Resistance of Sorghum Genotypes to Ergot (Claviceps Species)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Site
2.2. Plant Materials
2.3. Experimental Design and Field Layout
2.4. Data Collection and Analysis
3. Results
3.1. Analysis of Variance
3.2. Response of Genotypes to Ergot Infection
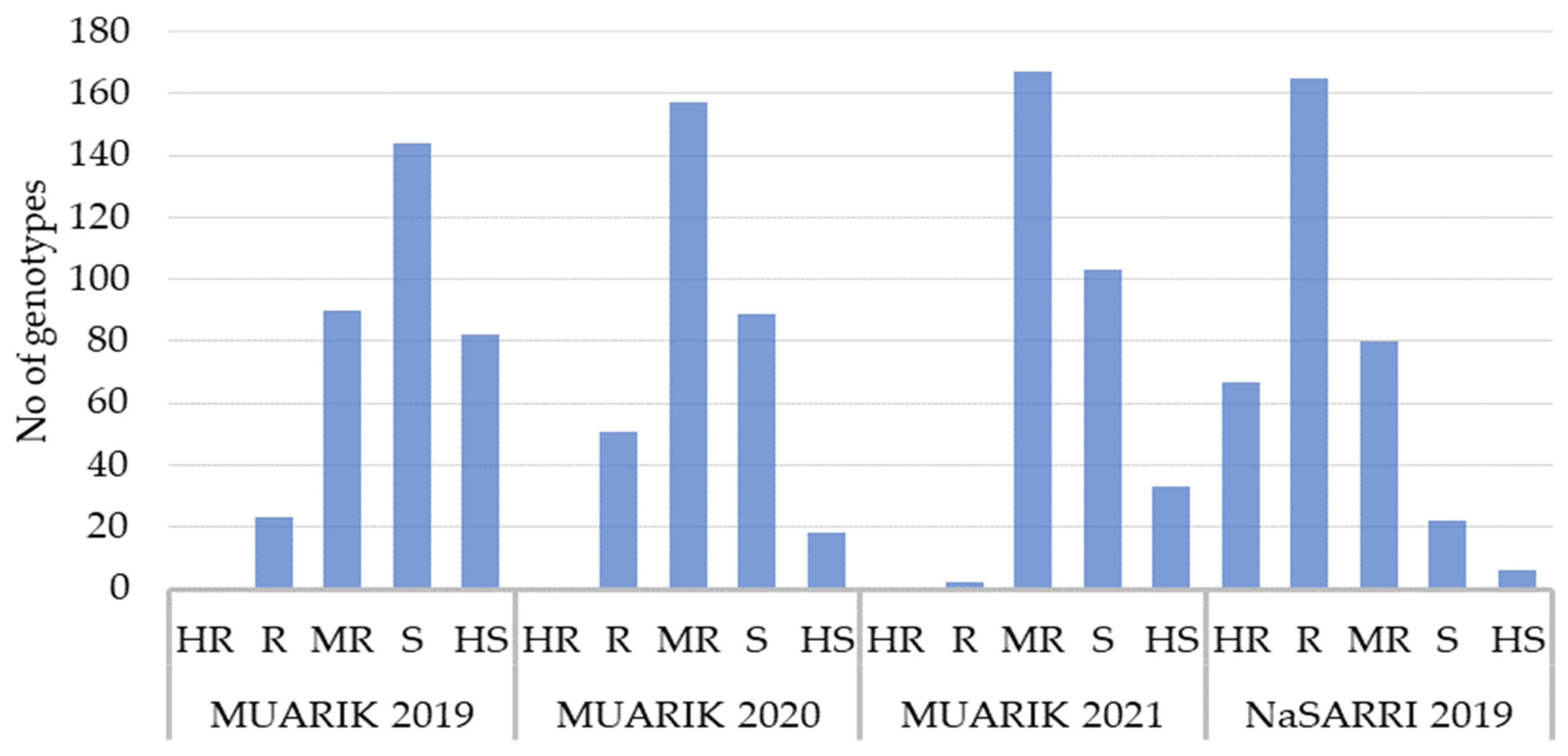
3.2.1. Disease Incidence
3.2.2. Disease Severity
3.2.3. Disease Severity vs. PQ and DTF
3.3. Variability for Agronomic Traits
3.4. Correlation between Resistance to Ergot and Agronomic Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. Statistics Division, Rome, Italy. Available online: https://www.fao.org/faostat/en/ (accessed on 20 May 2022).
