Lettuce Plant Trace-Element-Deficiency Symptom Identification via Machine Vision Methods
Abstract
1. Introduction
2. Materials and Methods
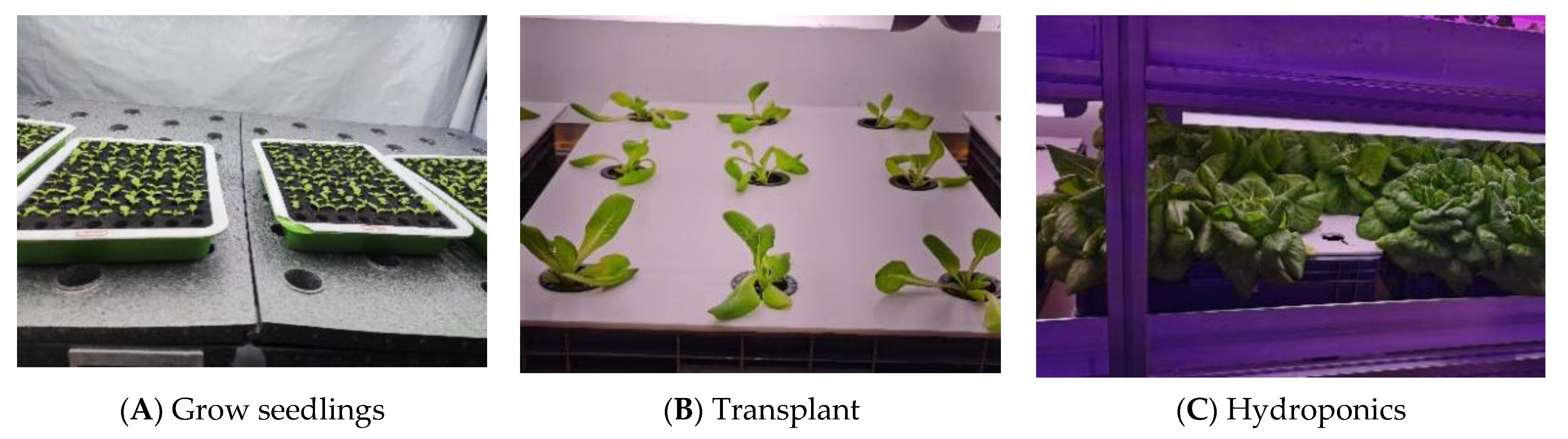
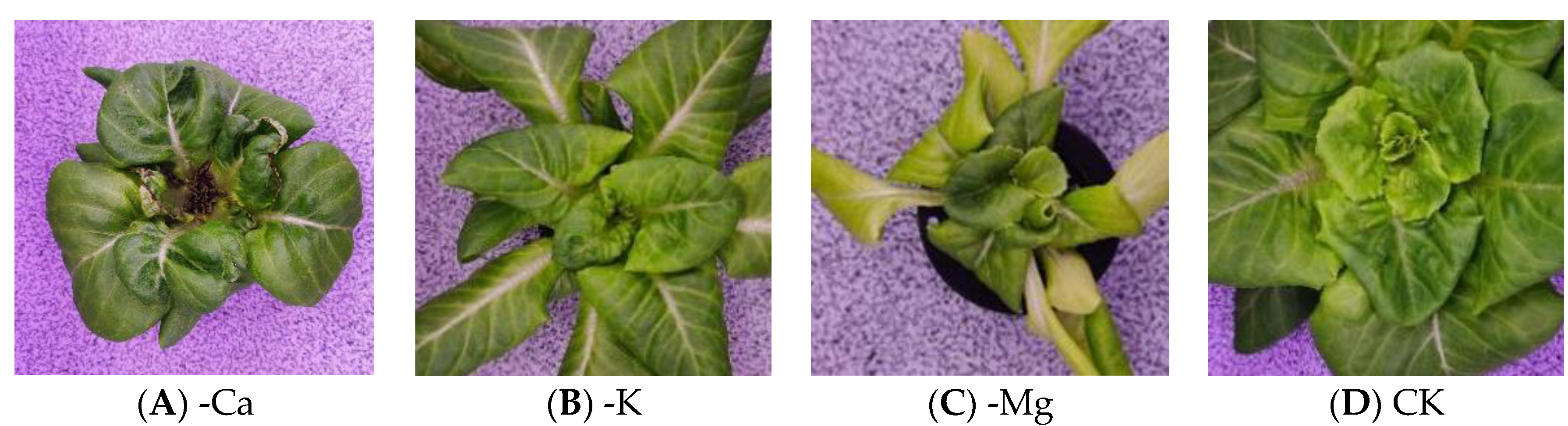
2.1. Deficiency Samples Preparation
2.2. Image Collection
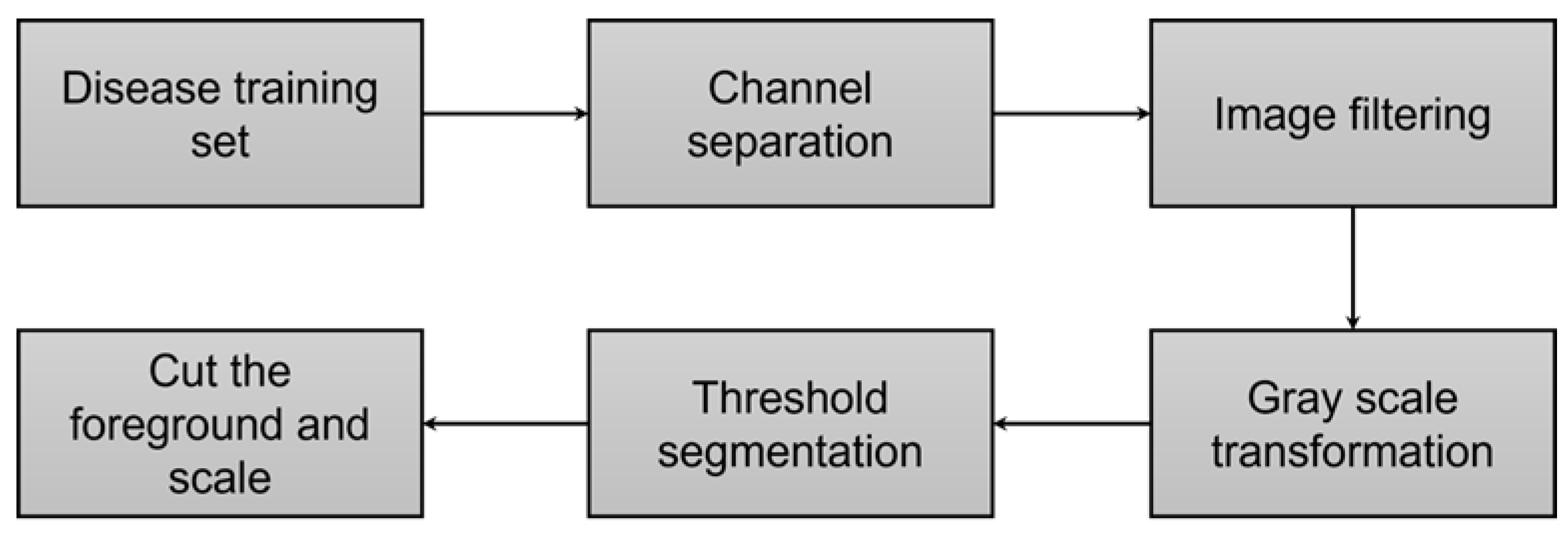
