Insights into Cottonseed Cultivar Identification Using Raman Spectroscopy and Explainable Machine Learning
Abstract
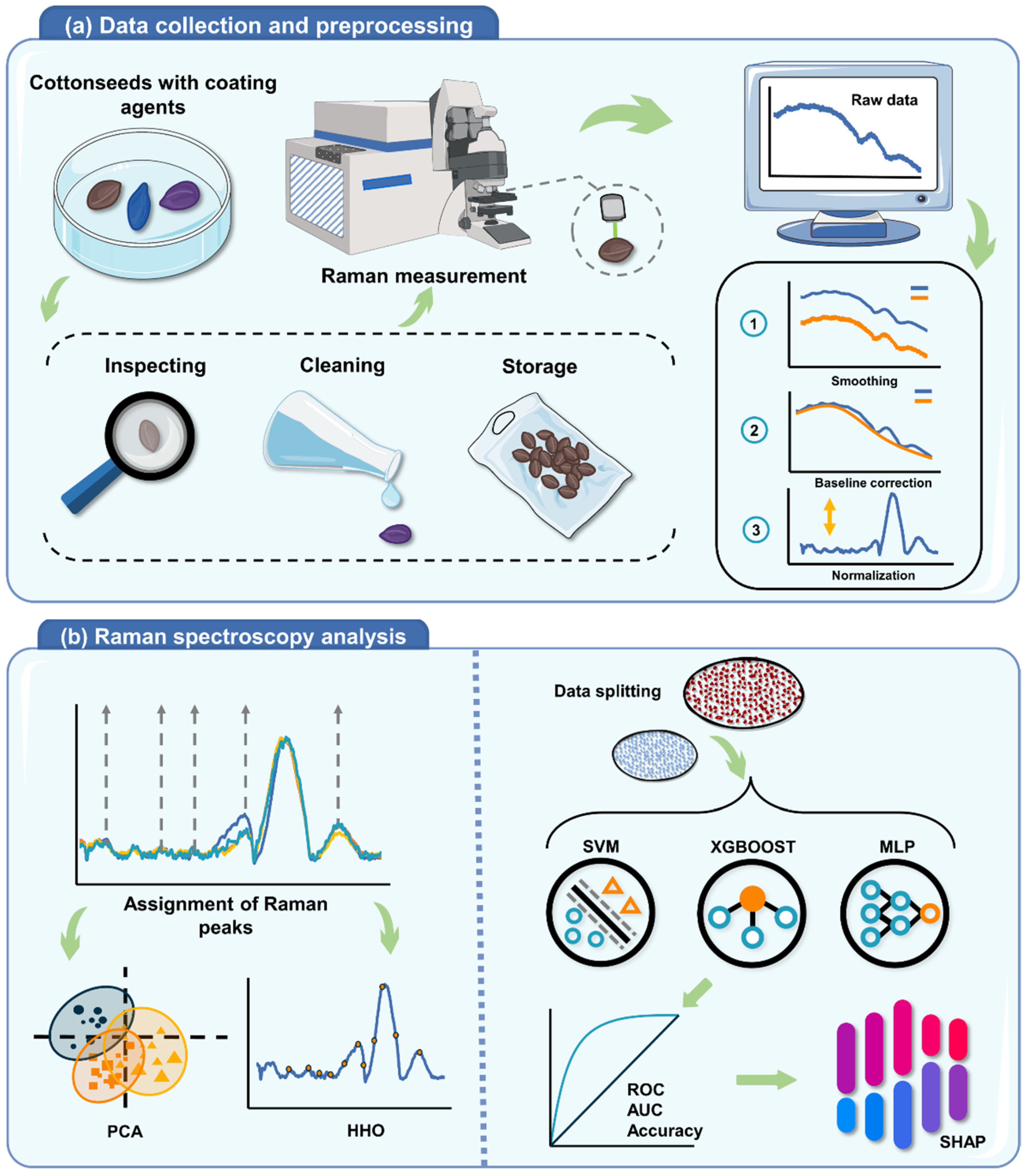
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Preparation
2.3. Raman Measurements of Cottonseed
2.4. Data Preprocessing
2.5. Exploratory Analysis of Features
2.6. Machine Learning Methods
2.7. Model Interpretation
3. Results and Discussion
3.1. Visual Characteristics Analysis of Cottonseed
3.2. Raman Analysis of Cottonseed
3.3. Exploratory Analysis of Raman Features
3.3.1. Exploratory Analysis of Clustering Trend
3.3.2. Exploratory Analysis of Critical Raman Wavenumbers
3.4. Model Analysis
3.5. Model Interpretation
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khan, M.A.; Wahid, A.; Ahmad, M.; Tahir, M.T.; Ahmed, M.; Ahmad, S.; Hasanuzzaman, M. World Cotton Production and Consumption: An Overview. In Cotton Production and Uses: Agronomy, Crop Protection, and Postharvest Technologies; Ahmad, S., Hasanuzzaman, M., Eds.; Springer: Singapore, 2020; pp. 1–7. ISBN 9789811514722. [Google Scholar]
- Feng, L.; Dai, J.; Tian, L.; Zhang, H.; Li, W.; Dong, H. Review of the Technology for High-Yielding and Efficient Cotton Cultivation in the Northwest Inland Cotton-Growing Region of China. Field Crop. Res. 2017, 208, 18–26. [Google Scholar] [CrossRef]
- Nations, U. World Cotton Day. Available online: https://www.un.org/en/observances/cotton-day (accessed on 18 February 2023).
