Abstract
Irrigation is important in many crop production systems. However, irrigation water can be a carrier of plant pathogens that can enter the system and spread to fields, resulting in crop damage and yield losses. The Lower Rio Grande Valley of South Texas is an important area for agricultural production which depends on the Rio Grande River as a source of water for irrigation. Thus, the presence of plant pathogens in the Rio Grande River could have important implications for crop productivity in the region. Cultured-based methods and molecular identification methods are used for monitoring plant pathogens in irrigation water. However, these methods are labor-intensive and just detect targeted pathogens. To overcome these limitations, in this study, the ITS2 amplicon metagenomic method was applied for evaluating the fungal diversity, composition, and presence of fungal plant pathogens in irrigation water from the Rio Grande River as it leaves the water reservoir (WR) and it arrives at an irrigation valve at a farm (FA). Results from the Shannon (WR = 4.6 ± 0.043, FA = 3.63 ± 0.13) and Simpson indices (WR = 4.6 ± 0.043, FA = 3.63 ± 0.13) showed that there are significant differences in the fungal diversity and community structure between the two locations and the PCA analysis showed a clear differentiation between both fungal communities. Several OTUs identified in both locations included potential plant pathogens from diverse genera including Cladosporium, Exserohilum, and Nigrospora, while others such as Colletotrichum and Plectosphaerella were found only in one of the two locations assessed. This work indicates that microbes, including plant pathogens, may enter or exit throughout the irrigation-water distribution system, thereby modifying the microbial community composition along the way. Understanding the dynamics of plant pathogen movement in irrigation water systems can help growers identify risk factors to develop measures to mitigate those risks. This study also shows the usefulness of the metagenomic approach for detecting and monitoring plant pathogen in irrigation water.
1. Introduction
In many regions of the world, rivers are used as a source of irrigation water to enhance agricultural production, economic development, urbanization, etc. In South Texas, USA, limited water resources based on the Rio Grande River (RGR) support irrigation-dependent agriculture, rapid population growth and urbanization, and sensitive wildlife and marine habitats, while facing frequent periods of severe drought [1]. The suitability of river water for irrigation is affected by factors such as the amount of minerals, salts, heavy metals, or other contaminants in the water [2]. The RGR experiences water quality issues such as excessive loading of sodium, minerals, and nutrients which can vary during the year based on environmental events [3]. Although the water in the RGR along the Lower Rio Grande Valley (LRGV) is ranked as permissible for irrigation by several water quality indices [3], it may contain plant fungal pathogens that could be distributed through the irrigation system to crops.
Fungal plant diseases are major problems in agriculture, resulting in crop damage and yield loss. Many fungal pathogens are dispersed through water from irrigation, increasing the risks of crop production losses [4]. Important phytopathogens, such as the Oomycetes Phytophthora and Pythium, produce swimming zoospores and can cause substantial agricultural problems [4]. Other fungal phytopathogens, such as Fusarium and Alternaria, can also be found in irrigation water, although their long-term survival and infectivity have not been determined [4]. In greenhouses or systems where water is recycled for irrigation, the risk of pathogen contamination is amplified. Intensive care of horticultural crops is needed in closed and open systems to manage diseases, especially when plant pathogens are introduced from irrigation water. Pathogen distribution via irrigation water is very rapid; thus, the risks associated with pathogen presence and abundance in irrigation water have been recognized as a major crop health concern [4]. Contaminated irrigation water with fungal plant pathogens can be a major factor in disease distribution and spread in open fields [4,5]. Water from many sources, such as open reservoirs and ponds, can carry fungal plant pathogen propagules which are introduced by runoff water filled with crop debris from agricultural fields [6,7,8,9,10]. Irrigation water from the RGR is distributed to crops in South Texas from settling basins in the irrigation district’s pump stations, and then transported through open or underground canals to farms [11,12]. Because the RGR, as well as its tributary rivers, Devils and Pecos, receive runoff water from agricultural fields, their water can have an increased level of plant pathogens that can be later distributed to new fields through irrigation water. Thus, it is important to detect the presence of fungal plant pathogens in RGR water used for irrigation.
There are several methods available for detecting, screening, surveilling, and identifying microorganisms in water samples. Cultured-based methods such as baiting are a common approach for monitoring plant fungal pathogens [13]. However, this method has many drawbacks, including the inability to detect slow-growing or unculturable microbes, which may cause inconclusive results. Molecular identification methods, including quantitative PCR (qPCR) and loop-mediated isothermal amplification (LAMP), which sometimes can overcome the limitations of cultured-based methods, are the most sensitive for detecting and quantifying microorganisms, but this method can detect only one or a few target microbes in a single reaction mixture [14,15]. High-throughput sequencing can overcome these drawbacks and has the potential to be an efficient method for the detection and monitoring of plant pathogens in water. It can be used for identifying potential harmful microorganisms, indicating the need for more specific analyses, while also providing comprehensive data on the microorganisms that are naturally occurring in an agricultural and environmental sample. Shortgun metagenomics is the untargeted high-throughput sequencing of the entire DNA of a sample. This technique has the significant advantage of detecting all microorganisms present in a sample, including both fungi and bacteria. In addition, shortgun metagenomics can also return genomes of the detected pathogens, making it ideal for sequencing unculturable ones [16]. To date, shortgun and amplicon-based metagenomics are widely applied for detecting and monitoring plant pathogens. Most studies have focused on surveillance of soilborne and airborne plant pathogens [17,18,19,20]; however, there are still few reports about applying the high-throughput sequencing approach for detecting plant pathogens in irrigation water [21]. Therefore, we hypothesized that a metagenomics approach may be useful for identifying potential pathogens in irrigation water.
