Genetic Diversity and Population Structure Analysis to Construct a Core Collection from Safflower (Carthamus tinctorius L.) Germplasm through SSR Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Genotyping through SSR Markers
2.3. Genetic Diversity and Population Structure Analysis
2.4. Analysis of Molecular Variance (AMOVA)
2.5. Establishment and Evaluation of Core Collection
3. Results
3.1. Genetic Diversity Analysis
3.2. Population Structure Analysis
3.3. Analysis of Molecular Variance
3.4. Development of Core Collection
3.5. Evaluation of Core Collection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fernández-Martinez, J.; del Rio, M.; de Haro, A. Survey of safflower (Carthamus tinctorius L.) germplasm for variants in fatty acid composition and other seed characters. Euphytica 1993, 69, 115–122. [Google Scholar] [CrossRef]
- Jhajharia, S.; Choudhary, P.; Jhajharia, A.; Meena, L.K.; Singh, D. Heterosis and combining ability in Safflower (Carthamus tinctorius) germplasm lines. Bioscan 2013, 8, 1453–1460. [Google Scholar]
- Dajue, L.; Mündel, H.H. Safflower Carthamus tinctorius L. Promoting the Conservation and Use of Underutilized and Neglected Crops; Institute of Plant Genetics and Crop Plant Research, Gatersleben, International Plant Genetic Resources Institute: Rome, Italy, 1996; Volume 7. [Google Scholar]
- Flider, F.J. Development and commercialization of GLA Safflower oil. Lipid Tech. 2013, 25, 227–229. [Google Scholar] [CrossRef]
- Carlsson, A.S.; Zhu, L.H.; Andersson, M.; Hofvander, P. Platform crops amenable to genetic engineering–a requirement for successful production of bio-industrial oils through genetic engineering. Biocatal. Agric. Biotechnol. 2014, 3, 58–64. [Google Scholar] [CrossRef]
- Zhou, X.; Tang, L.; Xu, Y.; Zhou, G.; Wang, Z. Towards a better understanding of medicinal uses of Carthamus tinctorius L. In traditional Chinese medicine: A phytochemical and pharmacological review. J. Ethnopharmacol. 2014, 151, 27–43. [Google Scholar] [CrossRef]
- Ilkılıç, C.; Aydın, S.; Behcet, R.; Aydin, H. Biodiesel from Safflower oil and its application in a diesel engine. Fuel Process. Technol. 2011, 92, 356–362. [Google Scholar] [CrossRef]
- Kaya, M.D.; Bayramin, S.; Kaya, G.; Uzun, O. Seed vigor and ion toxicity in Safflower (Carthamus tinctorius L.) seedlings produced by various seed sizes under NaCl stress. Arch. Biol. Sci. 2011, 63, 723–729. [Google Scholar] [CrossRef]
- Bahrami, F.; Arzani, A.; Karimi, V. Evaluation of yield-based drought tolerance indices for screening Safflower genotypes. Agron. J. 2014, 106, 1219–1224. [Google Scholar] [CrossRef] [Green Version]
- Yeilaghi, H.; Arzani, A.; Ghaderian, M. Evaluating the contribution of ionic and agronomic components toward salinity tolerance in Safflower. Agron. J. 2015, 107, 2205–2212. [Google Scholar] [CrossRef]
- Ebrahimi, F.; Majidi, M.M.; Arzani, A.; Mohammadi-Nejad, G. Oil and seed yield stability in a worldwide collection of Safflower under arid environments of Iran. Euphytica 2016, 212, 131–144. [Google Scholar] [CrossRef]
- Knowles, P.F. Centers of plant diversity and conservation of crop germplasm: Safflower. Econ. Bot. 1969, 23, 324–329. [Google Scholar] [CrossRef]
- Ashri, A. Evaluation of the Germ Plasm Collection of Safflower, Carthamus tinctorius L. V. Distribution and Regional Divergence for Morphological Characters. Euphytica 1975, 24, 651–659. [Google Scholar] [CrossRef]
- Chapman, M.A.; Hvala, J.; Strever, J.; Burke, J.M. Population genetic analysis of Safflower (Carthamus tinctorius; Asteraceae) reveals a near eastern origin and five centres of diversity. Am. J. Bot. 2010, 97, 831–840. [Google Scholar] [CrossRef]
- Food and Agriculture Organization (FAO) of United Nations. The Food and Agriculture Organization Corporate Statistical Database Production Quantities of Safflower Seed and Plantains for 2018. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 7 December 2022).