- Yang, Z.; Gao, C.; Zhang, Y.; Yan, Q.; Hu, W.; Yang, L.; Wang, Z.; Li, F. Recent Progression and Future Perspectives in Cotton Genomic Breeding. J. Integr. Plant Biol. 2023, 65, 548–569. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, J.; Qi, Z.; Long, Y.; Pei, L.; Huang, X.; Grover, C.E.; Du, X.; Xia, C.; Wang, P.; et al. Genomic Innovation and Regulatory Rewiring during Evolution of the Cotton Genus Gossypium. Nat. Genet. 2022, 54, 1959–1971. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Sajid, M.; Iqbal, D.; Sarwar, M.I.; Farooq, A.; Siddique, A.; Khan, M.Q.; Kim, I.-S. Impact of Novel Varietal and Regional Differences on Cotton Fiber Quality Characteristics. Materials 2022, 15, 3242. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.Q.; Li, Z.L.; Li, J.J.; Lin, H. Raman Spectroscopic Techniques for Nondestructive Analysis of Agri-Foods: A State-of-the-Art Review. Trends Food Sci. Technol. 2021, 118, 490–504. [Google Scholar] [CrossRef]
- Zareef, M.; Arslan, M.; Hassan, M.M.; Ahmad, W.; Ali, S.; Li, H.H.; Qin, O.Y.; Wu, X.Y.; Hashim, M.M.; Chen, Q.S. Recent Advances in Assessing Qualitative and Quantitative Aspects of Cereals Using Nondestructive Techniques: A Review. Trends Food Sci. Technol. 2021, 116, 815–828. [Google Scholar] [CrossRef]
- Salisu, I.B.; Shahid, A.A.; Yaqoob, A.; Ali, Q.; Bajwa, K.S.; Rao, A.Q.; Husnain, T. Molecular Approaches for High Throughput Detection and Quantification of Genetically Modified Crops: A Review. Front. Plant Sci. 2017, 8, 1670. [Google Scholar] [CrossRef] [Green Version]
- Bahadoran, Z.; Mirmiran, P.; Jeddi, S.; Azizi, F.; Ghasemi, A.; Hadaegh, F. Nitrate and Nitrite Content of Vegetables, Fruits, Grains, Legumes, Dairy Products, Meats and Processed Meats. J. Food Compos. Anal. 2016, 51, 93–105. [Google Scholar] [CrossRef]
- Payne, W.Z.; Kurouski, D. Raman Spectroscopy Enables Phenotyping and Assessment of Nutrition Values of Plants: A Review. Plant Methods 2021, 17, 78. [Google Scholar] [CrossRef]
- Weng, S.; Zhu, W.; Zhang, X.; Yuan, H.; Zheng, L.; Zhao, J.; Huang, L.; Han, P. Recent Advances in Raman Technology with Applications in Agriculture, Food and Biosystems: A Review. Artif. Intell. Agric. 2019, 3, 1–10. [Google Scholar] [CrossRef]
- Jentzsch, P.V.; Ciobota, V.; Salinas, W.; Kampe, B.; Aponte, P.M.; Rosch, P.; Popp, J.; Ramos, L.A. Distinction of Ecuadorian Varieties of Fermented Cocoa Beans Using Raman Spectroscopy. Food Chem. 2016, 211, 274–280. [Google Scholar] [CrossRef]
- Oo, T.K.; Arunrat, N.; Sereenonchai, S.; Ussawarujikulchai, A.; Chareonwong, U.; Nutmagul, W. Comparing Four Machine Learning Algorithms for Land Cover Classification in Gold Mining: A Case Study of Kyaukpahto Gold Mine, Northern My-anmar. Sustainability 2022, 14, 10754. [Google Scholar] [CrossRef]
- Balducci, F.; Impedovo, D.; Pirlo, G. Machine Learning Applications on Agricultural Datasets for Smart Farm Enhancement. Machines 2018, 6, 38. [Google Scholar] [CrossRef] [Green Version]