With the goal of identifying if water from the RGR used for irrigation was carrying plant pathogens, and if those plant pathogens were transported to farms, in this study, an amplicon-based (ITS) metagenomics approach was used for detecting fungal plant pathogens in irrigation water collected from two locations, the water reservoir and the irrigation valve in a farm. The detection of fungal plant pathogens in the water reservoir and at the irrigation site were recorded and the populations of fungal plant pathogens from both locations were compared to determine the abundance, similarities, and differences between fungal populations.
2. Materials and Methods
2.1. Collection of Irrigation Water Samples
Two locations were selected for the sampling of irrigation water from the RGR: irrigation water that was emerging from an irrigation valve at a citrus orchard located at the Texas A&M University Kingsville Citrus Center South Research Farm (FA) (air temperature = 27 °C, precipitation = 0 (in), humidity = 70%), and from the water reservoir of the Mercedes irrigation district (WR) (air temperature = 29 °C, precipitation = 0 (in), humidity = 79%) from the Lower Rio Grande Valley region in South Texas. Water samples were collected from running water using two new, disinfected 5-gallon buckets during the early morning hours at 10:00 a.m. To avoid cross contamination, the buckets were washed with 70% ethanol and rinsed with sterile water before use. Water samples were collected the same day in October 2020 from both sites, transported to the laboratory, and processed within 1 h of collection following Redekar et al.’s method (2019) with minor modifications [21]. Briefly, four and a half liters of water samples were divided into three repeat groups for filtering through a 75-micron nylon screen into sterile flasks to remove large debris. The samples collected from the water reservoir were labelled as WR1, WR2, and WR3, and samples from the farm were labeled FA1, FA2, and FA3. Each filtered water sample was then vacuum-infiltrated onto 5 cellulose ester membranes with 5 µm pore size (Millipore-Sigma, Burlington, MA, USA). The membrane filters were cut into small pieces and placed in a 15 mL centrifuge tube. Then, 800 µL of extraction buffer (3% CTAB, 100 mM Tris-HCl, 20 mM EDTA, 1.4 M NaCl, 5% PVP, pH 8.0) was transferred to the tubes and stored in a −20 °C freezer before continuing with DNA extraction.
2.2. DNA Extraction
Tubes containing the membranes in extraction buffer were incubated at 65 °C in a water bath for 30 min. After incubation, the filter papers and extraction buffer from each sample were divided and placed into two 2 mL tubes. 400 µL of 5M potassium acetate (KoAc)/acetic acid were added to each tube, mixed, and incubated for 5 min on ice. The tubes were then removed from the ice and centrifuged at 13,000 rpm for 5 min. The supernatant was removed from each tube and the two supernatants corresponding to each water sample were combined and transferred into a new 2 mL tube. Next, the samples were centrifuged for 5 min at maximum speed. An amount of 600 µL of supernatant was transferred to a new 2.0 mL tube, and 900 µL of ice-cold isopropanol was added and mixed gently for DNA precipitation. Then, 700 µL was pipetted onto a spin column (Genessee Scientific, San Diego, CA, USA), followed by centrifugation at 8000 rpm for 1 min. These steps were repeated until all the supernatant was processed via centrifugation. Once the last centrifugation was complete, the spin column was placed in a clean 2 mL tube with the cap open and all the flowthrough was discarded. To wash the column, 500 µL of 70% ethanol was added to the column and centrifuged at 10,000 rpm for 1 min, and the flowthrough was discarded. This was performed twice, and an extra spin in the centrifuge was performed to completely remove the ethanol from the column. The column was placed into new sterile 1.5 mL tubes and the DNA was eluted through adding 50 µL of AE buffer and centrifuging at 8000 rpm for 2 min. This step was repeated with 20 µL of AE buffer. The DNA was kept at −20 °C.
2.3. High-Throughput Amplicon Sequencing
Genomic DNA concentration and purity were determined on a NanoDrop ND-2000 (NanoDrop Technologies, Wilmington, DE, USA) spectrophotometer. For fungal community analysis, the internal Transcribed Spacer (ITS1-2) sequence was amplified with primer set ITS1 (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS-2 (5′-GCTGCGTTCTTCATCGATGC-3′) [22,23]. For multiplex sequencing, the primers were barcoded. PCR amplifications were performed in a 25 μL reaction with 5 μL 5× reaction buffer, 5 μL 5× GC buffer, 2.5 mM dNTPs, 10 μM of forward and reverse primers, 2 μL DNA template, 8.75 μL ddH2O, and 0.25 μL Q5 high-fidelity DNA polymerase (NEB, Ipswich, MA, USA). The PCR protocol was initiated with a denaturation step at 98 °C for 2 min, followed by 30 cycles at 98 °C for 15 s, 55 °C for 30 s, and 72 °C for 30 s, as well as a final extension at 72 °C for 5 min. To minimize bias, three PCR amplifications were pooled for analysis. PCR products of 350 bp were purified after gel electrophoresis with a DNA Gel Extraction Kit (Axygen, Union City, CA, USA). The sequencing of PCR amplicons of fungal DNA was carried out on the Illumina MiSeq sequencer from BGI Co., Ltd. (San Jose, CA, USA).
2.4. Data Processing
Raw sequences were assembled for each sample based on the unique barcode via the QIIME pipeline (Quantitative Insights Into Microbial Ecology, v1.8.0 qiime.org) [24]. Low-quality sequences were filtered following Gill’s method [25]. High-quality sequences were assembled into operational taxonomic units (OTUs) at 97% sequence identity via UCLUST [26]. Representative sequences from each OTU were used to establish the taxonomic classification through performing a search against the UNITE (Release 5.0, https://unite.ut.ee/ accessed on 4 November 2020) Database for fungal sequences [27].