- Nimbkar, N. “Issues in Safflower production in India,” In Safflower: Unexploited Potential and World Adaptability. In Proceedings of the of the Seventh International Safflower Conference, Wagga Wagga, NSW, Australia, 3–6 November 2008. [Google Scholar]
- Varshney, R.K.; Mahendar, T.; Aggarwal, R.K.; Borner, A. Genetic Molecular Markers in Plants: Development and Applications. In Genomics-Assisted Crop Improvement; Springer: Berlin/Heidelberg, Germany, 2007; pp. 13–29. [Google Scholar]
- Johnson, R.C.; Kisha, T.J.; Evans, M.A. Characterizing Safflower germplasm with AFLP molecular markers. Crop Sci. 2007, 47, 1728–1736. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Ambreen, H.; Murali, T.V.; Bali, S.; Agarwal, M.; Kumar, A.; Goel, S.; Jagannath, A. Assessment of genetic diversity and population structure in a global reference collection of 531 accessions of Carthamus tinctorius (Safflower) using AFLP markers. Plant Mol. Bio. Report 2014, 33, 1299–1313. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.A.; Witzke-Ehbrecht, S.V.; Maass, B.L.; Becker, H.C. Relationships among different geographical groups, agromorphology, fatty acid composition and RAPD marker diversity in Safflower (Carthamus tinctorius). Genet. Resour. Crop Evol. 2009, 56, 19–30. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.X.; Wu, W.; Zheng, Y.L.; Chen, L.; Liu, R.J.; Huang, C.Y. Genetic diversity and relationships among Safflower (Carthamus tinctorius L.) analyzed by inter-simple sequence repeats (ISSRs). Genet. Resour. Crop Evol. 2007, 54, 1043–1051. [Google Scholar] [CrossRef]
- Golkar, P.; Arzani, A.; Rezaei, M.A. Genetic variation in Safflower (Carthamus tinctorius L.) for seed quality related traits and inter-simple sequence repeat (ISSR) markers. Int. J. Mol. Sci. 2011, 12, 2664–2677. [Google Scholar] [CrossRef] [Green Version]
- Panahi, B.; Neghab, M.G. Genetic characterization of Iranian Safflower (Carthamus tinctorius) using inter-simple sequence repeats (ISSR) markers. Physiol. Mol. Biol. Plants 2013, 19, 239–243. [Google Scholar] [CrossRef] [Green Version]
- Majidi, M.M.; Zadhoush, S. Molecular and morphological variation in a world-wide collection of Safflower. Crop Sci. 2014, 54, 2109–2119. [Google Scholar] [CrossRef]
- Yaman, H.; Tarıkahya-Hacıoğlu, B.; Arslan, Y.; Subas, I. Molecular characterization of the wild relatives of Safflower (Carthamus tinctorius L.) in Turkey as revealed by ISSRs. Genet. Resour. Crop Evol. 2014, 61, 595–602. [Google Scholar] [CrossRef]
- Talebi, M.; Mokhtari, N.; Rahimmalek, M.; Sahhafi, S.R. Molecular characterization of Carthamus tinctorius and C. oxyacanthus germplasm using sequence related amplified polymorphism (SRAP) markers. Plant Omics J. 2012, 5, 136–142. [Google Scholar]
