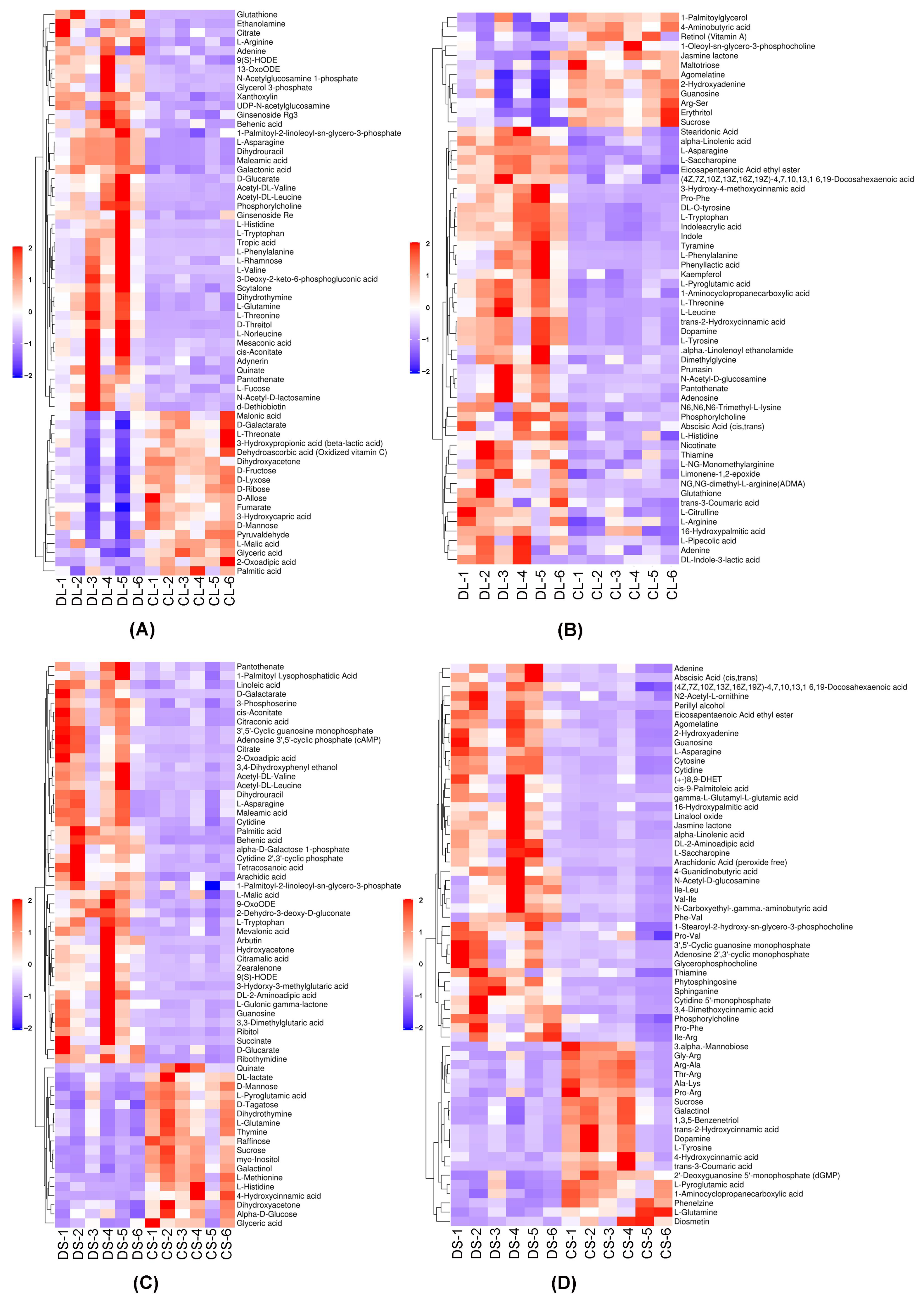
3.3. Heatmap of Differential Metabolites
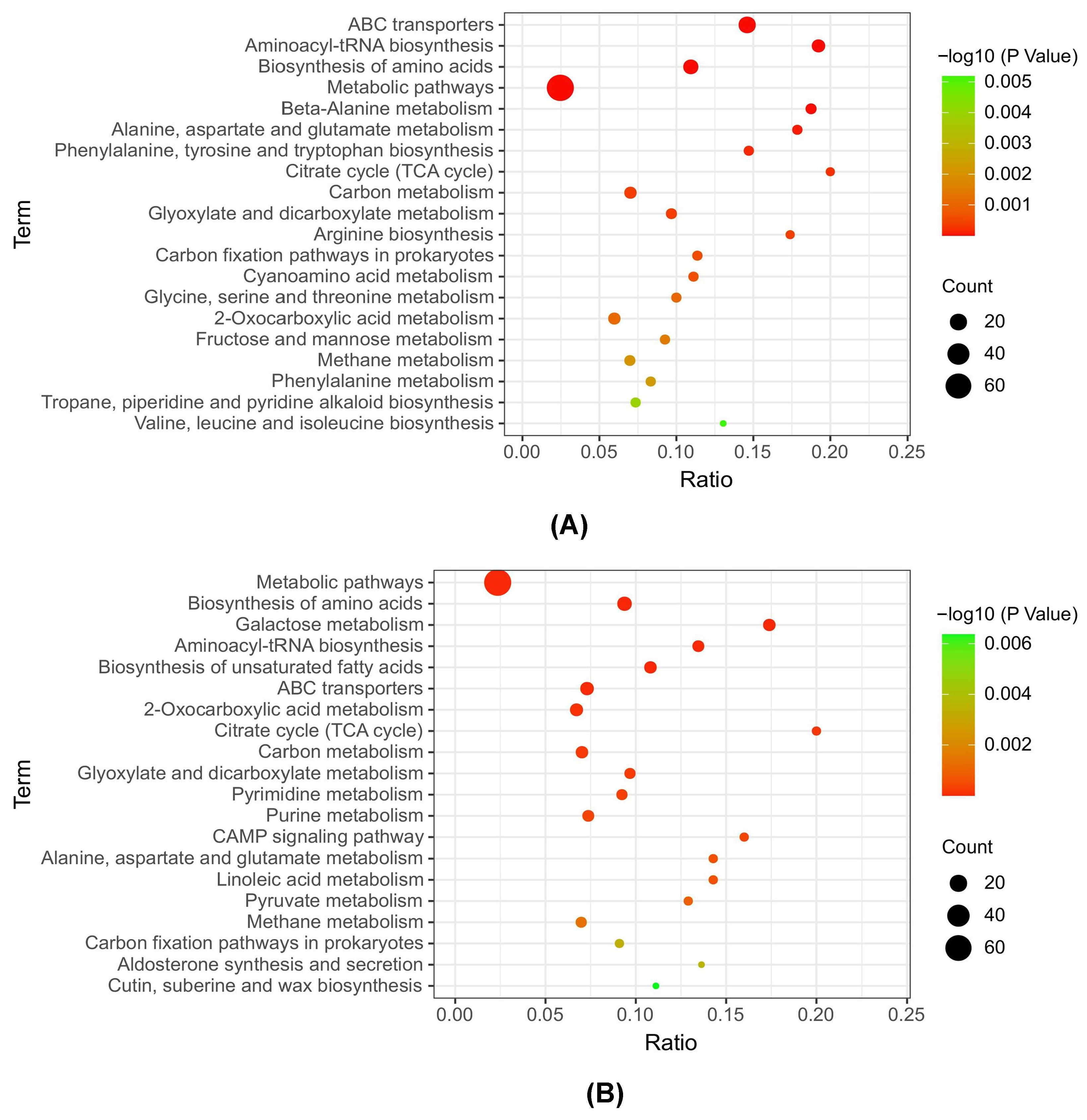
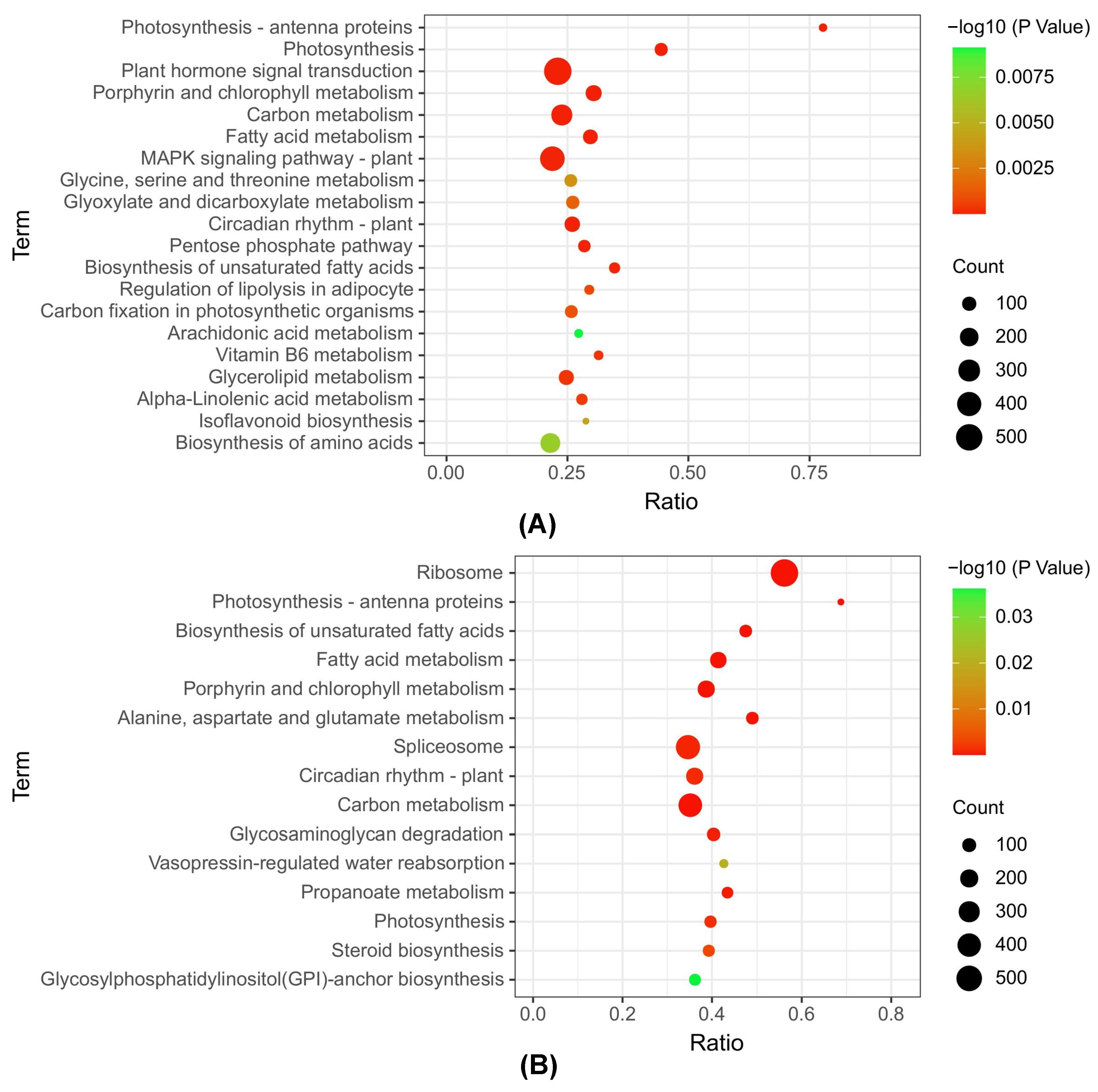
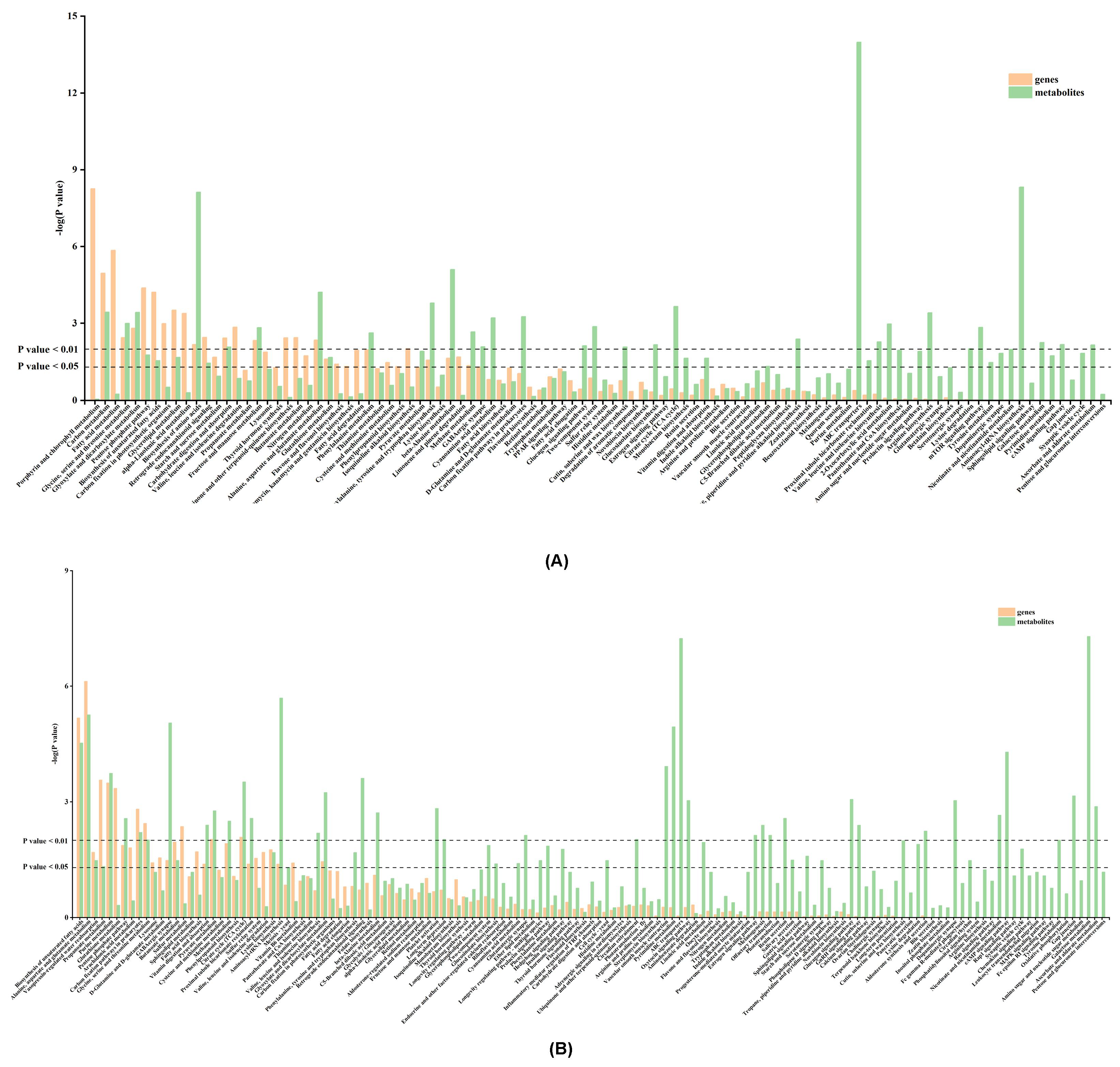
3.4. KEGG Metabolic Pathway Analysis
3.5. Quantitative Analysis of Plant Hormones
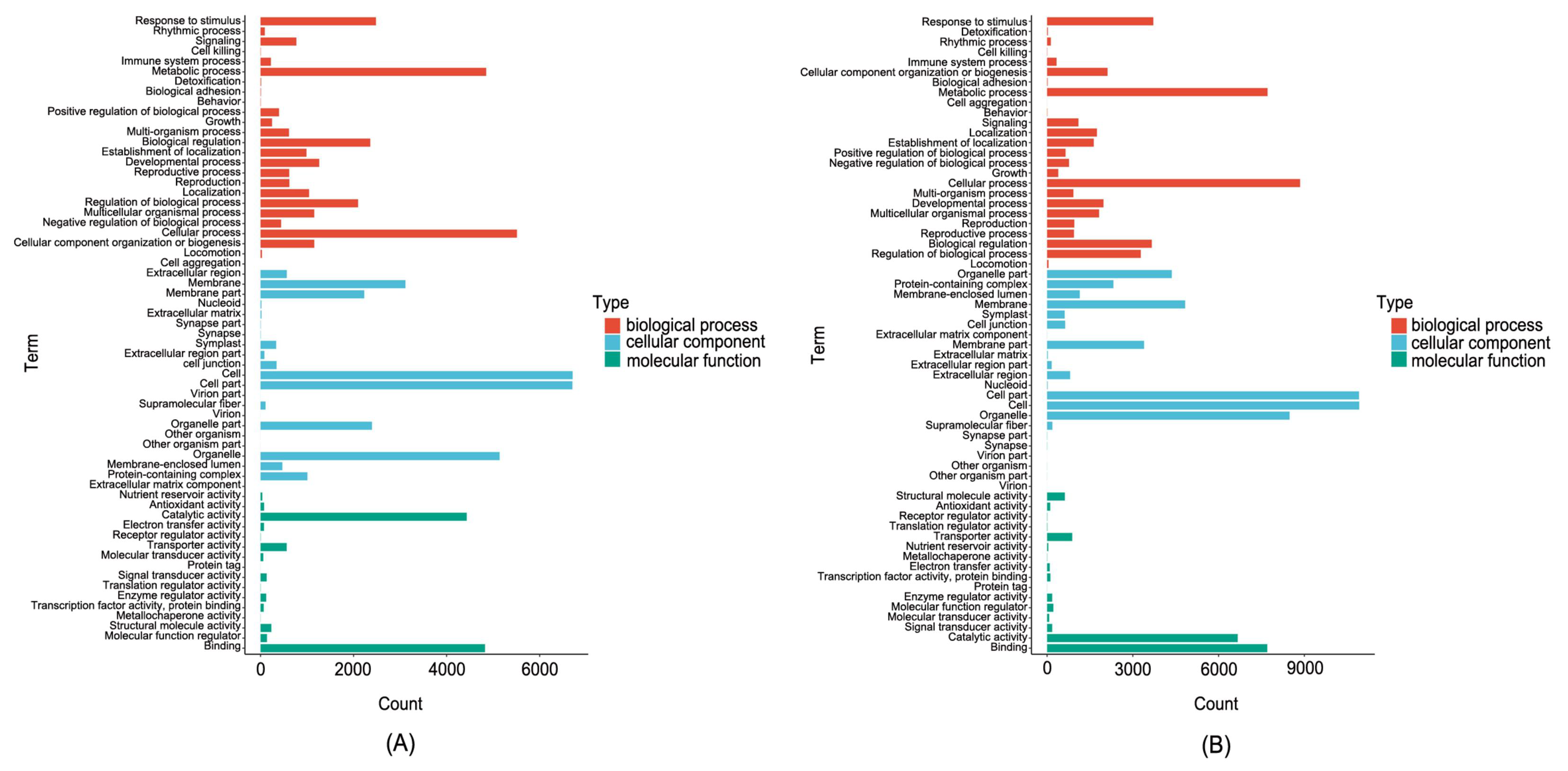
3.6. DEGs Analysis in Ginseng Related to P. cactorum
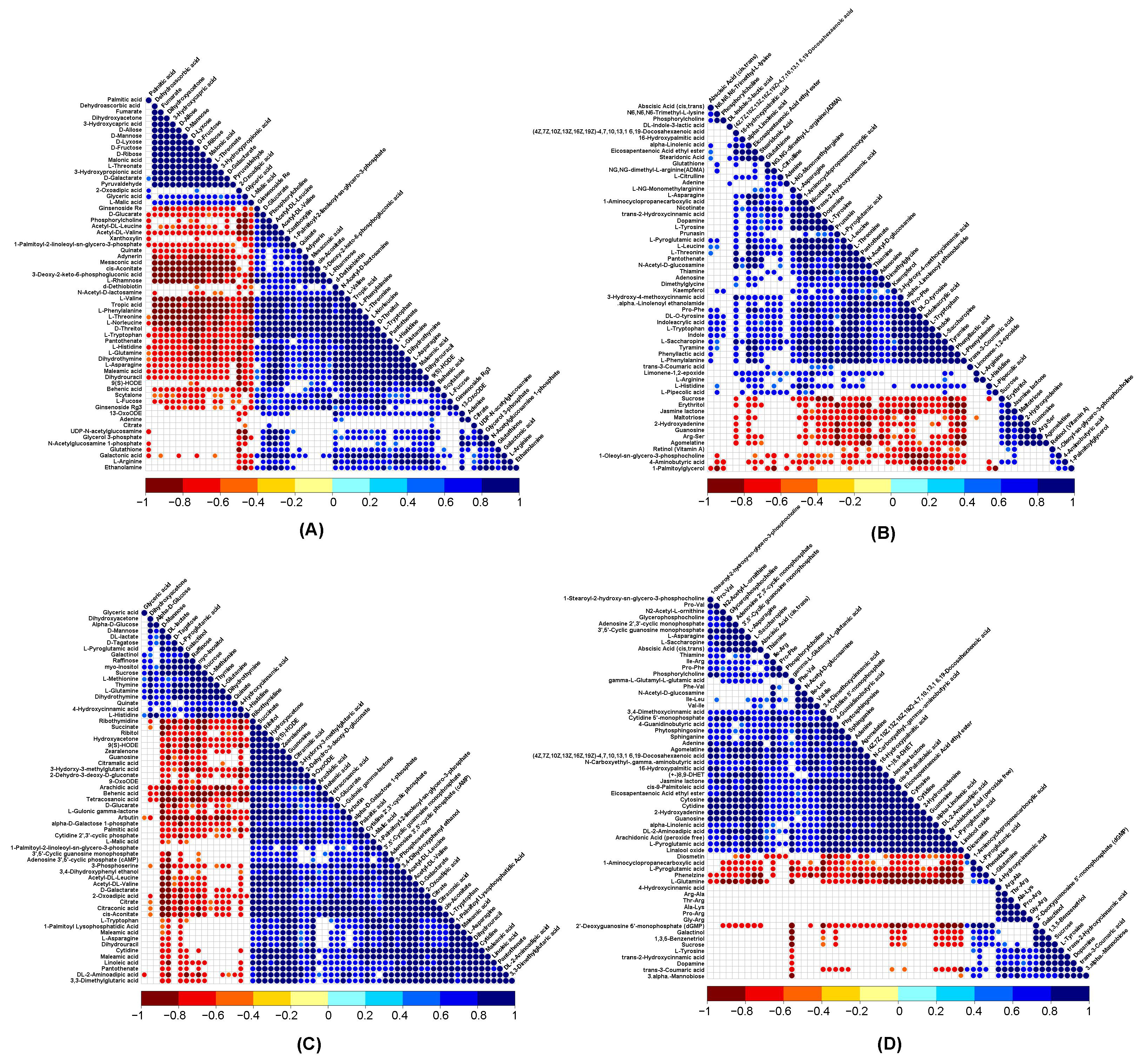
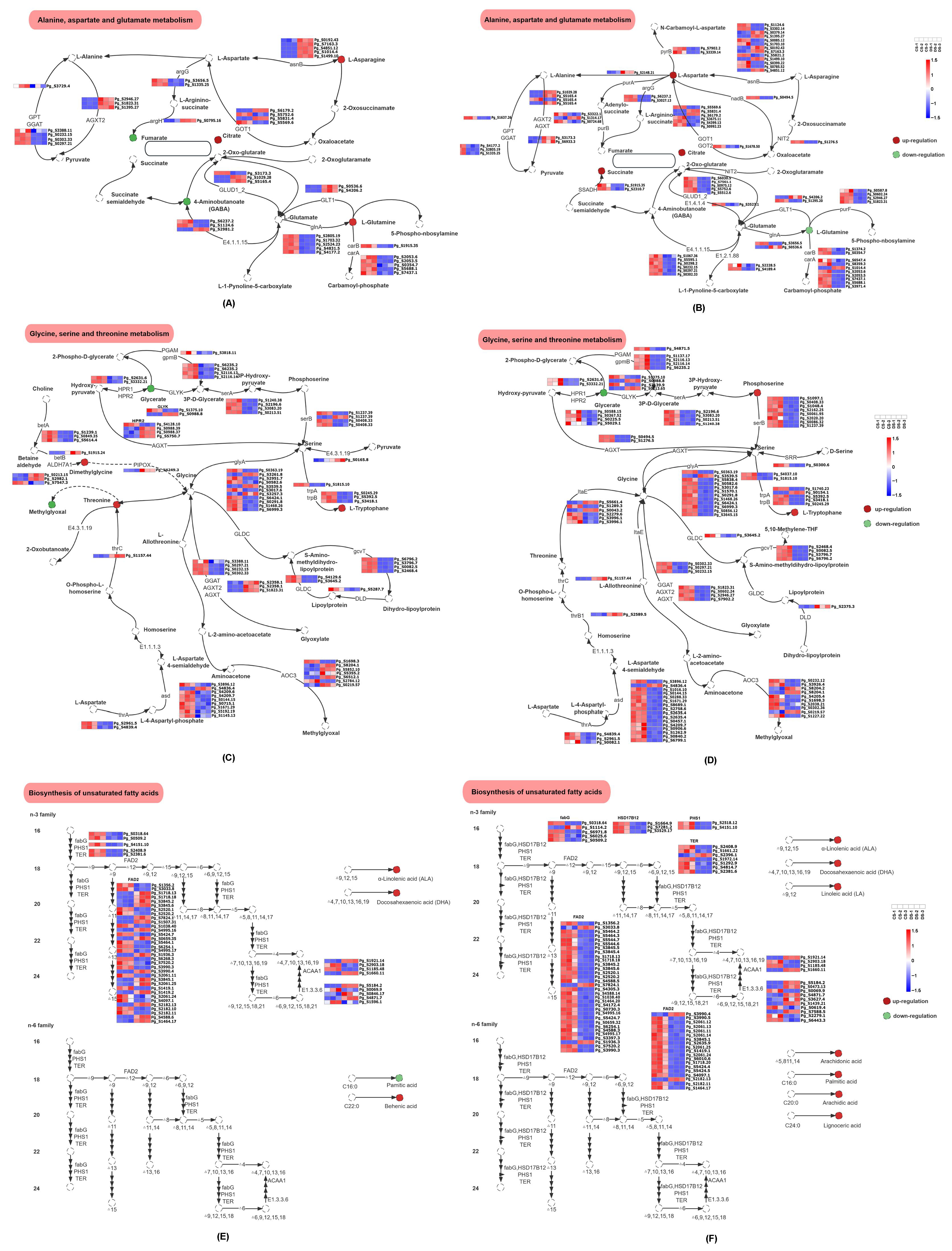
3.7. Correlation Analysis of DRMs and DEGs
3.8. Real-Time Quantitative Polymerase Chain Reaction (qRT-PCR) Validation Results
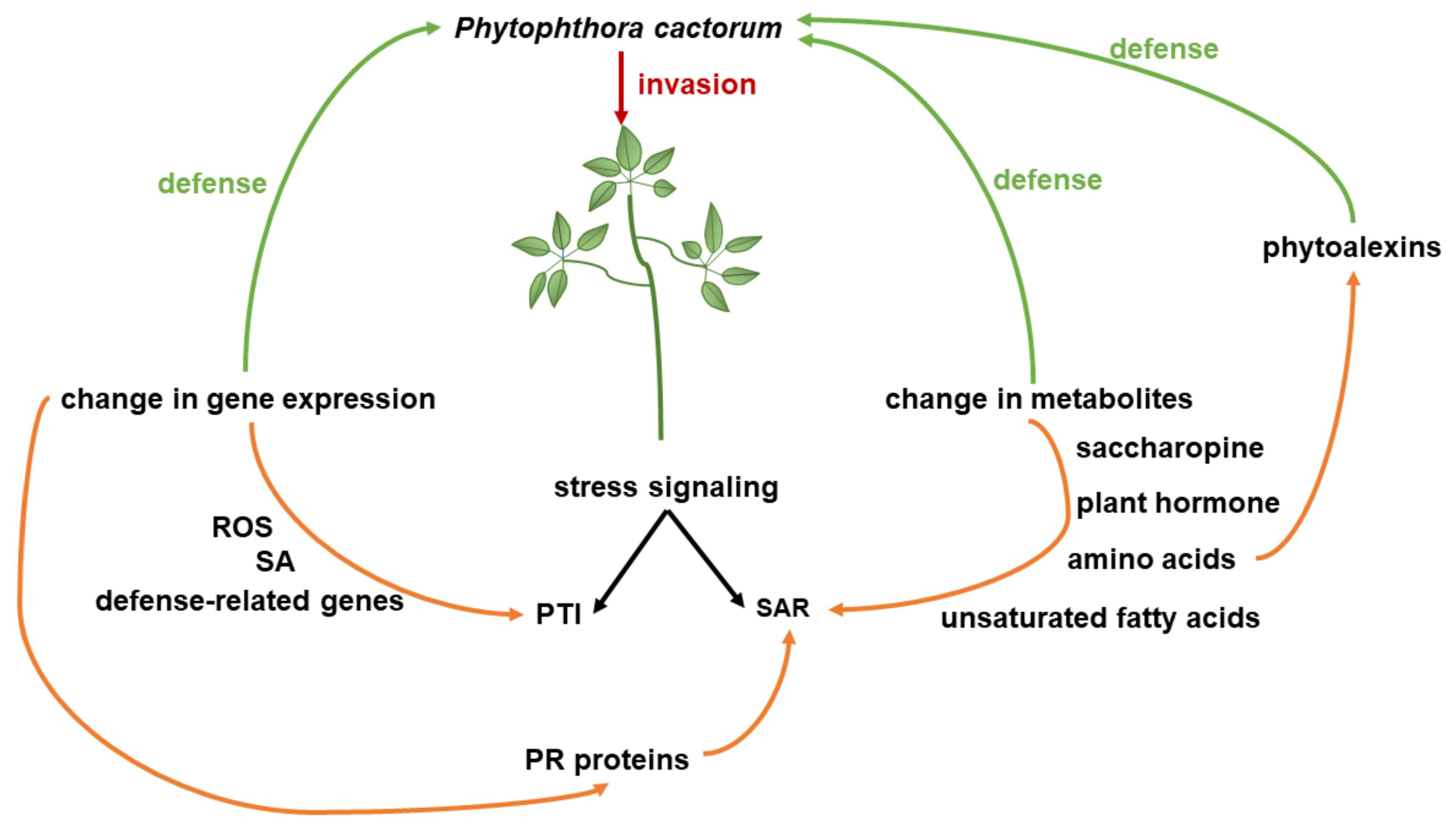
4. Discussion
4.1. Alteration in Biosynthesis of Unsaturated Fatty Acids
4.2. Alteration in Amino Acids Metabolism Pathway
4.3. Alteration in Plant Hormone
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhai, F.G.; Liang, Q.C.; Wu, Y.Y.; Liu, J.Q.; Liu, J.W. Red Ginseng Polysaccharide Exhibits Anticancer Activity Through GPX4 Downregulation-induced Ferroptosis. Pharm. Biol. 2022, 60, 909–914. [Google Scholar] [CrossRef] [PubMed]
- Abd Eldaim, M.A.A.; Abd El Latif, A.S.; Hassan, A.; El-Borai, N.B. Ginseng attenuates Fipronil-induced Hepatorenal Toxicity Via Its Antioxidant, Anti-apoptotic, and Anti-inflammatory Activities in Rats. Environ. Sci. Pollut. Res. Int. 2020, 27, 45008–45017. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Lai, S.; Fu, D.; Liu, J.; Wang, C.A.O.; Feng, H.; Liu, J.A.-O.X.; Li, Z.; Li, P. Neuroprotective Effects and Metabolomics Study of Protopanaxatriol (PPT) on Cerebral Ischemia/Reperfusion Injury In Vitro and In Vivo. Int. J. Mol. Sci. 2023, 24, 1789. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Fan, M.; Yin, S.; Xu, X.; Fu, B.; Jiang, R.; Sun, L. Ginseng Oligosaccharides (GSO) Inhibit C48/80-stimulated Pseudoallergic Mechanisms Through the PLC/Ca2+/PKC/MAPK/c-Fos Signaling Pathway. J. Funct. Foods 2022, 96, 105211. [Google Scholar] [CrossRef]
- Liu, Z.; Bian, X.; Gao, W.; Su, J.; Ma, C.; Xiao, X.; Yu, T.; Zhang, H.; Liu, X.; Fan, G. Rg3 Promotes the SUMOylation of SERCA2a and Corrects Cardiac Dysfunction in Heart Failure. Pharmacol. Res. 2021, 172, 105843. [Google Scholar] [CrossRef]
- Han, Y.; Su, Y.; Han, M.; Liu, Y.; Shi, Q.; Li, X.; Wang, P.; Li, W.; Li, W. Ginsenoside Rg1 Attenuates Glomerular Fibrosis by Inhibiting CD36/TRPC6/NFAT2 Signaling in Type 2 Diabetes Mellitus Mice. J. Ethnopharmacol. 2023, 302, 115923. [Google Scholar] [CrossRef]
- Ryu, H.; Park, H.; Suh, D.S.; Jung, G.H.; Park, K.; Lee, B.D. Biological Control of Colletotrichum Panacicola on Panax Ginseng by Bacillus Subtilis HK-CSM-1. J. Ginseng Res. 2014, 38, 215–219. [Google Scholar] [CrossRef] [Green Version]