2.5. Bioinformatics Analysis
Analysis of the sequences were conducted using QIIME (v1.8.0) and R packages (v3.2.0). OTU tables were separated according to WR and FA type and normalized using total sum scaling (TTS). Diversity and richness indices including alpha, Chao1, ACE, Shannon, and Simpson were calculated using the OTU table in QIIME (v1.8.0). Because the data did not follow a normal distribution, statistical differences were calculated using nonparametric Kruskal–Wallis one-way ANOVA followed by Dunn’s multiple-comparisons post hoc test (p < 0.05). To determine the variation amongst the fungal communities and structures from WR and FA samples, a weighted UniFrac distance was calculated in Mothur (v.1.25.1). PCoA (Principal Coordinate Analysis) was conducted on distance metrics, and coordinates were used to produce a 2D graph [28]. The Venn diagram was constructed using R packages (v3.2.0). Relative abundances of fungal OTUs between WR and FA samples were analyzed using one-way ANOVA (p < 0.05) using SPSS Version 16.0 (SPSS Inc., Chicago, IL, USA).
3. Results
3.1. Diversity of Fungal Communities in Irrigation Water from Two Locations: Mercedes Irrigation District (WR) and South Farm (FA)
High-quality sequences of 899,335 reads (average connect ratio = 96.30 ± 2.36%) for ITS2 were obtained for an average of 940,496 fungal ITS2 reads (Table S1). In total, 1793 OTUs of fungi from six samples in both locations were acquired after discarding OTUs less than 0.001% of total sequences cross all samples. In total, 1286 and 1265 OTUs were detected in three samples from WR and FA, respectively. In addition, there were no statistically significant differences in the number of OTUs, or in the richness indices Chao-1 and ACE, in fungal communities between WR and FA. However, significant differences were detected in the Shannon and Simpson diversity indices between locations (Table 1). The Shannon index was statistically higher in WR samples than the FA samples (p < 0.05). In contrast, the Simpson index was lower in WR than the FA samples (p < 0.05).

Table 1.
Fungal diversity and richness indices of irrigation water collected at the water reservoir (WR) and at the farm (FA). Different letters represent significant differences. All data were analyzed with Duncan’s multiple range tests using SPSS software.
3.2. Differences in Composition and Structure between WR and FA
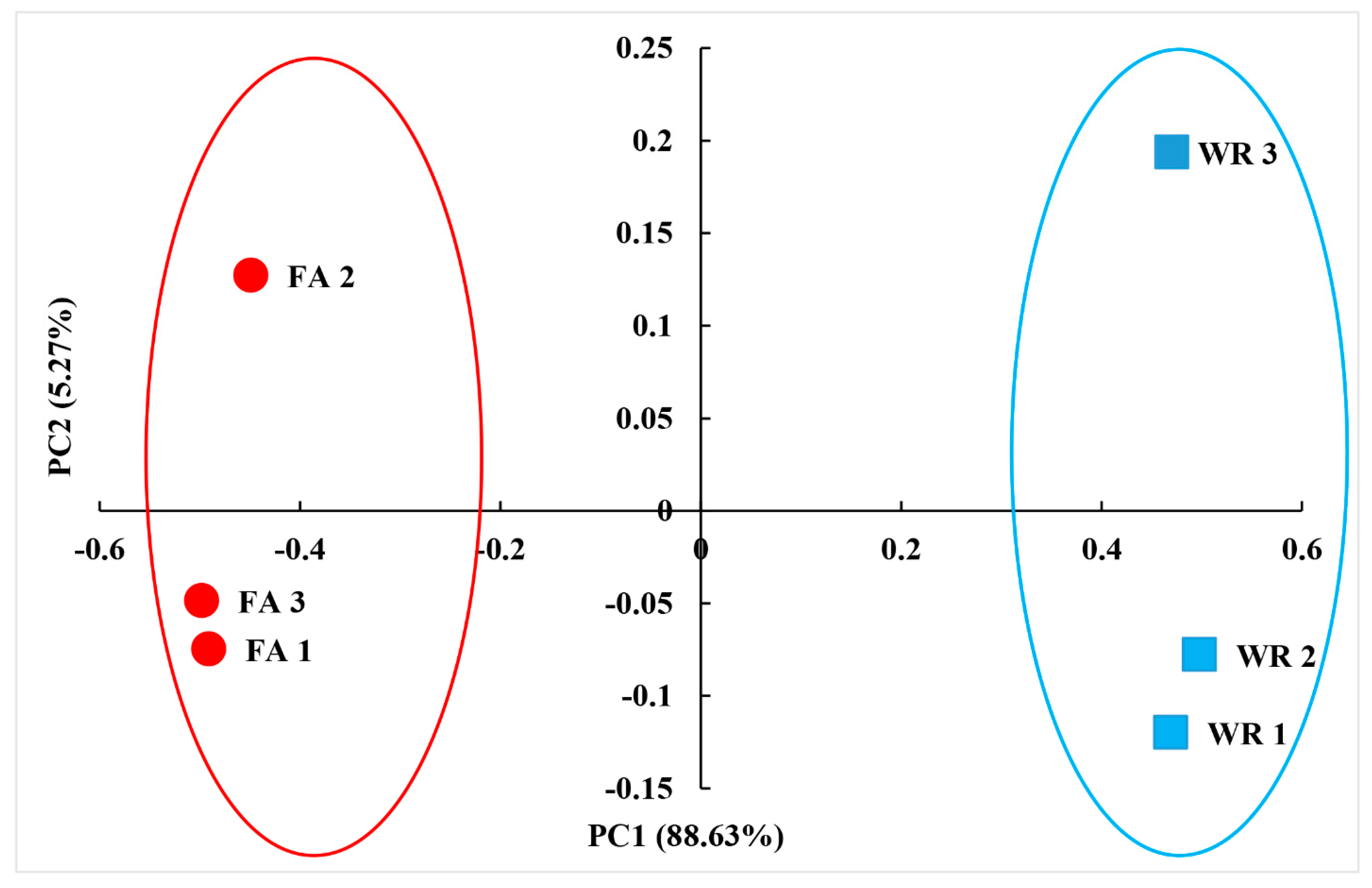
The similarity of the fungal communities in both two locations (WR and FA) was measured using weighted Unifrac PCoA. The results indicated that the fungal communities associated with the water from the water reservoir (WR) and the water collected at the farm (FA) were clearly separated (Figure 1) and statistically different from each other (analysis of similarity, R = 1, p = 0.014). The principal coordinates PC1 accounted for 88.63% of the variance, and PC2 represented 5.27% of the variance (Figure 1). The cumulative contribution of the variances represents 93.90% of the variance.