- Parida, S.K.; Kalia, S.K.; Kaul, S. Informative genomic microsatellite markers for efficient genotyping applications in Sugarcane. Theor. Appl. Genet. 2009, 118, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.A.; Hvala, J.; Strever, J.; Matvienko, M.; Kozik, A.; Michelmore, R.W.; Tang, S.; Knapp, S.J.; Burke, J.M. Development, polymorphism, and cross-taxon utility of EST–SSR markers from Saffower (Carthamus tinctorius L.). Theor. Appl. Genet. 2009, 120, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, Y.A.S.; Garcia-Moreno, M.; Redondo-Nevado, J.; Velasco, L.; Perez-Vich, B. Development and characterization of genomic microsatellite markers in Safflower (Carthamus tinctorius L.). Plant Breed. 2011, 130, 237–241. [Google Scholar] [CrossRef]
- Yamini, K.N.; Ramesh, K.; Naresh, V.; Rajendrakumar, P.; Anjani, K.; Dinesh, K.V. Development of EST–SSR markers and their utility in revealing cryptic diversity in Safflower (Carthamus tinctorius L.). J. Plant Biochem. Biotechnol. 2013, 22, 90–102. [Google Scholar] [CrossRef]
- Lee, G.A.; Sung, J.S.; Lee, S.Y.; Chung, J.W.; Yi, J.Y.; Kim, Y.Y.; Lee, M.C. Genetic assessment of Safflower (Carthamus tinctorius L.) collection with microsatellite markers acquired via pyrosequencing method. Mol. Ecol. Resour. 2014, 14, 69–78. [Google Scholar] [CrossRef]
- Barati, M.; Arzani, A. Genetic diversity revealed by EST– SSR markers in cultivated and wild Safflower. Biochem. Syst. Ecol. 2012, 44, 117–123. [Google Scholar] [CrossRef]
- Derakhshan, E.; Majidi, M.M.; Sharafi, Y.; Mirlohi, A. Discrimination and genetic diversity of cultivated and wild Safflowers (Carthamus spp.) using EST–microsatellites markers. Biochem. Syst. Ecol. 2014, 54, 130–136. [Google Scholar] [CrossRef]
- van Hintum, T.J.L.; Brown, A.H.D.; Spillane, C.; Hodgkin, T. Core collections of plant genetic resources. IPGRI Tech. Bull. 2000, 3, 1–49. [Google Scholar]
- Belaj, A.; Dominguez-García, M.d.C.; Atienza, S.G. Developing a core collection of Olive (Olea europaea L.) based on molecular markers (DArTs, SSRs, SNPs) and agronomic traits. Tree Genet. Genomes 2012, 8, 365–378. [Google Scholar] [CrossRef]
- Frankel, O.H. Genetic Perspectives of Germplasm Conservation. In Genetic Manipulation: Impact on Man and Society; Werner, A., Ed.; Cambridge University Press: Cambridge, UK, 1984; pp. 161–170. [Google Scholar]
- Charmet, G.; Balfourier, F.; Monestiez, P. Hierarchical Clustering of Perennial Ryegrass Populations with Geographic Contiguity Constraint. Theor. Appl. Genet. 1994, 88, 42–48. [Google Scholar] [CrossRef]
- Yonezawa, K.; Nomura, T.; Morishima, H. Sampling strategies for use in stratified germplasm collections. In Core Collections of Plant Genetic Resources; Hodgkin, T., Brown, A.H.D., van Hintum, T.J.L., Morales, E.A.V., Eds.; John Wiley and Sons: Chichester, UK, 1995; pp. 35–53. [Google Scholar]