- Mhlongo, M.I.; Piater, L.A.O.; Steenkamp, P.A.O.; Labuschagne, N.; Dubery, I.A.O. Concurrent Metabolic Profiling and Quantification of Aromatic Amino Acids and Phytohormones in Solanum Lycopersicum Plants Responding to Phytophthora Capsici. Metabolites 2020, 10, 466. [Google Scholar] [CrossRef]
- Wang, C. Studies on Safety Application of Fungicides for Main Diseases of Ginseng in Jilin Province. Master’s Thesis, Jilin Agricultural University, Changchun, China, 2011. [Google Scholar]
- Wang, Y.Y. Studies on the Key Techniques for Safe and Efficient Pesticides Control to Main Diseases on Ginseng. Master’s Thesis, Jilin Agricultural University, Changchun, China, 2013. [Google Scholar]
- Arora, R.K.; Sharma, S.; Singh, B.P. Late Blight Disease of Potato and Its Management. Potato J. 2014, 41, 16–40. [Google Scholar]
- Lee, B.D.; Park, H. Control of Phytophthora Blight of Panax Ginseng Caused by Phytophthora Cactorum Using Phosphonate Under the Controlled Condition. J. Ginseng Res. 2009, 33, 311–315. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.D.; Dutta, S.; Ryu, H.; Yoo, S.J.; Suh, D.S.; Park, K. Induction of Systemic Resistance in Panax Ginseng Against Phytophthora Cactorum by Native Bacillus Amyloliquefaciens HK34. J. Ginseng Res. 2015, 39, 213–220. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.-J.; Joo, S.C.; Shi, J.; Hu, C.; Quan, S.; Hu, J.; Sukweenadhi, J.; Mohanan, P.; Yang, D.-C.; Zhang, D. Metabolic Dynamics and Physiological Adaptation of Panax Ginseng During Development. Plant Cell Rep. 2018, 37, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Jeong, J.; Alves, A.C.; Han, S.T.; In, G.; Kim, E.H.; Jeong, W.S.; Hong, Y.S. Metabolomic Understanding of Intrinsic Physiology in Panax Ginseng During Whole Growing Seasons. J. Ginseng Res. 2019, 43, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Yoon, D.; Shin, W.C.; Oh, S.M.; Choi, B.R.; Young Lee, D. Integration of Multiplatform Metabolomics and Multivariate Analysis for Geographical Origin Discrimination of Panax Ginseng. Food Res. Int. 2022, 159, 111610. [Google Scholar] [CrossRef] [PubMed]
- Attard, A.; Evangelisti, E.; Kebdani-Minet, N.; Panabières, F.; Deleury, E.; Maggio, C.; Ponchet, M.; Gourgues, M. Transcriptome Dynamics of Arabidopsis Thaliana Root Penetration by the Oomycete Pathogen Phytophthora Parasitica. BMC Genom. 2014, 15, 538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khare, S.; Singh, N.B.; Singh, A.; Hussain, I.; Niharika, K.; Yadav, V.; Bano, C.; Yadav, R.K.; Amist, N. Plant Secondary Metabolites Synthesis and Their Regulations Under Biotic and Abiotic Constraints. J. Plant Biol. 2020, 63, 203–216. [Google Scholar] [CrossRef]
- Saddhe, A.A.O.; Manuka, R.; Penna, S.A.O. Plant sugars: Homeostasis and Transport Under Abiotic Stress in Plants. Physiol. Plant 2021, 171, 739–755. [Google Scholar] [CrossRef]