Figure 1.
UnFrac-weighted principal coordinate (PC) analysis of fungal community structures in irrigation water from water reservoirs (WR) and irrigation valve (FA) in South Texas, USA. Analysis of similarities was conducted to evaluate the significant differences in microbial community using QIIME (Version 1.8.0) (p < 0.05).
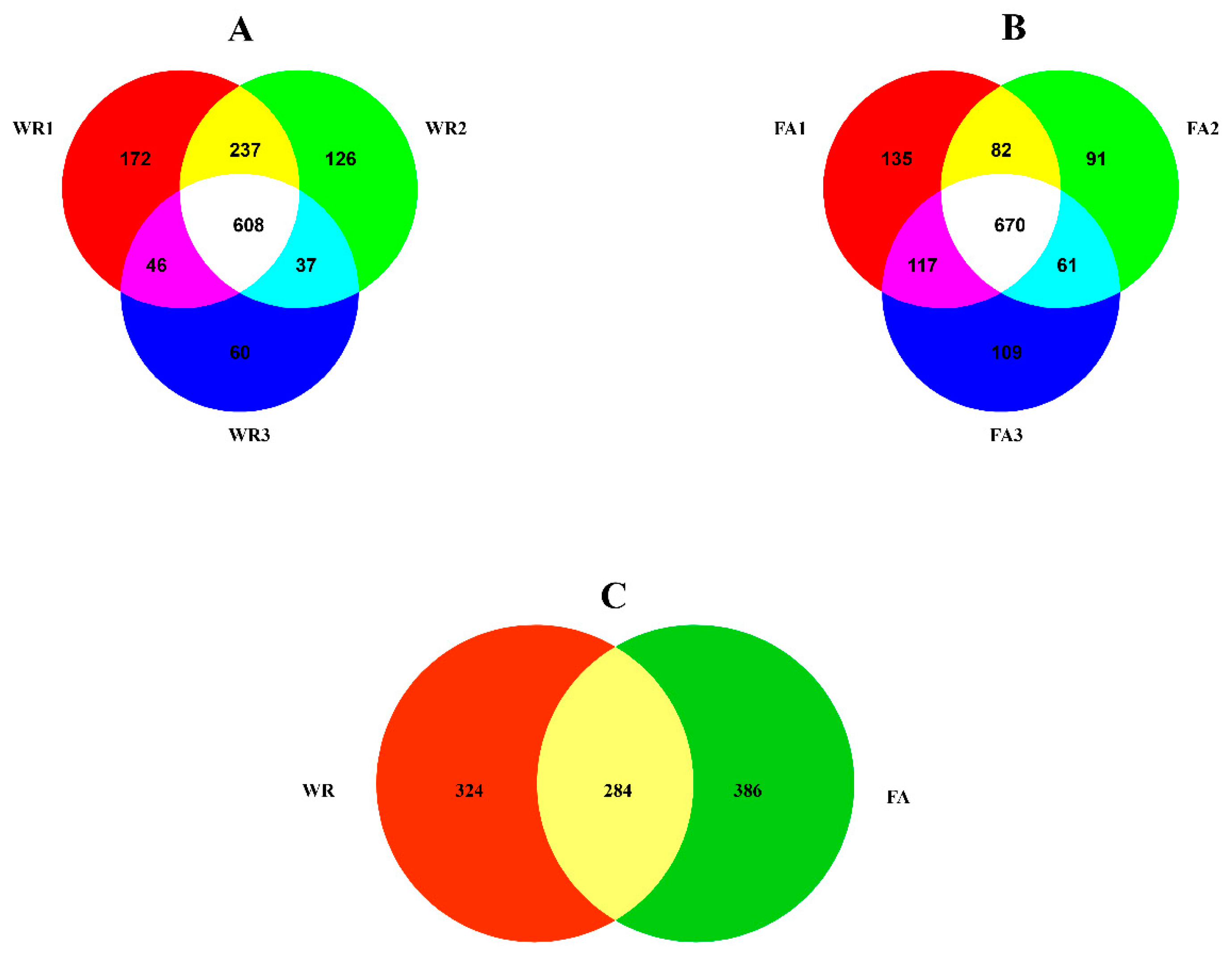
In this work, 608 and 670 OTUs were detected in WR and FA samples, respectively (Figure 2A,B). In total, 284 OTUs were shared between WR and FA samples, while 324 and 386 OTUs were just present in WR or FA samples, respectively (Figure 2C). Of the 284 OTUs that were present in both WR and FA samples, 8 were identified at the species level (OTU672, OTU456, OTU34, OTU199, OTU213, OTU251, OTU112, OTU589), 2 at the genus level (OTU381, OTU162), 1 at the order level (OTU45), and 2 at the phylum level (OTU294, OTU125) (Table S2). The relative abundance of six OTUs (including OTU456, OTU294, OTU125, OTU672, OTU45, and OTU381) in FA samples was significantly higher than in WR samples (Table S2). In WR samples, 30 of the 324 special OTUs were identified at least at the phylum level, and 50 of the 386 OTUs in FA samples were confirmed at least at the phylum level (Tables S3 and S4).