- Lussier, F.; Thibault, V.; Charron, B.; Wallace, G.Q.; Masson, J.-F. Deep Learning and Artificial Intelligence Methods for Raman and Surface-Enhanced Raman Scattering. Trac-Trends Anal. Chem. 2020, 124, 115796. [Google Scholar] [CrossRef]
- Adadi, A.; Berrada, M. Peeking Inside the Black-Box: A Survey on Explainable Artificial Intelligence (XAI). IEEE Access 2018, 6, 52138–52160. [Google Scholar] [CrossRef]
- Jia, S.Q.; An, D.; Liu, Z.; Gu, J.C.; Li, S.M.; Zhang, X.D.; Zhu, D.H.; Guo, T.T.; Yan, Y.L. Variety Identification Method of Coated Maize Seeds Based on Near-Infrared Spectroscopy and Chemometrics. J. Cereal Sci. 2015, 63, 21–26. [Google Scholar] [CrossRef]
- Engel, J.; Gerretzen, J.; Szymanska, E.; Jansen, J.J.; Downey, G.; Blanchet, L.; Buydens, L.M.C. Breaking with Trends in Pre-Processing? Trac-Trends Anal. Chem. 2013, 50, 96–106. [Google Scholar] [CrossRef]
- Chen, S.; Lin, X.; Yuen, C.; Padmanabhan, S.; Beuerman, R.W.; Liu, Q. Recovery of Raman Spectra with Low Signal-to-Noise Ratio Using Wiener Estimation. Opt. Express 2014, 22, 12102–12114. [Google Scholar] [CrossRef]
- Bocklitz, T.; Walter, A.; Hartmann, K.; Rösch, P.; Popp, J. How to Pre-Process Raman Spectra for Reliable and Stable Models? Anal. Chim. Acta 2011, 704, 47–56. [Google Scholar] [CrossRef]
- Morais, C.L.M.; Lima, K.M.G.; Singh, M.; Martin, F.L. Tutorial: Multivariate Classification for Vibrational Spectroscopy in Biological Samples. Nat. Protoc. 2020, 15, 2143–2162. [Google Scholar] [CrossRef]
- Heidari, A.A.; Mirjalili, S.; Faris, H.; Aljarah, I.; Mafarja, M.; Chen, H. Harris Hawks Optimization: Algorithm and Applications. Future Gener. Comput. Syst.-Int. J. Esci. 2019, 97, 849–872. [Google Scholar] [CrossRef]
- Elgamal, Z.M.; Yasin, N.B.M.; Tubishat, M.; Alswaitti, M.; Mirjalili, S. An Improved Harris Hawks Optimization Algorithm with Simulated Annealing for Feature Selection in the Medical Field. IEEE Access 2020, 8, 186638–186652. [Google Scholar] [CrossRef]
- Abbott, D. Foreword 2 for 1st Edition. In Handbook of Statistical Analysis and Data Mining Applications, 2nd ed.; Nisbet, R., Miner, G., Yale, K., Eds.; Academic Press: Boston, MA, USA, 2018; pp. xv–xvi. ISBN 978-0-12-416632-5. [Google Scholar]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Cichosz, P. Assessing the Quality of Classification Models: Performance Measures and Evaluation Procedures. Open Eng. 2011, 1, 132–158. [Google Scholar] [CrossRef]
- Vilone, G.; Longo, L. Explainable Artificial Intelligence: A Systematic Review. arXiv 2020, arXiv:2006.00093. [Google Scholar]
- Aas, K.; Jullum, M.; Loland, A. Explaining Individual Predictions When Features Are Dependent: More Accurate Approximations to Shapley Values. Artif. Intell. 2021, 298, 103502. [Google Scholar] [CrossRef]
- Alabi, R.O.; Almangush, A.; Elmusrati, M.; Leivo, I.; Makitie, A.A. An Interpretable Machine Learning Prognostic System for Risk Stratification in Oropharyngeal Cancer. Int. J. Med. Inf. 2022, 168, 104896. [Google Scholar] [CrossRef]