- Liu, S.; Wang, X.; Zhang, R.; Song, M.; Zhang, N.; Li, W.; Wang, Y.; Xu, Y.; Zhang, L. Amino Acid, Fatty Acid, and Carbohydrate Metabolomic Profiles With Ginsenoside-induced Insecticidal Efficacy Against Ostrinia Furnacalis (Guenee). J. Ginseng Res. 2020, 44, 544–551. [Google Scholar] [CrossRef]
- Castro-Moretti, F.R.; Gentzel, I.N.; Mackey, D.; Alonso, A.P. Metabolomics as an Emerging Tool for the Study of Plant-Pathogen Interactions. Metabolites 2020, 10, 52. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Guo, R.; Li, M.; Liu, Y.; Wang, X.; Fu, H.; Wang, S.; Liu, X.; Shi, L. Physiological Characteristics and Metabolomics Reveal the Tolerance Mechanism to Low Nitrogen in Glycine Soja Leaves. Physiol. Plant 2020, 168, 819–834. [Google Scholar] [CrossRef]
- Peng, Z.; Wang, Y.; Zuo, W.T.; Gao, Y.R.; Li, R.Z.; Yu, C.X.; Liu, Z.Y.; Zheng, Y.; Shen, Y.Y.; Duan, L.S. Integration of Metabolome and Transcriptome Studies Reveals Flavonoids, Abscisic Acid, and Nitric Oxide Comodulating the Freezing Tolerance in Liriope Spicata. Front. Plant Sci. 2022, 27, 764625. [Google Scholar] [CrossRef] [PubMed]
- Kusano, M.A.O.; Fukushima, A.A.O.; Tabuchi-Kobayashi, M.; Funayama, K.; Kojima, S.A.O.; Maruyama, K.; Yamamoto, Y.A.O.; Nishizawa, T.; Kobayashi, M.; Wakazaki, M.; et al. Cytosolic GLUTAMINE SYNTHETASE1;1 Modulates Metabolism and Chloroplast Development in Roots. Plant Physiol. 2020, 182, 1894–1909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, K.; Cho, K.S.; Sohn, H.B.; Ha, I.J.; Hong, S.Y.; Lee, H.; Kim, Y.M.; Nam, M.H. Network Analysis of the Metabolome and Transcriptome Reveals Novel Regulation of Potato Pigmentation. J. Exp. Bot. 2016, 67, 1519–1533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, X.A.O.; Luo, T.A.O.; Han, D.; Wu, Z.A.O. Analysis of Metabolomics Associated With Quality Differences Between Room-temperature- and low-temperature-stored Litchi Pulps. Food Sci. Nutr. 2019, 7, 3560–3569. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Yu, W.; Li, J.; Liu, M.; Sang, Y.; Wang, X. Characterization and Discrimination of Chilli Peppers Based on Multi-element and Non-targeted Metabolomics Analysis. LWT 2020, 131, 109742. [Google Scholar] [CrossRef]
- Shao, Y.; Zhou, H.-Z.; Wu, Y.; Zhang, H.; Lin, J.; Jiang, X.; He, Q.; Zhu, J.; Li, Y.; Yu, H.; et al. OsSPL3, an SBP-Domain Protein, Regulates Crown Root Development in Rice. Plant Cell 2019, 31, 1257–1275. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.A.O.; Zhan, Y.; Xu, Y.; Zhang, L.; Di, P.; Lu, B.A.O.; Chen, C. Deciphering the Transcriptomic Response of Ilyonectria Robusta in Relation to Ginsenoside Rg1 Treatment and the Development of Ginseng Rusty Root Rot. FEMS Microbiol. Lett. 2022, 369, fnac075. [Google Scholar] [CrossRef]