Figure 2.
Venn diagrams indicating the number of OTUs obtained from water samples. (A): Number of OTUs obtained in samples from the water reservoir (WR). (B): Number of OTUs obtained in water samples collected at the irrigation valve at the farm (FA). (C): Number of OTUs co-present in irrigation water samples collected at the water reservoir (WR) and irrigation valve at the farm (FA).
The phyla that were more abundantly represented in both locations were Chytridiomycota followed by Ascomycota, Basidiomycota, and Monoblepharomycota. The Chytridiomycota phylum showed large relative abundances in both locations, with 4.4% in WR and 1.9% in FA. Relative abundance between both locations showed that there was greater relative abundance of Basidiomycota in the FA as opposed to the WR location, which shows a minimal representation from the Basidiomycota at less than 0.1%. The classes with relative abundance higher than 0.01% are listed on Table 2 for both locations. Two classes belonging to Chytridiomycota, GS13 (p = 0.0161) and Rhizophydiomycetes (p = 0.0373) in WR were significantly higher than in FA (Table 2). Moreover, the classes Cladochytriomycetes and Tremellomycetes were only present in WR and FA, respectively (Table 2).

Table 2.
Dominant classes (relative abundance more than 0.01%) in irrigation water collected at the water reservoir (WR) and at the farm (FA).
At the family level, five dominant families (relative abundance of more than 0.01%), including Chytridiaceae, Cladosporiaceae, Pleosporaceae, Sanchytriaceae, and Stachybotryaceae, were present in both WR and FA, without significant differences between locations (Table 3). In WR samples, Aquamycetaceae, Lentitheciaceae, Nowakowskiellaceae, Plectosphaerellaceae, and Trichosphaeriaceae were the dominant families, while in FA samples, Alphamycetaceae, Aureobasidiaceae, Bulleribasidiaceae, Metschnikowiaceae, and Microdochiaceae were the dominant families (Table 3).

Table 3.
Dominant families (relative abundance more than 0.01%) in irrigation water collected at the water reservoirs (WR) and at the farm (FA).
Based on the 48 OTUs identified at the species level, the most abundant species was the Chytridiomycota Zygophlyctis melosirae, which was associated with 17 OTUs (Tables S2–S4). Moreover, eight OTUs were associated with plant pathogenic fungi representing six species (Table 4). Three of these OTUs were present in both locations, while the other five were present either at WR or FA (Table 4). The relative abundance of OTU456 was significantly different between the WR and FA samples (Table 4).

Table 4.
OTUs associated with plant pathogenic fungi in irrigation water collected at the water reservoir (WR) and at the farm (FA).
4. Discussion
To assess the risks that fungal phytopathogens in river water pose to agriculture, we focused our work on the irrigation water that is supplied by the Rio Grande River to the LRGV agricultural system. In this study, we used ITS2 amplicon metagenomic analysis to evaluate the fungal diversity and composition of the irrigation water from the Rio Grande River as it leaves the water reservoir and it arrives at an irrigation valve at the farm level. This is the first report that utilizes a metagenomic approach to assess fungal microorganisms in river water, with the aim to identify fungal phytopathogens that could pose a risk to agricultural production. This work shows that the communities and structure of the fungal microbiome found in the water reservoir (WR) change substantially from the microbiome found at the irrigation valve at the farm (FA). This study showed a significant difference in the Shannon and Simpson indices between locations, indicating differences in the diversity of species present. The Shannon index of the water reservoir (WR) had a higher value than the water collected at the farm (FA). This could indicate that WR water has lower diversity than FA water. The Simpson index showed opposite results. The WR had lower Simpson diversity values than that of the FA, indicating that there are more species identified at the WR (higher richness), but that higher species evenness was present in the water collected at the farm. The PCA analysis showed a clear differentiation of the fungal communities between the locations at the WR and the FA, indicating loss or gain of species during the distribution of water from the source (WR) to the farm (FA). These results suggest that microbes may enter water systems at several points along the distribution path and change the water microbial communities and structures. Any irrigation method in which the water comes in contact with soil or plant debris has an in an increased potential to acquire new microbes into its water path [4]. Furrow flooding and runoff from other irrigation methods can carry microbes from soil throughout the water area [4]. Therefore, several fungal or bacterial microbes may enter irrigation water from WR to FA through these pathways, which resulted in the change of the communities and structure of the fungal microbiome from the WR to the FA. Moreover, the water from the WR traveled 10 miles before reaching the irrigation valve at the farm (FA), being in contact with lines with plants and debris that could trap or release microbes into the water.