- Noirot, M.; Hamon, S.; Anthony, F. The Principal Component Scoring: A New Method of Constituting a Core Collection Using Quantitative Data. Genet. Resour. Crop Evol. 1996, 43, 1–6. [Google Scholar] [CrossRef]
- Mongkolporn, O.; Hanyong, S.; Chunwongse, J.; Wasee, S. Establishment of a Core Collection of Chilli Germplasm Using Microsatellite Analysis. Plant Genet. Resour. 2015, 13, 104–110. [Google Scholar] [CrossRef]
- Ebana, K.; Kojima, Y.; Fukuoka, S.; Nagamine, T.; Kawase, M. Development of Mini Core Collection of Japanese Rice Landrace. Breed. Sci. 2008, 58, 281–291. [Google Scholar] [CrossRef] [Green Version]
- Kaga, A.; Shimizu, T.; Watanabe, S.; Tsubokura, Y.; Katayose, Y.; Harada, K.; Vaughan, D.A.; Tomooka, N. Evaluation of Soybean Germplasm Conserved in NIAS Genebank and Development of Mini Core Collections. Breed. Sci. 2012, 61, 566–592. [Google Scholar] [CrossRef] [Green Version]
- Balfourier, F.; Roussel, V.; Strelchenko, P.; Exbrayat-Vinson, F.; Sourdille, P.; Boutet, G.; Koenig, J.; Ravel, C.; Mitrofanova, O.; Beckert, M.; et al. A Worldwide Bread Wheat Core Collection Arrayed in a 384-Well Plate. Theor. Appl. Genet. 2007, 114, 1265–1275. [Google Scholar] [CrossRef]
- Upadhyaya, H.D.; Bramel, P.J.; Singh, S. Development of a Chickpea Core Subset Using Geographic Distribution and Quantitative Traits. Crop Sci. 2001, 41, 206–210. [Google Scholar] [CrossRef]
- Amalraj, V.A.; Balakrishnan, R.; Jebadhas, A.W.; Balasundaram, N. Constituting a Core Collection of Saccharum spontaneum L. and Comparison of Three Stratified Random Sampling Procedures. Genet. Resour. Crop Evol. 2006, 53, 1563–1572. [Google Scholar] [CrossRef]
- Quenouille, J.; Saint-Felix, L.; Moury, B.; Palloix, A. Diversity of Genetic Backgrounds Modulating the Durability of a Major Resistance Gene. Analysis of a Core Collection of Pepper Landraces Resistant to Potato Virus Y. Mol. Plant. Pathol. 2016, 17, 296–302. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Chen, X.S.; Zhang, Y.M.; Yuan, Z.H.; Liu, Z.C.; Wang, Y.L.; Lin, Q. A method for constructing core collection of Malus sieversii using molecular markers. Agric. Sci. China 2009, 42, S1671–S2927. [Google Scholar] [CrossRef]
- Liu, W.; Shahid, M.Q.; Bai, L.; Lu, Z.; Chen, Y.; Jiang, L.; Diao, M.; Liu, X.; Lu, Y. Evaluation of Genetic Diversity and Development of a Core Collection of Wild Rice (Oryza rufipogon Griff.) Populations in China. PLoS ONE 2015, 10, e0145990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Escribano, P.; Viruel, M.A.; Hormaza, J.I. Comparison of different methods to sequence repeat markers. A case study in cherimoya (Annona cherimola, Annonaceae), an underutilised subtropical fruit tree species. Ann. Appl. Biol. 2008, 153, 25–32. [Google Scholar] [CrossRef]