- Bian, X.; Zhao, Y.; Xiao, S.; Yang, H.; Han, Y.; Zhang, L. Metabolome and Transcriptome Analysis Reveals the Molecular Profiles Underlying the Ginseng Response to Rusty Root Symptoms. BMC Plant Biol. 2021, 21, 215. [Google Scholar] [CrossRef]
- Che, X.; Zhao, R.; Xu, H.; Liu, X.; Zhao, S.; Ma, H. Differently Expressed Genes (DEGs) Relevant to Type 2 Diabetes Mellitus Identification and Pathway Analysis Via Integrated Bioinformatics Analysis. Med. Sci. Monit. 2019, 25, 9237–9244. [Google Scholar] [CrossRef] [PubMed]
- Farh, M.E.; Kim, Y.J.; Abbai, R.; Singh, P.; Jung, K.H.; Kim, Y.J.; Yang, D.C. Pathogenesis Strategies and Regulation of Ginsenosides by Two Species of Ilyonectria in Panax Ginseng: Power of Speciation. J. Ginseng Res. 2020, 44, 332–340. [Google Scholar] [CrossRef]
- Wan, L.; Li, B.; Lei, Y.; Yan, L.; Ren, X.; Chen, Y.; Dai, X.; Jiang, H.; Zhang, J.; Guo, W.; et al. Mutant Transcriptome Sequencing Provides Insights Into Pod Development in Peanut (Arachis hypogaea L.). Front. Plant Sci. 2017, 8, 1900. [Google Scholar] [CrossRef] [Green Version]
- Chen, K.; Liu, J.; Ji, R.; Chen, T.; Zhou, X.; Yang, J.; Tong, Y.; Jiang, C.; Zhou, J.; Zhao, Y.; et al. Biogenic Synthesis and Spatial Distribution of Endogenous Phytohormones and Ginsenosides Provide Insights on Their Intrinsic Relevance in Panax Ginseng. Front. Plant Sci. 2019, 9, 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metab. 2013, 1, 92–107. [Google Scholar] [CrossRef]
- Zhao, G.; Zhao, W.; Han, L.; Ding, J.; Chang, Y. Metabolomics Analysis of Sea Cucumber (Apostichopus Japonicus) in Different Geographical Origins Using UPLC-Q-TOF/MS. Food Chem. 2020, 333, 127453. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Hu, K.; Qu, F.; Ni, D.; Zhang, H.; Liu, S.; Chen, Y. Metabolomics Analysis Reveals Major Differential Metabolites and Metabolic Alterations in Tea Plant Leaves (Camellia sinensis L.) Under Different Fluorine Conditions. J. Plant Growth Regul. 2021, 40, 798–810. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopaedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Mishra, J.; Srivastava, R.A.O.; Trivedi, P.A.-O.; Verma, P.A.O. Effect of Virus Infection on the Secondary Metabolite Production and Phytohormone Biosynthesis in Plants. 3 Biotech 2020, 10, 547. [Google Scholar] [CrossRef]
- Alazem, M.; Lin, N.S. Antiviral Roles of Abscisic Acid in Plants. Front. Plant Sci. 2017, 8, 1760–1769. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.G.; Zhu, T.; Peng, X.J.; Xi, D.H.; Guo, H.; Yin, Y.; Zhang, D.W.; Lin, H.-H. Role of Brassinosteroid Signaling in Modulating Tobacco mosaic virus Resistance in Nicotiana benthamiana. Sci. Rep. 2016, 6, 20579. [Google Scholar] [CrossRef] [Green Version]