Several OTUs associated with fungal plant pathogens were detected and identified in the water reservoir (WR) and the irrigation valve at the farm (FA). In both sampling locations, OTU112, OTU213, and OTU456 were present (Table 4). OTU112 was identified as Cladosporium cladosporioides, which is associated with strawberry blossom blight [29] and Cladosporium rot of grapevine disease [30]. OTU213 was identified as Exserohilum rostratum, causing Exserohilum leaf spot on tiger grass [31] and pineapple leaf spot disease [32]. OTU456 was identified as Nigrospora oryzae, which causes leaf spots and rot diseases in many important crops including rice, wheat, sorghum, barley, maize, and cotton [33,34,35,36,37]. OUT316 was only detected in the WR samples and was associated with Plectosphaerella cucumerina, which causes root and collar rots in melon, pepper, and tomato [38,39]. Other potential fungal plant pathogens were only detected in FA samples, including Curvularia verruculosa and Colletotrichum sublineola. Curvularia verruculosa is a common plant pathogen that can cause leaf spot disease in many plants including bean, cotton, and grape [40,41,42]. Anthracnose caused by Colletotrichum sublineola is an important sorghum disease worldwide [43,44]. Therefore, in this study, the amplicon-based metagenomics method was efficient for detecting fungal plant pathogens, compared with culture-based methods [13] and molecular identification methods [14,45,46]. However, metagenomics approaches still have limitations that should be taken into account in future research. For instance, the presence of DNA from dead cells can detect the presence of specific genera, but that does not mean viability. A novel DNA-binding dye, propidium monoazide (PMA), can differentiate living and dead cells, and PMA coupled with metagenomics approaches has been applied for investigating microbiomes to exclude nonviable microorganisms [47,48]. Moreover, two different versions of the database, UNITE Version 5.0 and UNITE Version 8.0, were used for analyzing metagenomics data in this study. In total, 1793 OTUs were detected in all water samples using Version 5.0, while just 639 OTUs were found in Version 8.0. Meanwhile, the number of OTUs in the WR (OTUs = 515.33 ± 39.1) and FA (OTUs = 500 ± 13.61) samples were also smaller using Version 8.0, relative to the 940.67 ± 166.54 and 955 ± 50.03 OTUs obtained for WR and FA, respectively, using Version 5.0. Although UNITE Version 8.0 comprises more fungal ITS sequences than UNITE Version 5.0, they may be incomplete or yield inconsistent results due to taxonomic reclassifications [49]. This may be reason why more OTUs were detected using Version 5.0 compared with Version 8.0. In addition, the objective of this study was to identify potential fungal plant pathogens in irrigation water, and if fewer OTUs are obtained from the metagenomics data, several OTUs related to fungal pathogens may be lost. Acquiring more OTUs and more information from metagenomics data was beneficial for identifying plant fungal pathogens in irrigation water. Therefore, UNITE Version 5.0 was selected for analyzing data in this study. For metagenomics approaches, the taxonomic attribution of each sequence depends upon alignment to a database; thus, the most comprehensive database is Genbank Uniprot, which is not revised and not always reliable [50]. Therefore, users must ensure the accuracy of sequences regarding the target plant pathogens in databases. In addition, the application of metagenomics approaches is also limited by the expertise required to properly manage sequencing data. Even if much software, such as QIME and Mothur [24,51], can be used remotely and with a user-friendly interface through the Galaxy web platform [52], an in-depth knowledge of the commands and operations necessary to perform the analyses remains necessary to choose the best methods and avoid mistakes.
5. Conclusions
To overcome the limitations of traditional methods for identifying plant pathogens in water, the metagenomic approach was useful for detecting fungal pathogens in river water used for irrigation. This study showed that there is a high diversity of fungal species present in the irrigation water coming from the Rio Grande River, including plant pathogens, which could also hold true for other rivers that supply water for irrigation. Many of the plant pathogen genera that were found in the water can be of economic importance in the region, as they can be pathogenic to the crops planted in the area. However, the threshold for their abundance in irrigation water is unknown. Moreover, it is important to acknowledge that the water source used for irrigation is very dynamic and the presence of plant pathogens that can affect agricultural crops can fluctuate between seasons and increase without warning. Further studies that assess the abundance of plant pathogens in the water in different seasons would be helpful to growers of the region so that they can assess the risks and continue producing important crops in the LRGV.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agriculture13071401/s1, Table S1: Statistics of Illumina sequencing of internal transcribed spacer (ITS) amplicons for irrigation water collected at the water reservoir (WR) and at the farm (FA). Table S2: The abundance and classification of OTUs that co-presented in irrigation water from water reservoirs (WR) and irrigation valve (FA). Table S3: The abundance and classification of OTUs that presented in irrigation water from water reservoirs (WR). Table S4: The abundance and classification of OTUs that presented in irrigation water from irrigation valve (FA).