- Yan, H.; Hua, Z.; Qian, G.; Wang, M.; Du, G.; Chen, J. Analysis of the Chemical Composition of Cotton Seed Coat by Fourier-Transform Infrared (FT-IR) Microspectroscopy. Cellulose 2009, 16, 1099–1107. [Google Scholar] [CrossRef]
- Gao, W.; Zhou, L.; Liu, S.; Guan, Y.; Gao, H.; Hui, B. Machine Learning Prediction of Lignin Content in Poplar with Raman Spectroscopy. Bioresour. Technol. 2022, 348, 126812. [Google Scholar] [CrossRef]
- Lupoi, J.S.; Gjersing, E.; Davis, M.F. Evaluating Lignocellulosic Biomass, Its Derivatives, and Downstream Products with Raman Spectroscopy. Front. Bioeng. Biotechnol. 2015, 3, 50. [Google Scholar] [CrossRef] [Green Version]
- Bock, P.; Gierlinger, N. Infrared and Raman Spectra of Lignin Substructures: Coniferyl Alcohol, Abietin, and Coniferyl Aldehyde. J. Raman Spectrosc. 2019, 50, 778–792. [Google Scholar] [CrossRef]
- Macmillan, C.; Birke, H.; Bedon, F.; Pettolino, F. Lignin Deposition in Cotton Cells—Where Is the Lignin? J. Plant Biochem. Physiol. 2014, 1, e106. [Google Scholar] [CrossRef]
- Rygula, A.; Majzner, K.; Marzec, K.M.; Kaczor, A.; Pilarczyk, M.; Baranska, M. Raman Spectroscopy of Proteins: A Review. J. Raman Spectrosc. 2013, 44, 1061–1076. [Google Scholar] [CrossRef]
- da Mata, M.M.; Rocha, P.D.; Teles de Farias, I.K.; Brasil da Silva, J.L.; Medeiros, E.P.; Silva, C.S.; Simoes, S.d.S. Distinguishing Cotton Seed Genotypes by Means of Vibrational Spectroscopic Methods (NIR and Raman) and Chemometrics. Spectrochim. Acta Part-Mol. Biomol. Spectrosc. 2022, 266, 120399. [Google Scholar] [CrossRef]
- Afzal, I.; Javed, T.; Amirkhani, M.; Taylor, A.G. Modern Seed Technology: Seed Coating Delivery Systems for Enhancing Seed and Crop Performance. Agriculture 2020, 10, 526. [Google Scholar] [CrossRef]
- Ren, G.; Wang, Y.; Ning, J.; Zhang, Z. Highly Identification of Keemun Black Tea Rank Based on Cognitive Spectroscopy: Near Infrared Spectroscopy Combined with Feature Variable Selection. Spectrochim. Acta Part-Mol. Biomol. Spectrosc. 2020, 230, 118079. [Google Scholar] [CrossRef]
- Du, K.-L.; Leung, C.-S.; Mow, W.H.; Swamy, M.N.S. Perceptron: Learning, Generalization, Model Selection, Fault Tolerance, and Role in the Deep Learning Era. Mathematics 2022, 10, 4730. [Google Scholar] [CrossRef]
- Li, Y.; Jia, J.; Zhang, L.; Khattak, A.M.; Sun, S.; Gao, W.; Wang, M. Soybean Seed Counting Based on Pod Image Using Two-Column Convolution Neural Network. IEEE Access 2019, 7, 64177–64185. [Google Scholar] [CrossRef]
- Loddo, A.; Loddo, M.; Di Ruberto, C. A Novel Deep Learning Based Approach for Seed Image Classification and Retrieval. Comput. Electron. Agric. 2021, 187, 106269. [Google Scholar] [CrossRef]
- Uzal, L.C.; Grinblat, G.L.; Namias, R.; Larese, M.G.; Bianchi, J.S.; Morandi, E.N.; Granitto, P.M. Seed-per-Pod Estimation for Plant Breeding Using Deep Learning. Comput. Electron. Agric. 2018, 150, 196–204. [Google Scholar] [CrossRef]
- Ma, T.; Tsuchikawa, S.; Inagaki, T. Rapid and Non-Destructive Seed Viability Prediction Using near-Infrared Hyperspectral Imaging Coupled with a Deep Learning Approach. Comput. Electron. Agric. 2020, 177, 9. [Google Scholar] [CrossRef]