- Richards, C.M.; Volk, G.M.; Reeves, P.A.; Reilley, A.A.; Henk, A.D.; Forsline, P.L.; Aldwinckle, H.S. Selection of Stratified Core Sets Representing Wild Apple (Malus Sieversii). J. Am. Soc. Hortic. Sci. 2009, 134, 228–235. [Google Scholar] [CrossRef] [Green Version]
- De Beukelaer, H.; Davenport, G.F.; Fack, V. Core Hunter 3: Flexible Core Subset Selection. BMC Bioinform. 2018, 19, 203. [Google Scholar] [CrossRef]
- Soleimani, B.; Lehnert, H.; Keilwagen, J.; Plieske, J.; Ordon, F.; Naseri Rad, S.; Ganal, M.; Beier, S.; Perovic, D. Comparison Between Core Set Selection Methods Using Different Illumina Marker Platforms: A Case Study of Assessment of Diversity in Wheat. Front. Plant Sci. 2020, 11, 1040. [Google Scholar] [CrossRef]
- Xu, Q.; Zeng, X.; Lin, B.; Li, Z.; Yuan, H.; Wang, Y.; Tashi, N. A Microsatellite Diversity Analysis and the Development of Core-Set Germplasm in a Large Hulless Barley (Hordeum vulgare L.) Collection. BMC Genet. 2017, 18, 102. [Google Scholar] [CrossRef] [Green Version]
- Dwivedi, S.; Upadhyaya, H.; Hegde, D. Development of Core Collection Using Geographic Information and Morphological Descriptors in Safflower (Carthamus tinctorius L.) Germplasm. Genet. Resour. Crop Evol 2005, 52, 821–830. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Ambreen, H.; Variath, M.T.; Rao, A.R.; Agarwal, M.; Kumar, A.; Goel, S.; Jagannath, A. Utilization of Molecular, Phenotypic, and Geographical Diversity to Develop Compact Composite Core Collection in the Oilseed Crop, Safflower (Carthamus tinctorius L.) through Maximization Strategy. Front. Plant Sci. 2016, 7, 1554. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Muse, S.V. PowerMarker: An Integrated Analysis Environment for Genetic Marker Analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [Green Version]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic Analysis in Excel. Population Genetic Software for Teaching and Research—An Update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef] [Green Version]
- Hammer, O.; Harper, D.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Pritchard, J.K.; Wen, X.; Falush, D. Documentation for Structure Software, version 2.3; University of Chicago: Chicago, IL, USA, 2010. [Google Scholar]
- Earl, D.; Vonholdt, B.; Earl, D.A.; Von Holdt, B.M. Structure Harvester: A Website and Program for Visualizing STRUCTURE Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Beukelaer, H.D.; Smýkal, P.; Davenport, G.F.; Fack, V. Core Hunter II: Fast core subset selection based on multiple genetic diversity measures using Mixed Replica search. BMC Bioinform. 2012, 13, 312. [Google Scholar] [CrossRef] [Green Version]