- Caroline, O.; Evans, O.; Samuel, G. Screening of Selected Sorghum Genotypes for Resistance to Covered Kernel Smut Disease in Western Kenya. J. Agric. Sci. 2021, 13, 63–73. [Google Scholar] [CrossRef]
- Mofokeng, M.; Shimelis, H.; Laing, M.; Shargie, N. Sorghum [Sorghum bicolor (L.) Moench] breeding for resistance to leaf and stalk anthracnose, Colletotrichum sublineolum, and improved yield: Progress and prospects. Aust. J. Crop Sci. 2017, 11, 1078–1085. [Google Scholar] [CrossRef]
- Gebrekorkos, G.; Egziabher, Y.G.; Habtu, S. Response of Sorghum (Sorghum bicolor (L.) Moench) Varieties to Blended Fertilizer on yield, yield component and nutritional content under Irrigation in Raya Valley, Northern Ethiopia. Int. J. Agric. Biosci. 2019, 6, 153–162. Available online: www.ijagbio.com (accessed on 1 November 2019).
- Beyene, A.; Hussien, S.; Pangirayi, T.; Mark, L. Physiological mechanisms of drought tolerance in sorghum, genetic basis, and breeding methods: A review. Afr. J. Agric. Res. 2015, 10, 3029–3040. [Google Scholar] [CrossRef]
- Tenywa, M.M.; Nyamwaro, S.O.; Kalibwani, R.; Mogabo, J.; Buruchara, R.; Oluwole, F. Innovation Opportunities in Sorghum Production in Uganda. FARA Res. Rep. 2018, 2, 20. [Google Scholar]
- Njoroge, S.M.; Takan, J.P.; Letayo, E.A.; Okoth, P.S.; Ajaku, D.O.; Kumar, A.; Rathore, A.; Ojulong, H.; Manyasa, E. Survey of Fungal Foliar and Panicle Diseases of Sorghum in Important Survey of Fungal Foliar and Panicle Diseases of Sorghum in Important Agroecological Zones of Tanzania and Uganda. Plant Health Prog. 2018, 19, 265–271. [Google Scholar] [CrossRef]
- Amelework, B.A.; Shimelis, H.A.; Tongoona, P.; Laing, M.D.; Ayele, D.G. Sorghum production systems and constraints, and coping strategies under drought-prone agro-ecologies of Ethiopia. S. Afr. J. Plant Soil 2016, 33, 207–217. [Google Scholar] [CrossRef]
- Muturi, P.W.; Mgonja, M.; Rubaihayo, P. Identification of new sorghum genotypes resistant to the African and spotted stemborers. Int. J. Trop. Insect Sci. 2014, 34, 260–268. [Google Scholar] [CrossRef]
- Thakur, R.P.; Reddy, B.V.; Mathur, K. Screening Techniques for Sorghum Diseases. Information Bulletin No. 76; International Crops Research Institute for the Semi-Arid Tropics: Patancheru, India, 2007; pp. 1–92. ISBN 978-92-9066-504-5. [Google Scholar]
- Miedaner, T.; Geiger, H.H. Biology, Genetics, and Management of Ergot (Claviceps spp.) in Rye, Sorghum, and Pearl Millet. Toxins 2015, 7, 659–678. [Google Scholar] [CrossRef]
- Kazungu, F.K.; Mundi, E.M.; Mulinge, J.M. Overview of Sorghum (Sorghum bicolor L.), its Economic Importance, Ecological Requirements and Production Constraints in Kenya. Int. J. Plant Soil Sci. 2023, 35, 62–71. [Google Scholar] [CrossRef]
- Frederickson, D.E.; Mantle, P.G.; DE Milliano, W.A.J. Susceptibility to ergot in Zimbabwe of sorghums that remained unaffected in their native climates in Ethiopia and Rwanda. Plant Pathol. 1994, 43, 27–32. [Google Scholar] [CrossRef]
- Bandyopadhyay, R.; Frederickson, D.E.; Mclaren, N.W.; Odvody, G.N.; Ryley, M.J. Ergot: A New Disease Threat to Sorghum in the Americas and Australia. Plant Dis. 1998, 82, 356–367. [Google Scholar] [CrossRef] [PubMed]