- Dehkordi, A.N.; Rubio, M.; Babaeian, N.; Albacete, A.; Martínez-Gómez, P. Phytohormone Signaling of the Resistance to Plum Pox Virus (PPV, Sharka Disease) Induced by Almond (Prunus dulcis (Miller) Webb) Grafting to Peach (P. persica L. Batsch). Viruses 2018, 10, 238. [Google Scholar] [CrossRef] [Green Version]
- De Haro, L.A.; Arellano, S.M.; Novák, O.; Feil, R.; Dumón, A.D.; Mattio, M.F.; Tarkowská, D.; Llauger, G.; Strnad, M.; Lunn, J.E.; et al. Mal de Río Cuarto Virus Infection Causes Hormone Imbalance and Sugar Accumulation in Wheat Leaves. BMC Plant Biol. 2019, 19, 112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, P.D. The Gene Ontology and the Meaning of Biological Function. In The Gene Ontology Handbook; Springer: Berlin/Heidelberg, Germany, 2017; Volume 1446. [Google Scholar] [CrossRef] [Green Version]
- Volynchikova, E.A.O.X.; Kim, K.A.O. Biological Control of Oomycete Soilborne Diseases Caused by Phytophthora capsici, Phytophthora infestans, and Phytophthora nicotianae in Solanaceous Crops. Mycobiology 2022, 50, 269–293. [Google Scholar] [CrossRef] [PubMed]
- Farh, M.E.-A.; Kim, Y.-J.; Kim, Y.-J.; Yang, D.-C. Mini Review: Cylindrocarpon Destructans/Ilyonectria Radicicola-Species Complex: Causa-tive Agent of Ginseng Root-rot Disease and Rusty Symptoms. J. Ginseng Res. 2018, 42, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Rojas, C.; Senthil-Kumar, M.; Tzin, V.; Mysore, K. Regulation of Primary Plant Metabolism During Plant-pathogen Interactions and its Contribution to Plant Defense. Front. Plant Sci. 2014, 5, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Li, S.P.; Cui, H.T.; Zou, S.H.; Wang, W. Molecular Genetic Mechanisms of Interaction Between Host Plants and Pathogens. Yi Chuan 2020, 42, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Ninkuu, V.; Yan, J.; Zhang, L.; Fu, Z.; Yang, T.; Li, S.; Li, B.; Duan, J.; Ren, J.; Li, G.; et al. Hrip1 Mediates Rice Cell Wall Fortification and Phytoalexins Elicitation to Confer Immunity Against Magnaporthe oryzae. Front. Plant Sci. 2022, 13, 3180. [Google Scholar] [CrossRef]
- Li, B.; Meng, X.; Shan, L.; He, P. Transcriptional Regulation of Pattern-Triggered Immunity in Plants. Cell Host Microbe 2016, 19, 641–650. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Monsalvo, I.; Ly, M.; Jahan, M.A.; Wi, D.; Martirosyan, I.; Kovinich, N.A.O. RNA-Seq Dissects Incomplete Activation of Phytoalexin Biosynthesis by the Soybean Transcription Factors GmMYB29A2 and GmNAC42-1. Plants 2023, 12, 545. [Google Scholar] [CrossRef]
- Pagán, I.A.O.; García-Arenal, F.A.O. Cucumber Mosaic Virus-Induced Systemic Necrosis in Arabidopsis Thaliana: Determinants and Role in Plant Defense. Viruses 2022, 14, 2790. [Google Scholar] [CrossRef]
- Li, J. Expression and Purification of Tobacco PR-1a Protein for Punction Analysis. Asian J. Chem. 2009, 21, 3697–3707. [Google Scholar]