- D’hoop, B.B.; Paulo, M.J.; Kowitwanich, K.; Sengers, M.; Visser, R.G.F.; van Eck, H.J.; van Eeuwijk, A.F. Population structure and linkage disequilibrium unravelled in tetraploid potato. Theor. Appl. Genet. 2010, 121, 1151–1170. [Google Scholar] [CrossRef] [Green Version]
- Odong, T.L.; van Heerwaarden, J.; Jansen, J.; van Hintum, T.J.L.; van Eeuwijk, F.A. Determination of Genetic Structure of Germplasm Collections: Are Traditional Hierarchical Clustering Methods Appropriate for Molecular Marker Data? Theor. Appl. Genet. 2011, 123, 195–205. [Google Scholar] [CrossRef] [Green Version]
- Wada, T.; Noguchi, Y.; Isobe, S.; Kunihisa, M.; Sueyoshi, T.; Shimomura, K. Development of a Core Collection of Strawberry Cultivars Based on SSR and CAPS Marker Polymorphisms. Hortic. J. 2017, 86, 365–378. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, E.J.; Ferreira, C.F.; Santos, V.S.; Oliveira, G.A.F. Development of a Cassava Core Collection Based on Single Nucleotide Polymorphism Markers. Genet. Mol. Res. 2014, 13, 6472–6485. [Google Scholar] [CrossRef]
- Zhu, Y.; Liang, D.; Song, Z.; Tan, Y.; Guo, X.; Wang, D. Genetic Diversity Analysis and Core Germplasm Collection Construction of Camellia Oleifera Based on Fruit Phenotype and SSR Data. Genes 2022, 13, 2351. [Google Scholar] [CrossRef]
- Ambreen, H.; Kumar, S.; Variath, M.T.; Joshi, G.; Bali, S.; Agarwal, M.; Kumar, A.; Jagannath, A.; Goel, S. Development of Genomic Microsatellite Markers in Carthamus tinctorius L. (Safflower) Using Next Generation Sequencing and Assessment of Their Cross-Species Transferability and Utility for Diversity Analysis. PLoS ONE 2015, 10, e0135443. [Google Scholar] [CrossRef] [Green Version]
- Golkar, P.; Mokhtari, N. Molecular Diversity Assessment of a World Collection of Safflower Genotypes by SRAP and SCoT Molecular Markers. Physiol. Mol. Biol. Plants 2018, 24, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Mishra, M.; Dixit, S.; Sahoo, S. In Silico Identification of Microsatelite Markers and Its SSR-FDM Analysis from Ests Sequence in Asteraceae Family. World J. Pharm. Res. 2017, 6, 977–997. [Google Scholar] [CrossRef]
- Nabloussi, A.; Velasco, L.; Fernández-Martínez, J. Cross Pollination of Safflower under Moroccan Environmental Conditions. Int. J. Plant Breed. 2013, 7, 145–147. [Google Scholar]
- Gomashe, S.; Ingle, K.; Sarap, Y.; Subramani, R.; Chand, D. Safflower (Carthamus tinctorius L.): An Underutilized Crop with Potential Medicinal Values. Ann. Phytomedicine Int. J. 2021, 1, 242–248. [Google Scholar] [CrossRef]