- Zhu, S.; Zhou, L.; Gao, P.; Bao, Y.; He, Y.; Feng, L. Near-Infrared Hyperspectral Imaging Combined with Deep Learning to Identify Cotton Seed Varieties. Molecules 2019, 24, 3268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bannigan, P.; Bao, Z.; Hickman, R.J.; Aldeghi, M.; Häse, F.; Aspuru-Guzik, A.; Allen, C. Machine Learning Models to Accelerate the Design of Polymeric Long-Acting Injectables. Nat. Commun. 2023, 14, 35. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, S.; Singh, R.; Taneja, B.; Mande, S.S. MarkerML—Marker Feature Identification in Metagenomic Datasets Using Interpretable Machine Learning. J. Mol. Biol. 2022, 434, 167589. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Verma, V.K. Cycocel Induced Lignin Deposition in Cotton Cells and Its Role in Crop Growth. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 1567–1573. [Google Scholar] [CrossRef]
- Xu, L.; Zhu, L.; Tu, L.; Liu, L.; Yuan, D.; Jin, L.; Long, L.; Zhang, X. Lignin Metabolism Has a Central Role in the Resistance of Cotton to the Wilt Fungus Verticillium Dahliae as Revealed by RNA-Seq-Dependent Transcriptional Analysis and Histochemistry. J. Exp. Bot. 2011, 62, 5607–5621. [Google Scholar] [CrossRef] [Green Version]







| Cultivar | Breeding | Growth Period (Days) | Micronaire (MIC) | Resistance/ Tolerance | Growing Region |
|---|---|---|---|---|---|
| ZH-75 | Transgenic Bt hybrid | 123 | 5.1 | Fusarium wilt (t) Verticillium wilt (t) Cotton bollworm (r) | Yellow River basin |
| ZH-70 | Transgenic Bt + CpTI hybrid | 121 | 4.3 | Fusarium wilt (t) Verticillium wilt (t) Cotton bollworm (r) | Yellow River basin |
| ZH-88 | Hybridization | 145 | 4.1 | Fusarium wilt (r) | Northwestern inland |
| Methods | Training Accuracy | Testing Accuracy | |
|---|---|---|---|
| Machine learning | SVM + PCA | 0.90 | 0.78 |
| SVM + HHO | 0.67 | 0.63 | |
| XGBoost + PCA | 1.00 | 0.94 | |
| XGBoost + HHO | 1.00 | 0.89 | |
| Deep learning | MLP + PCA | 0.98 | 0.89 |
| MLP + HHO | 1.00 | 0.78 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chi, J.; Bu, X.; Zhang, X.; Wang, L.; Zhang, N. Insights into Cottonseed Cultivar Identification Using Raman Spectroscopy and Explainable Machine Learning. Agriculture 2023, 13, 768. https://doi.org/10.3390/agriculture13040768
Chi J, Bu X, Zhang X, Wang L, Zhang N. Insights into Cottonseed Cultivar Identification Using Raman Spectroscopy and Explainable Machine Learning. Agriculture. 2023; 13(4):768. https://doi.org/10.3390/agriculture13040768
Chicago/Turabian StyleChi, Jianan, Xiangxin Bu, Xiao Zhang, Lijun Wang, and Nannan Zhang. 2023. "Insights into Cottonseed Cultivar Identification Using Raman Spectroscopy and Explainable Machine Learning" Agriculture 13, no. 4: 768. https://doi.org/10.3390/agriculture13040768
APA StyleChi, J., Bu, X., Zhang, X., Wang, L., & Zhang, N. (2023). Insights into Cottonseed Cultivar Identification Using Raman Spectroscopy and Explainable Machine Learning. Agriculture, 13(4), 768. https://doi.org/10.3390/agriculture13040768