- Shin, S.H.; Pak, J.H.; Kim, M.J.; Kim, H.J.; Oh, J.S.; Choi, H.K.; Jung, H.W.; Chung, Y.S. An Acidic PATHOGENESIS-RELATED1 Gene of Oryza grandiglumis is Involved in Disease Resistance Response Against Bacterial Infection. Plant Pathol. J. 2014, 30, 208–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging Insights Into the Functions of Pathogenesis-Related Protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY Transcription Factors in Plant Responses to Stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.B.; Luan, Y.S.; Liu, Z. Overexpression of SpWRKY1 Promotes Resistance to Phytophthora Nicotianae and Tolerance to Salt and Drought Stress in Transgenic Tobacco. Physiol. Plant 2015, 155, 248–266. [Google Scholar] [CrossRef]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP Kinase Signalling Cascade in Arabidopsis Innate Immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef]
- Wang, Z.; Wei, X.; Wang, Y.; Sun, M.; Zhao, P.; Wang, Q.; Yang, B.; Li, J.; Jiang, Y.Q. WRKY29 Transcription Factor Regulates Ethylene Biosynthesis and Response in Arabidopsis. Plant Physiol. Biochem. 2023, 194, 134–145. [Google Scholar] [CrossRef]
- Gao, J.; Bi, W.; Li, H.; Wu, J.; Yu, X.; Liu, D.; Wang, X. WRKY Transcription Factors Associated With NPR1-Mediated Acquired Resistance in Barley Are Potential Resources to Improve Wheat Resistance to Puccinia triticina. Front. Plant. Sci. 2018, 9, 1486. [Google Scholar] [CrossRef]
- De Carvalho, C.; Caramujo, M.J. The Various Roles of Fatty Acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef] [Green Version]
- Mu, Q.; Zhang, M.; Li, Y.; Feng, F.; Yu, X.; Nie, J.A.O.X. Metabolomic Analysis Reveals the Effect of Insecticide Chlorpyrifos on Rice Plant Metabolism. Metabolites 2022, 12, 1289. [Google Scholar] [CrossRef]
- Per, T.S.; Khan, M.I.R.; Anjum, N.A.; Masood, A.; Hussain, S.J.; Khan, N.A. Jasmonates in Plants Under Abiotic Stresses: Crosstalk With Other Phytohormones Matters. Environ. Exp. Bot. 2018, 145, 104–120. [Google Scholar] [CrossRef]
- Zeier, J. New Insights Into the Regulation of Plant Immunity by Amino Acid Metabolic Pathways. Plant Cell Environ. 2013, 36, 2085–2103. [Google Scholar] [CrossRef] [PubMed]
- Nehela, Y.; Killiny, N.A.O. The Unknown Soldier in Citrus Plants: Polyamines-based Defensive Mechanisms Against Biotic and Abiotic Stresses and Their Relationship With Other Stress-associated Metabolites. Plant. Signal. Behav. 2020, 15, 1761080. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Alam, M.U.; Hossain, M.S.; Mahmud, J.A.; Nahar, K.; Fujita, M.; Hasanuzzaman, M. Exogenous Gallic Acid Confers Salt Tolerance in Rice Seedlings: Modulation of Ion Homeostasis, Osmoregulation, Antioxidant Defense, and Methylglyoxal Detoxification Systems. Agronomy 2023, 13, 16. [Google Scholar] [CrossRef]