Author Contributions
Conceived and designed the experiment: V.A. Performed the experiments: M.C. and C.Y. Analyzed the data: M.C. and C.Y. Contributed reagents/materials/analysis tools: V.A. Wrote and revised the manuscript: C.Y. and V.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Science Foundation Centers for Research Excellence in Science and Technology—Center for Sustainable Water Use (CREST-SWU) (Grant Number: 1914745) to V.A.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rister, M.E.; Sturdivant, A.W.; Lacewell, R.D.; Michelsen, A.M. Challenges and opportunities for water of the Rio Grande. J. Agric. Appl. Econ. 2011, 43, 367–378. [Google Scholar] [CrossRef]
- Malakar, A.; Snow, D.D.; Ray, C. Irrigation water quality—A contemporary perspective. Water 2019, 11, 1482. [Google Scholar] [CrossRef]
- De La Garza, M.N.; Ren, J.; Ancona, V. Spatiotemporal variations of hydrochemical characteristics of irrigation water: A case study of the Lower Rio Grande Valley, USA. Water Supply 2023, 23, 2001–2013. [Google Scholar] [CrossRef]
- Hong, C.; Moorman, G. Plant pathogens in irrigation water: Challenges and opportunities. Crit. Rev. Plant Sci. 2005, 24, 189–208. [Google Scholar] [CrossRef]
- Bewley, W.; Buddin, W. On the Fangus Flora of glass-house water supplies in relation to Plant Diseases. Ann. Appl. Biol. 1921, 8, 10. [Google Scholar] [CrossRef]
- Ali-Shtayeh, M.; MacDonald, J. Occurrence of Phytophthora species in irrigation water in the Nablus area (West Bank of Jordan). Phytopathol. Mediterr. 1991, 30, 143–150. [Google Scholar]
- McIntosh, D. The occurrence of Phytophthora spp. in irrigation systems in British Columbia. Can. J. Bot. 1966, 44, 1591–1596. [Google Scholar] [CrossRef]
- Pittis, J.; Colhoun, J. Isolation and identification of pythiaceous fungi from irrigation water and their pathogenicity to Antirrhinum, tomato and Chamaecyparis lawsoniana. J. Phytopathol. 1984, 110, 301–318. [Google Scholar] [CrossRef]
- Pottorff, L.P.; Panter, K.L. Survey of Pythium and Phytophthora spp. in irrigation water used by Colorado commercial greenhouses. HortTechnology 1997, 7, 153–155. [Google Scholar] [CrossRef]
- Shokes, F.; McCarter, A.M. Occurrence, dissemination, and survival of plant pathogens in surface irrigation ponds in southern Georgia. Phytopathology 1979, 69, 510–516. [Google Scholar] [CrossRef]
- Knight, L. A Field Guide to Irrigation in the Lower Rio Grande Valley; Texas Department of Transportation: Austin, TX, USA, 2009.
- Fernandez, L.; Robinson, J.R.; Lacewell, R.D.; Rister, M.E.; Ellis, J.R.; Sturdivant, A.W.; Stubbs, M.J. Evolution of Irrigation Districts and Operating Institutions: Texas, Lower Rio Grande Valley; Texas Water Resources Institute: College Station, TX, USA, 2003. [Google Scholar]
- Erwin, D.C.; Ribeiro, O.K. Phytophthora Diseases Worldwide; American Phytopathological Society Press: St. Paul, MN, USA, 1996. [Google Scholar]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Schena, L.; Ippolito, A.; Nigro, F. Real-Time PCR Detection and QUantification of Soilborne Fungal Pathogens: The Case of “Rosellinia necatrix”, “Phytophthora nicotianae”, “P. citrophthora” and “Verticillium dahlia”. Real Time PCR Detect. QUantification Soilborne Fungal Pathog. 2004, 43, 1000–1008. [Google Scholar]
- Duan, Y.; Zhou, L.; Hall, D.G.; Li, W.; Doddapaneni, H.; Lin, H.; Liu, L.; Vahling, C.M.; Gabriel, D.W.; Williams, K.P. Complete genome sequence of citrus huanglongbing bacterium,‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol. Plan Microbe Interact. 2009, 22, 1011–1020. [Google Scholar] [CrossRef] [PubMed]
- Franco Ortega, S.; Ferrocino, I.; Adams, I.; Silvestri, S.; Spadaro, D.; Gullino, M.L.; Boonham, N. Monitoring and surveillance of aerial mycobiota of rice paddy through DNA metabarcoding and qPCR. J. Fungi 2020, 6, 372. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, É.D.; Duceppe, M.-O.; Bérubé, J.A.; Kimoto, T.; Lemieux, C.; Bilodeau, G.J. Screening for exotic forest pathogens to increase survey capacity using metagenomics. Phytopathology 2018, 108, 1509–1521. [Google Scholar] [CrossRef]
- Prigigallo, M.I.; Abdelfattah, A.; Cacciola, S.O.; Faedda, R.; Sanzani, S.M.; Cooke, D.E.; Schena, L. Metabarcoding analysis of Phytophthora diversity using genus-specific primers and 454 pyrosequencing. Phytopathology 2016, 106, 305–313. [Google Scholar] [CrossRef]
- Català, S.; Perez-Sierra, A.; Abad-Campos, P. The use of genus-specific amplicon pyrosequencing to assess Phytophthora species diversity using eDNA from soil and water in northern Spain. PLoS ONE 2015, 10, e0119311. [Google Scholar] [CrossRef]
- Redekar, N.R.; Eberhart, J.L.; Parke, J.L. Diversity of Phytophthora, Pythium, and Phytopythium species in recycled irrigation water in a container nursery. Phytobiomes J. 2019, 3, 31–45. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zhang, D.; Cai, X.; Xia, L.; Luo, Y.; Cheng, X.; An, S. Significant alterations in soil fungal communities along a chronosequence of Spartina alterniflora invasion in a Chinese Yellow Sea coastal wetland. Sci. Total Environ. 2019, 693, 133548. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.R.; Pop, M.; DeBoy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic analysis of the human distal gut microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef]