- Zhao, W.; Cho, G.-T.; Ma, K.-H.; Chung, J.-W.; Gwag, J.-G.; Park, Y.-J. Development of an Allele-Mining Set in Rice Using a Heuristic Algorithm and SSR Genotype Data with Least Redundancy for the Post-Genomic Era. Mol. Breed. 2010, 26, 639–651. [Google Scholar] [CrossRef]
- Ambreen, H.; Kumar, S.; Kumar, A.; Agarwal, M.; Jagannath, A.; Goel, S. Association Mapping for Important Agronomic Traits in Safflower (Carthamus tinctorius L.) Core Collection Using Microsatellite Markers. Front. Plant Sci. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
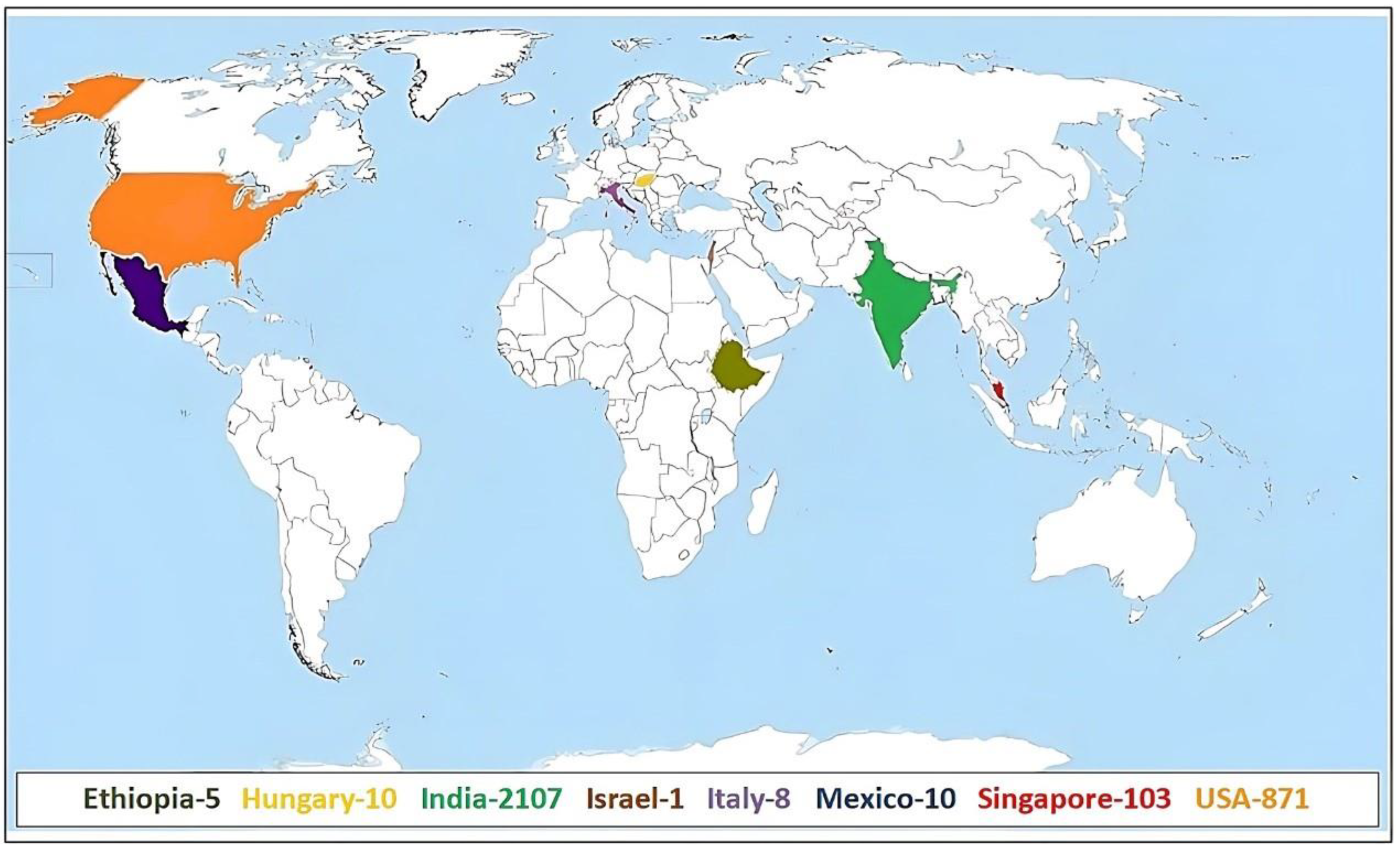


| Entire Collection (3115) (IC = 2107, EC = 1008) | |
|---|---|
| Country | No. of Accessions |
| India (Andhra Pradesh) | 90 |
| India (Bihar) | 3 |
| India (Delhi) | 4 |
| India (Gujarat) | 1 |
| India (Haryana) | 1 |
| India (Karnataka) | 37 |
| India (Maharashtra) | 1022 |
| India (Madhya Pradesh) | 78 |
| India (Odisha) | 8 |
| India (Punjab) | 4 |
| India (Rajasthan) | 1 |
| India (Tamil Nadu) | 2 |
| India (Telangana) | 6 |
| India (Uttar Pradesh) | 254 |
| India (West Bengal) | 1 |
| India (Others) | 595 |
| Ethiopia | 5 |
| Hungary | 10 |
| Israel | 1 |
| Italy | 8 |
| Mexico | 10 |
| Singapore | 103 |
| USA | 871 |
| SSR Marker Name | Forward (5’ to 3’) | Reverse (5’to 3’) |
|---|---|---|
| CAT-3 | CAATTAGAAAAACCCTTGTAGA | GACATCAACTTCCACTGCTG |
| CAT-5 | GGATATGGGTTGAGGGACGA | GTCCAGCCATCGCCACACTC |
| CAT-6 | AGGTTGGAGAAAGCTGGTTAG | GCTAACCTATAGCTTACCACC |
| CAT-8 | GATCAGATGAAAATACAAC | GTAAAGATAACTTGCCTTC |
| CAT-15 | GAATGACATAGAGGTAGACGTTC | AGGGTCAGGAGAAATATCAACAG |
| CAT-23 | CAAATGAGTGTTGAGAGTTG | TCTAAATAGTGGCAAACTCA |
| CAT-29 | TAGTATAAAGAGACACTTCCCA | AACGGCTATGATTGGCTTGTA |
| CAT-38 | GAGGAAGCTAGCTAATGAAATG | ATGATGATATCCTTGCGGAATC |
| CAT-43 | AGCTTGGTCTAGATGAACAC | GCAGTAGTAACCGATATGCTA |
| CAT-46 | CAAATAGGTGCTAGAAAACAC | ACTCAATCCTCATAGCAATTG |
| CAT-48 | GAAATCCGATGGTAGCCGGA | CTTCAACCTTCATCCCTCCC |
| CAT-52 | GAAACCCTAGATTCATTCA | CGCATGATTACAGTCTGAG |
| CAT-57 | GTTGGCCGAATAATCCTTCAC | TATGCGTATATATGGAGAGATG |
| CAT-58 | CATATGATAAAATATCACTAACA | TAAGATGATGCCATTGTGAC |
| CAT-64 | CTAAAGCAATCCTAAGCAAATCC | CTAGGGTTCTTACCAAATTGGGA |
| CAT-65 | AGAAGGTAAATCCATTGTGGAAG | TGCAAGAGTCCCTCAAGAGTC |
| CAT-91 | GAAGGTGTGTAGCCCAGATAC | GTAATGATTCACACGATAAACAG |
| CAT-96 | CATGCAATCATCAAGGGGTG | GTGCTCAAGTGTGTTTAATCA |
| Marker Name | MAF | Na | He | PIC |
|---|---|---|---|---|
| CAT-3 | 0.084 | 50 | 0.952 | 0.95 |
| CAT-5 | 0.100 | 67 | 0.966 | 0.97 |
| CAT-6 | 0.093 | 48 | 0.949 | 0.95 |
| CAT-8 | 0.085 | 80 | 0.969 | 0.97 |
| CAT-15 | 0.062 | 86 | 0.975 | 0.98 |
| CAT-23 | 0.081 | 84 | 0.970 | 0.97 |
| CAT-29 | 0.066 | 66 | 0.965 | 0.97 |
| CAT-38 | 0.071 | 68 | 0.965 | 0.96 |
| CAT-43 | 0.080 | 93 | 0.976 | 0.98 |
| CAT-46 | 0.054 | 79 | 0.972 | 0.97 |
| CAT-48 | 0.082 | 68 | 0.965 | 0.96 |