- Bogo, A.; Mantle, P.G.; Harthmann, O.E.L. Screening of sweet sorghum accessions for inhibition of secondary sporulation and saccharide measurements in honeydew of Claviceps africana. Fitopatol. Bras. 2004, 29, 86–90. [Google Scholar] [CrossRef]
- Prom, L.K.; Erpelding, J.E.; Montes-Garcia, N. Evaluation of Sorghum Germplasm from China against Claviceps africana, Causal Agent of Sorghum Ergot. Plant Health Prog. 2008, 9, 1–7. [Google Scholar] [CrossRef]
- Parh, D.K.; Jordan, D.R.; Aitken, E.A.B.; Mace, E.S.; Jun-ai, P.; McIntyre, C.L.; Godwin, I.D. QTL analysis of ergot resistance in sorghum. Theor. Appl. Genet. 2008, 117, 369–382. [Google Scholar] [CrossRef]
- Shivakumar, B.Y. Studies on Sorghum Ergot Caused by Claviceps africana Frederickson Mantle and de Milliano. Master’s Thesis, University of Agricultural Sciences, Dharwad, India, 2011. [Google Scholar]
- Mendoza-Onofre, L.E.; Hernández-Martínez, M.; Cárdenas-Soriano, E.; Ramírez-Vallejo, P. El germoplasma de sorgo tolerante al frío como fuente potencial de tolerancia al ergot (Claviceps africana Frederickson, Mantle & de Milliano). Agrociencia 2006, 40, 593–603. [Google Scholar]
- Dahlberg, J.A.; Bandyopadhyay, R.; Rooney, W.L.; Odvody, G.N.; Madera-Torres, P. Evaluation of sorghum germplasm used in US breeding programmes for sources of sugary disease resistance. Plant Pathol. 2001, 50, 681–689. [Google Scholar] [CrossRef]
- Sorghum Diseases Sorghum Ergot (Claviceps africana). Fusarium Stalk Rot (Fusarium spp.). Charcoal Rot (Macrophomina phaseolina). Rust (Puccinia purpurea). Johnson Grass Mosaic Virus|Head Smut (Sporisorium reilianum). Leaf Blight (Exserohilum turcic). Australia. Available online: https://grdc.com.au/about/who-we-are/corporate-governance/annual-reports (accessed on 22 December 2022).
- Kebede, D.; Rubaihayo, P.; Dramadri, I.; Odong, T.; Edema, R. Mechanisms associated with resistance to ergot in sorghum. Res. Agric. Vet. Sci. 2022, 6, 127–140. [Google Scholar]
- Beshir, M.M. Genetic Analysis of Dual Resistance to Anthracnose and Turcicum Leaf Blight in Sorghum. Ph.D. Thesis, Makerere University, Kampala, Uganda, 2016. [Google Scholar]
- Morris, G.P.; Ramu, P.; Deshpande, S.P.; Hash, C.T.; Shah, T.; Upadhyaya, H.D. Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc. Natl. Acad. Sci. USA 2013, 110, 453–458. [Google Scholar] [CrossRef]
- Musabyimana, T.; Sehene, C.; Bandyopadhyay, R. Ergot resistance in Sorghum in relation to flowering, inoculation technique and disease development. Plant Pathol. 1995, 44, 109–115. [Google Scholar] [CrossRef]
- Parh, D.K.; Jordan, D.R.; Aitken, E.A.; Gogel, B.J.; McIntyre, C.L.; Godwin, I.D. Genetic components of variance and the role of pollen traits in Sorghum ergot resistance. Crop Sci. 2006, 46, 2387–2395. [Google Scholar] [CrossRef]
- Awori, E.; Kiryowa, M.; Basirika, A.; Dradiku, F.; Kahunza, R.; Oriba, A.; Mukalazi, J. Performance of elite grain sorghum varieties in the West Nile Agro-ecological Zones. Uganda J. Agric. Sci. 2015, 16, 139–148. [Google Scholar] [CrossRef]
- RStudio: Integrated Development for R; RStudio, PBC: Boston, MA, USA; Available online: http://www.rstudio.com/ (accessed on 3 December 2022).