- Jinal, H.N.; Sakthivel, K.; Amaresan, N. Characterisation of Antagonistic Bacillus Paralicheniformis (strain EAL) by LC-MS, Antimicrobial Peptide Genes, and ISR Determinants. Antonie Van Leeuwenhoek 2020, 113, 1167–1177. [Google Scholar] [CrossRef]
- Eccleston, L.; Brambilla, A.; Vlot, A.A.O. New Molecules in Plant Defence Against Pathogens. Essays Biochem. 2022, 66, 683–693. [Google Scholar] [CrossRef]
- Arruda, P.; Barreto, P. Lysine Catabolism Through the Saccharopine Pathway: Enzymes and Intermediates Involved in Plant Responses to Abiotic and Biotic Stress. Front. Plant Sci. 2020, 11, 587. [Google Scholar] [CrossRef]
- Hartmann, M.; Zeier, T.; Bernsdorff, F.; Reichel-Deland, V.; Kim, D.; Hohmann, M.; Scholten, N.; Schuck, S.; Bräutigam, A.; Hölzel, T.; et al. Flavin Monooxygenase- Generated N-Hydroxypipecolic Acid Is a Critical Element of Plant Systemic Immunity. Cell 2018, 173, 456–469. [Google Scholar] [CrossRef] [Green Version]
- Oliva, M.; Guy, A.; Galili, G.; Dor, E.; Schweitzer, R.; Amir, R.; Hacham, Y. Enhanced Production of Aromatic Amino Acids in Tobacco Plants Leads to Increased Phenylpropanoid Metabolites and Tolerance to Stresses. Front. Plant Sci. 2021, 11, 2020. [Google Scholar] [CrossRef]
- Schenck, C.A.; Maeda, H.A. Tyrosine Biosynthesis, Metabolism, and Catabolism in Plants. Phytochemistry 2018, 149, 82–102. [Google Scholar] [CrossRef]
- Jiang, N.; Doseff, A.I.; Grotewold, E. Flavones: From Biosynthesis to Health Benefits. Plants 2016, 5, 27. [Google Scholar] [CrossRef] [Green Version]
- Dong, N.Q.; Lin, H.X. Contribution of Phenylpropanoid Metabolism to Plant Development and Plant-environment Interactions. J. Integr. Plant Biol. 2021, 63, 180–209. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant Hormone-mediated Regulation of Stress Responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Q.; Tan, W.; Zhao, Y.; Yang, F.; Yao, X.; Lin, H.; Zhang, D.A.-O. Salicylic Acid-activated BIN2 Phosphorylation of TGA3 Promotes Arabidopsis PR Gene Expression and Disease Resistance. EMBO J. 2022, 41, e110682. [Google Scholar] [CrossRef] [PubMed]
- Ton, J.; Flors, V.; Mauch-Mani, B. The Multifaceted Role of ABA in Disease Resistance. Trends Plant Sci. 2009, 14, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Corot, A.; Roman, H.; Douillet, O.; Autret, H.; Perez-Garcia, M.D.; Citerne, S.; Bertheloot, J.; Sakr, S.; Leduc, N.; Demotes-Mainard, S. Cytokinins and Abscisic Acid Act Antagonistically in the Regulation of the Bud Outgrowth Pattern by Light Intensity. Front. Plant Sci. 2017, 8, 1724. [Google Scholar] [CrossRef]