- Gubler, W.; Feliciano, A.; Bordas, A.; Civerolo, E.; Melvin, J.; Welch, N. First report of blossom blight of strawberry caused by Xanthomonas fragariae and Cladosporium cladosporioides in California. Plant Dis. 1999, 83, 400. [Google Scholar] [CrossRef]
- Briceño, E.X.; Latorre, B.A. Characterization of Cladosporium rot in grapevines, a problem of growing importance in Chile. Plant Dis. 2008, 92, 1635–1642. [Google Scholar] [CrossRef]
- Brunings, A.M.; Datnoff, L.E.; Palmateer, A.J.; Locke, J.C.; Krause, C.R. Exserohilum leaf spot on tiger grass. Plant Health Prog. 2009, 10, 1. [Google Scholar] [CrossRef]
- Luo, Z.; He, F.; Fan, H.; Wang, X.; Hua, M.; Hu, F.; Li, X.; Liu, Z.; Yu, N. First report of leaf spot disease caused by Exserohilum rostratum on pineapple in Hainan province, China. Plant Dis. 2012, 96, 458. [Google Scholar] [CrossRef]
- Palmateer, A.; McLean, K.; Van Santen, E.; Morgan-Jones, G. Occurrence of Nigrospora lint rot caused by Nigrospora oryzae on cotton in Alabama. Plant Dis. 2003, 87, 873. [Google Scholar] [CrossRef]
- Zhang, L.; Li, S.; Tan, G.; Shen, J.; He, T. First report of Nigrospora oryzae causing leaf spot of cotton in China. Plant Dis. 2012, 96, 1379. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhao, K.; Zhao, Y.; Zhang, Y.; Fu, Q.; Huang, S. Nigrospora oryzae causing panicle branch rot disease on Oryza sativa (rice). Plant Dis. 2021, 105, 2724. [Google Scholar] [CrossRef] [PubMed]
- Fakhrunnisa, M.H.; Ghaffar, A. Seed-borne mycoflora of wheat, sorghum and barley. Pak. J. Bot. 2006, 38, 185–192. [Google Scholar]
- Standen, J. Nigrospora oryzae (B. and Br.) Fetch on Maize. Phytopathology 1945, 35, 552–564. [Google Scholar]
- Xu, J.; Xu, X.-D.; Cao, Y.-Y.; Zhang, W.-M. First report of greenhouse tomato wilt caused by Plectosphaerella cucumerina in China. Plant Dis. 2014, 98, 158. [Google Scholar] [CrossRef]
- Carlucci, A.; Raimondo, M.; Santos, J.; Phillips, A. Plectosphaerella species associated with root and collar rots of horticultural crops in southern Italy. Pers. Mol. Phylogeny Evol. Fungi 2012, 28, 34–48. [Google Scholar] [CrossRef]
- Wei, T.; Luo, M.; Zhang, H.; Jia, W.; Zeng, Y.; Jiang, Y. Curvularia verruculosa as new causal pathogen of common bean leaf spot disease in China. Crop Prot. 2022, 162, 106091. [Google Scholar] [CrossRef]
- Rajput, N.A.; Huo, C.; Cao, J.; Atiq, M.; Atif, R.M.; Lodhi, A.M.; Syed, R.N.; Sarfraz, S.; Hameed, A.; Zhao, Z. First report of Curvularia verruculosa causing leaf spot disease of grape (Vitis vinifera) in Afghanistan. J. Plant Pathol. 2020, 102, 1337. [Google Scholar] [CrossRef]
- Shirsath, L.P.; Patil, S.P.; Patil, U.K. Incidence of leaf spot disease on cotton caused by Curvularia verruculosa and role of its hydrolytic enzymes in pathogenesis. Physiol. Mol. Biol. Plants 2018, 24, 711–714. [Google Scholar] [CrossRef]
- Ali, M.; Warren, H. Physiological races of Colletotrichum graminicola on sorghum. Plant Dis. 1987, 71, 402–404. [Google Scholar] [CrossRef]
- Chala, A.; Tronsmo, A.; Brurberg, M. Genetic differentiation and gene flow in Colletotrichum sublineolum in Ethiopia, the centre of origin and diversity of sorghum, as revealed by AFLP analysis. Plant Pathol. 2011, 60, 474–482. [Google Scholar] [CrossRef]
- Sanzani, S.M.; Li Destri Nicosia, M.G.; Faedda, R.; Cacciola, S.O.; Schena, L. Use of quantitative PCR detection methods to study biocontrol agents and phytopathogenic fungi and oomycetes in environmental samples. J. Phytopathol. 2014, 162, 1–13. [Google Scholar] [CrossRef]
- Schena, L.; Li Destri Nicosia, M.; Sanzani, S.; Faedda, R.; Ippolito, A.; Cacciola, S. Development of quantitative PCR detection methods for phytopathogenic fungi and oomycetes. J. Plant Pathol. 2013, 95, 7–24. [Google Scholar]
- Stinson, L.F.; Keelan, J.A.; Payne, M.S. Characterization of the bacterial microbiome in first-pass meconium using propidium monoazide (PMA) to exclude nonviable bacterial DNA. Lett. Appl. Microbiol. 2019, 68, 378–385. [Google Scholar] [CrossRef]
- Erkus, O.; de Jager, V.C.; Geene, R.T.; van Alen-Boerrigter, I.; Hazelwood, L.; van Hijum, S.A.; Kleerebezem, M.; Smid, E.J. Use of propidium monoazide for selective profiling of viable microbial cells during Gouda cheese ripening. Int. J. Food Microbiol. 2016, 228, 1–9. [Google Scholar] [CrossRef]
- Weaver, D.; Gago, S.; Bromley, M.; Bowyer, P. The human lung mycobiome in chronic respiratory disease: Limitations of methods and our current understanding. Curr. Fungal Infect. Rep. 2019, 13, 109–119. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Ryberg, M.; Kristiansson, E.; Abarenkov, K.; Larsson, K.-H.; Kõljalg, U. Taxonomic reliability of DNA sequences in public sequence databases: A fungal perspective. PLoS ONE 2006, 1, e59. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Blankenberg, D.; Gordon, A.; Von Kuster, G.; Coraor, N.; Taylor, J.; Nekrutenko, A.; Team, G. Manipulation of FASTQ data with Galaxy. Bioinformatics 2010, 26, 1783–1785. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).