- Mirdita, V.; Dhillon, B.S.; Geiger, H.H.; Miedaner, T. Genetic variation for resistance to ergot (Claviceps purpurea [Fr.] Tul.) among full-sib families of five populations of winter rye (Secale cereale L.). Theor. Appl. Genet. 2008, 118, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Cisneros-lópez, M.E.; Mendoza-onofre, L.E.; González-hernández, V.A.; Mora-aguilera, G.; Hernández-Martínez, M.; Zavaleta-Mancera, H.A.; Córdova-Téllez, A.L. Floral traits, ergot resistance and grain yield relationships in infected male-sterile sorghum A-lines. Seed Sci. Technol. 2010, 38, 114–124. [Google Scholar] [CrossRef]
- Kodisch, A.; Wilde, P.; Schmiedchen, B.; Fromme, F.J.; Rodemann, B.; Tratwal, A.; Oberforster, M.; Wieser, F.; Schiemann, A.; Jørgensen, L.N.; et al. Ergot infection in winter rye hybrids shows differential contribution of male and female genotypes and environment. Euphytica 2020, 216, 65. [Google Scholar] [CrossRef]

| Score | Infection (%) | Disease Reaction |
|---|---|---|
| 1 | No infection | Highly resistant (HR) |
| 2 | 1–10% infection | Resistant (R) |
| 3 | 11–25% infection | Moderately resistant (MR) |
| 4 | 26–50% infection | Susceptible (S) |
| 5 | >50% infection | Highly susceptible (HS) |
| SOV | DF | Incidence (%) | Severity (1–5) | DTF (Days) | Pollen Quantity (1–10) | Seedling Vigour (1–5) |
|---|---|---|---|---|---|---|
| Location | 1 | 290,823 ** | 110 ** | 3438 ** | 0.3 ns | 0.06 ns |
| Rep (Rep × Loc) | 2 | 7095 ** | 0.53 ** | 240 * | 4.5 ** | 2.6 ** |
| Genotype | 339 | 1059 ** | 0.17 ** | 225 ** | 0.7 ** | 0.9 ** |
| Genotype × Loc | 339 | 592 ** | 0.09 ** | 28.3 ns | 0.8 ** | 0.5 ** |
| Error | 664 | 166 | 0.02 | 24.8 | 0.2 | 0.4 |
| SOV | DF | Incidence (%) | Severity (1–5) | HSW (g) | DTF (Days) | PQ (1–10) | PH (cm) | PL (cm) | SV (1–5) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 14WAP | 15WAP | 16WAP | 14WAP | 15WAP | 16WAP | ||||||||
| Season | 2 | 48,869 ** | 121 ns | 8049 ** | 26.7 ** | 32 ** | 8.4 ** | 1.66 ** | 34,828 ** | 10.6 ** | 50,662 ** | 96 ** | 615 ** |
| Rep (Rep × Season) | 3 | 3037 ** | 4261 ** | 2517 ** | 0.17 ** | 0.4 ns | 0.18 ** | 0.14 ns | 454 ** | 5.1 ** | 535 ns | 29.5 * | 1.26 ** |
| Genotype | 356 | 549 ** | 779 ** | 1115 ** | 0.09 ** | 1.1 ** | 0.15 ** | 1.15 ** | 187 ** | 0.78 ** | 4316 ** | 105 ** | 0.78 ** |
| Genotype × Season | 607 | 261 ** | 316 ** | 415 ** | 0.03 ** | 0.35 ** | 0.05 ** | 0.12 ns | 71 ** | 0.55 ** | 435 ** | 14.5 ** | 0.48 ** |
| Error | 841 | 198 | 206 | 173 | 0.02 | 0.16 | 0.014 | 0.14 | 26 | 0.19 | 193 | 7.8 | 0.32 |
| Genotype | MUARIK | NaSARRI | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2019 | 2020 | 2021 | 2019 | |||||||||