- Davies, W.J.; Kudoyarova, G.; Hartung, W. Long-distance ABA Signaling and Its Relation to Other Signaling Pathways in the Detection of Soil Drying and the Mediation of the Plant’s Response to Drought. J. Plant Growth Regul. 2005, 24, 285–295. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Jin, Y.; Liu, C.; Vonapartis, E.; Liang, J.; Wu, W.; Gazzarrini, S.; He, J.; Yi, M. GhNAC83 Inhibits Corm Dormancy Release by Regulating ABA Signaling and Cytokinin Biosynthesis in Gladiolus Hybridus. J. Exp. Bot. 2019, 70, 1221–1237. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Wang, Y.; Guo, B.; Jing, M.; Zhou, H.; Li, Y.; Wang, H.; Huang, J.; Wang, Y.; Ye, W.; et al. The Phytophthora Sojae RXLR Effector Avh238 Destabilizes Soybean Type2 GmACSs to Suppress Ethylene Biosynthesis and Promote Infection. New Phytol. 2019, 222, 425–437. [Google Scholar] [CrossRef]
- Shekhawat, K.; Fröhlich, K.; García-Ramírez, G.X.; Trapp, M.A.; Hirt, H. Ethylene: A Master Regulator of Plant-Microbe Interactions Under Abiotic Stresses. Cells 2023, 12, 31. [Google Scholar] [CrossRef]
- Li, F.; Duan, P.; Zhang, H.; Lu, X.; Shi, Z.; Cui, J. Genome-wide Identification of CmaGH3 Family Genes, and Expression Analysis in Response to Cold and Hormonal Stresses in Cucurbita Maxima. Sci. Hortic. 2022, 304, 111256. [Google Scholar] [CrossRef]

| Groups | Treatments | Culture Time After Treatment | Harvested Tissue |
|---|---|---|---|
| CL1 | sterile water | 10 days | leaves |
| DL2 | P. cactorum | 10 days | leave |
| CS3 | sterile water | 10 days | stems |
| DS4 | P. cactorum | 10 days | stems |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kan, H.; Qu, S.; Dong, K.; Wang, S.; Xu, C.; Wang, Y.; Hua, S. Integrated Metabolome and Transcriptome Analysis Unveils the Underlying Molecular Response of Panax ginseng Plants to the Phytophthora cactorum Infection. Agriculture 2023, 13, 509. https://doi.org/10.3390/agriculture13020509
Kan H, Qu S, Dong K, Wang S, Xu C, Wang Y, Hua S. Integrated Metabolome and Transcriptome Analysis Unveils the Underlying Molecular Response of Panax ginseng Plants to the Phytophthora cactorum Infection. Agriculture. 2023; 13(2):509. https://doi.org/10.3390/agriculture13020509
Chicago/Turabian StyleKan, Hong, Shuai Qu, Kai Dong, Shihan Wang, Chen Xu, Yingping Wang, and Shuang Hua. 2023. "Integrated Metabolome and Transcriptome Analysis Unveils the Underlying Molecular Response of Panax ginseng Plants to the Phytophthora cactorum Infection" Agriculture 13, no. 2: 509. https://doi.org/10.3390/agriculture13020509
APA StyleKan, H., Qu, S., Dong, K., Wang, S., Xu, C., Wang, Y., & Hua, S. (2023). Integrated Metabolome and Transcriptome Analysis Unveils the Underlying Molecular Response of Panax ginseng Plants to the Phytophthora cactorum Infection. Agriculture, 13(2), 509. https://doi.org/10.3390/agriculture13020509