| CAT-52 | 0.092 | 78 | 0.968 | 0.97 |
| CAT-57 | 0.085 | 69 | 0.949 | 0.95 |
| CAT-58 | 0.089 | 88 | 0.972 | 0.97 |
| CAT-64 | 0.081 | 45 | 0.947 | 0.95 |
| CAT-65 | 0.064 | 62 | 0.960 | 0.96 |
| CAT-91 | 0.083 | 59 | 0.955 | 0.96 |
| CAT-96 | 0.080 | 78 | 0.957 | 0.96 |
| MEAN | 0.080 | 71 | 0.964 | 0.96 |
| Source | df | SS | MS | Est. Var. |
|---|---|---|---|---|
| Among collections | 1 | 1518.199 | 1518.199 | 0.550 |
| Among accessions | 3113 | 54,000.979 | 17.347 | 8.673 |
| Within Indiv | 3115 | 0.000 | 0.000 | 0.000 |
| Total | 6229 | 55,519.178 | 9.224 |
| Source | df | SS | MS | Est. Var. | % |
|---|---|---|---|---|---|
| Among model-based populations | 2 | 1596.746 | 798.373 | 0.380 | 4% |
| Among accessions | 3112 | 53,791.391 | 17.285 | 8.643 | 96% |
| Within Indiv | 3115 | 0.000 | 0.000 | 0.000 | 0% |
| Core Collection 311 (IC = 200, EC = 111) | |
|---|---|
| Country | No. of Accessions |
| India (Andhra Pradesh) | 10 |
| India (Karnataka) | 4 |
| India (Maharashtra) | 10 |
| India (Madhya Pradesh) | 81 |
| India (Tamil Nadu) | 1 |
| India (Telangana) | 1 |
| India (Uttar Pradesh) | 29 |
| India (West Bengal) | 1 |
| India (Others) | 63 |
| Hungary | 1 |
| Mexico | 1 |
| Singapore | 16 |
| USA | 93 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, G.P.; Pathania, P.; Goyal, N.; Gupta, N.; Parimalan, R.; Radhamani, J.; Gomashe, S.S.; Kadirvel, P.; Rajkumar, S. Genetic Diversity and Population Structure Analysis to Construct a Core Collection from Safflower (Carthamus tinctorius L.) Germplasm through SSR Markers. Agriculture 2023, 13, 836. https://doi.org/10.3390/agriculture13040836
Kumar GP, Pathania P, Goyal N, Gupta N, Parimalan R, Radhamani J, Gomashe SS, Kadirvel P, Rajkumar S. Genetic Diversity and Population Structure Analysis to Construct a Core Collection from Safflower (Carthamus tinctorius L.) Germplasm through SSR Markers. Agriculture. 2023; 13(4):836. https://doi.org/10.3390/agriculture13040836
Chicago/Turabian StyleKumar, Gaddam Prasanna, Pooja Pathania, Nitu Goyal, Nishu Gupta, R. Parimalan, J. Radhamani, Sunil Shriram Gomashe, Palchamy Kadirvel, and S. Rajkumar. 2023. "Genetic Diversity and Population Structure Analysis to Construct a Core Collection from Safflower (Carthamus tinctorius L.) Germplasm through SSR Markers" Agriculture 13, no. 4: 836. https://doi.org/10.3390/agriculture13040836
APA StyleKumar, G. P., Pathania, P., Goyal, N., Gupta, N., Parimalan, R., Radhamani, J., Gomashe, S. S., Kadirvel, P., & Rajkumar, S. (2023). Genetic Diversity and Population Structure Analysis to Construct a Core Collection from Safflower (Carthamus tinctorius L.) Germplasm through SSR Markers. Agriculture, 13(4), 836. https://doi.org/10.3390/agriculture13040836