| Incidence (%) | Severity (1–5) | RC | Incidence (%) | Severity (1–5) | RC | Incidence (%) | Severity (1–5) | RC | Incidence (%) | Severity (1–5) | RC | |
| E313 | 10.0 | 2.0 | R | 54.0 | 1.6 | R | 32.0 | 2.2 | MR | 4.4 | 1.3 | R |
| E351 | 14.0 | 1.5 | R | 14.5 | 1.8 | R | 8.0 | 1.8 | R | 0 | 1.0 | HR |
| E200 | 10.5 | 2.0 | R | 44.5 | 1.5 | R | 40.0 | 2.4 | MR | 6.0 | 1.3 | R |
| E352 | 38.5 | 1.5 | R | 22.5 | 2.0 | R | 15.5 | 2.1 | MR | 4.6 | 1.3 | R |
| E354 | 16.0 | 2.0 | R | 46.0 | 2.4 | MR | 14.5 | 1.9 | R | 6.8 | 1.8 | R |
| E138 | 28.0 | 3.0 | MR | 31.0 | 2.1 | MR | 36.0 | 2.6 | MR | 33.4 | 3.0 | MR |
| E187 | 58.0 | 2.5 | MR | 67.0 | 2.1 | MR | 54.0 | 2.8 | MR | 0 | 1.0 | HR |
| E230 | 19.0 | 3.0 | MR | 37.0 | 1.9 | MR | 67.5 | 3.0 | MR | 17.7 | 2.5 | MR |
| E304 | 41.0 | 3.0 | MR | 34.3 | 2.3 | MR | 55.5 | 2.9 | MR | 6.3 | 1.5 | MR |
| E017 | 32.0 | 3.0 | MR | 39.5 | 2.8 | MR | 46.5 | 2.9 | MR | 33.4 | 2.3 | MR |
| E163 | 54.3 | 4.0 | S | 100 | 3.8 | S | 63.5 | 3.8 | S | 76.0 | 4.0 | S |
| E213 | 82.0 | 4.0 | S | 100 | 4.5 | HS | 80 | 3.5 | S | 64.7 | 4.0 | S |
| E147 | 100 | 5.0 | HS | 75.0 | 3.5 | S | 66.5 | 3.2 | S | 100 | 4.5 | HS |
| E169 | 100 | 5.0 | HS | 71.5 | 4.0 | S | 58.5 | 3.1 | S | 100 | 5.0 | HS |
| CV | 24.0 | 10.8 | 24.8 | 16.5 | 23.1 | 12.3 | 57.0 | 22 | ||||
| LSD | 29.3 | 0.79 | 36.9 | 1.01 | 32.7 | 0.73 | 32.0 | 0.84 | ||||
| SE | 1.57 | 0.05 | 1.52 | 0.05 | 1.32 | 0.04 | 1.9 | 0.05 | ||||
| Trait | Days to 50% Flowering (Days) | Severity (1–5) | Incidence (%) | Hundred Seed Weight (g) | Plant Height (cm) | Panicle Length (cm) | Seedling Vigour (1–5) |
|---|---|---|---|---|---|---|---|
| Severity (1–5) | 0.11 * | ||||||
| Incidence (%) | 0.24 ** | 0.6 ** | |||||
| Hundred seed weight (g) | −0.21 ** | −0.1 * | 0.01 ns | ||||
| Plant height (cm) | −0.11 ns | −0.28 ** | −0.32 ** | 0.09 ns | |||
| Panicle length (cm) | 0.17 ** | −0.05 ns | −0.07 ns | −0.11 ns | 0.39 ** | ||
| Seedling vigour (1–5) | 0.14 * | −0.24 ** | 0.27 ** | 0.04 ns | −0.04 ns | −0.03 ns | |
| Pollen quantity (1–10) | 0.02 ns | −0.18 ** | −0.16 ** | −0.08 ns | −0.12 * | −0.02 ns | 0.01 ns |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kebede, D.; Dramadri, I.O.; Rubaihayo, P.; Odong, T.; Edema, R. Resistance of Sorghum Genotypes to Ergot (Claviceps Species). Agriculture 2023, 13, 1100. https://doi.org/10.3390/agriculture13051100
Kebede D, Dramadri IO, Rubaihayo P, Odong T, Edema R. Resistance of Sorghum Genotypes to Ergot (Claviceps Species). Agriculture. 2023; 13(5):1100. https://doi.org/10.3390/agriculture13051100
Chicago/Turabian StyleKebede, Dejene, Isaac Onziga Dramadri, Patrick Rubaihayo, Thomas Odong, and Richard Edema. 2023. "Resistance of Sorghum Genotypes to Ergot (Claviceps Species)" Agriculture 13, no. 5: 1100. https://doi.org/10.3390/agriculture13051100