2.3. Image Pre-Processing
2.4. Disease Image Features Extraction
2.4.1. Texture Feature
2.4.2. Color Features
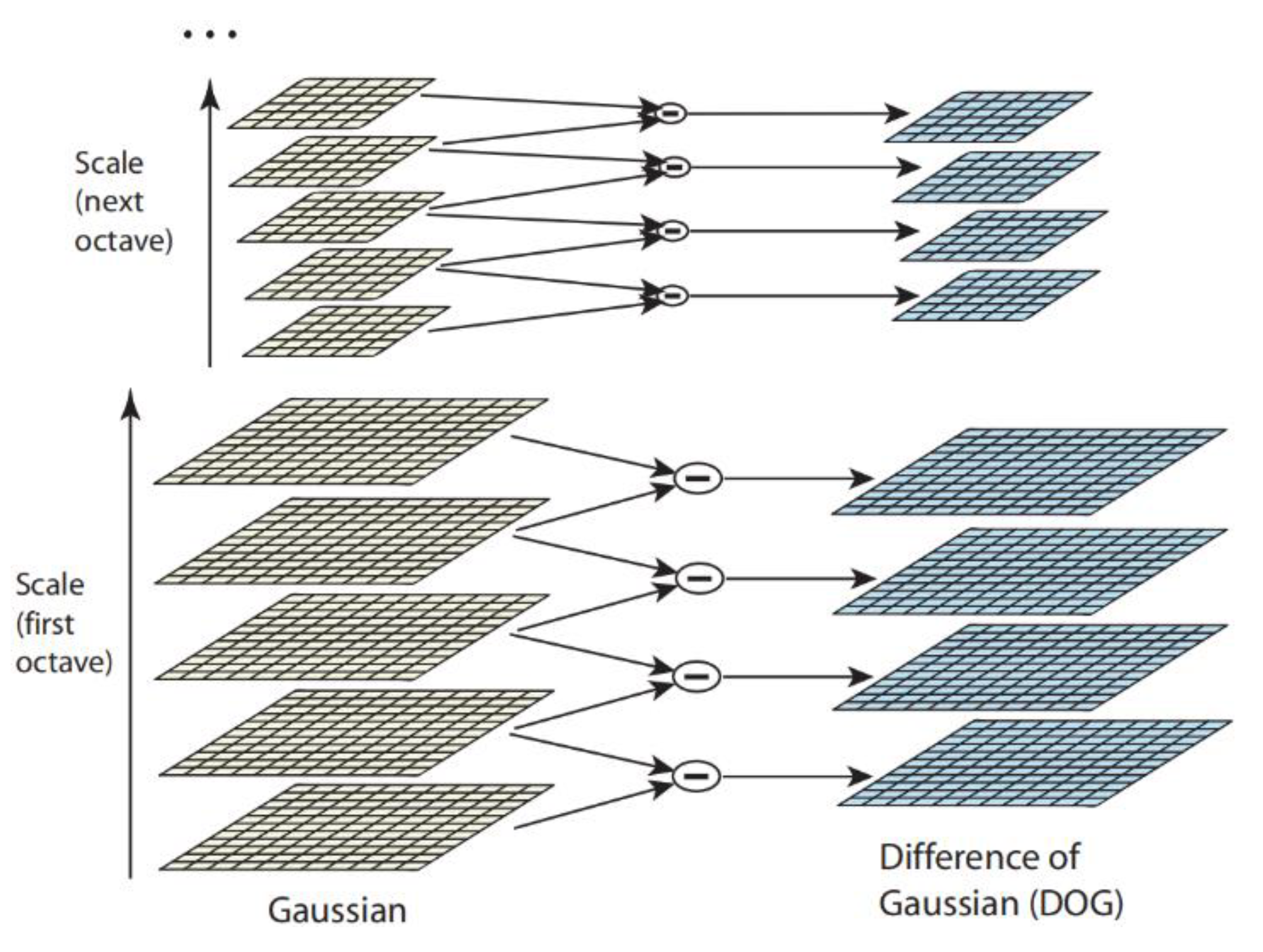
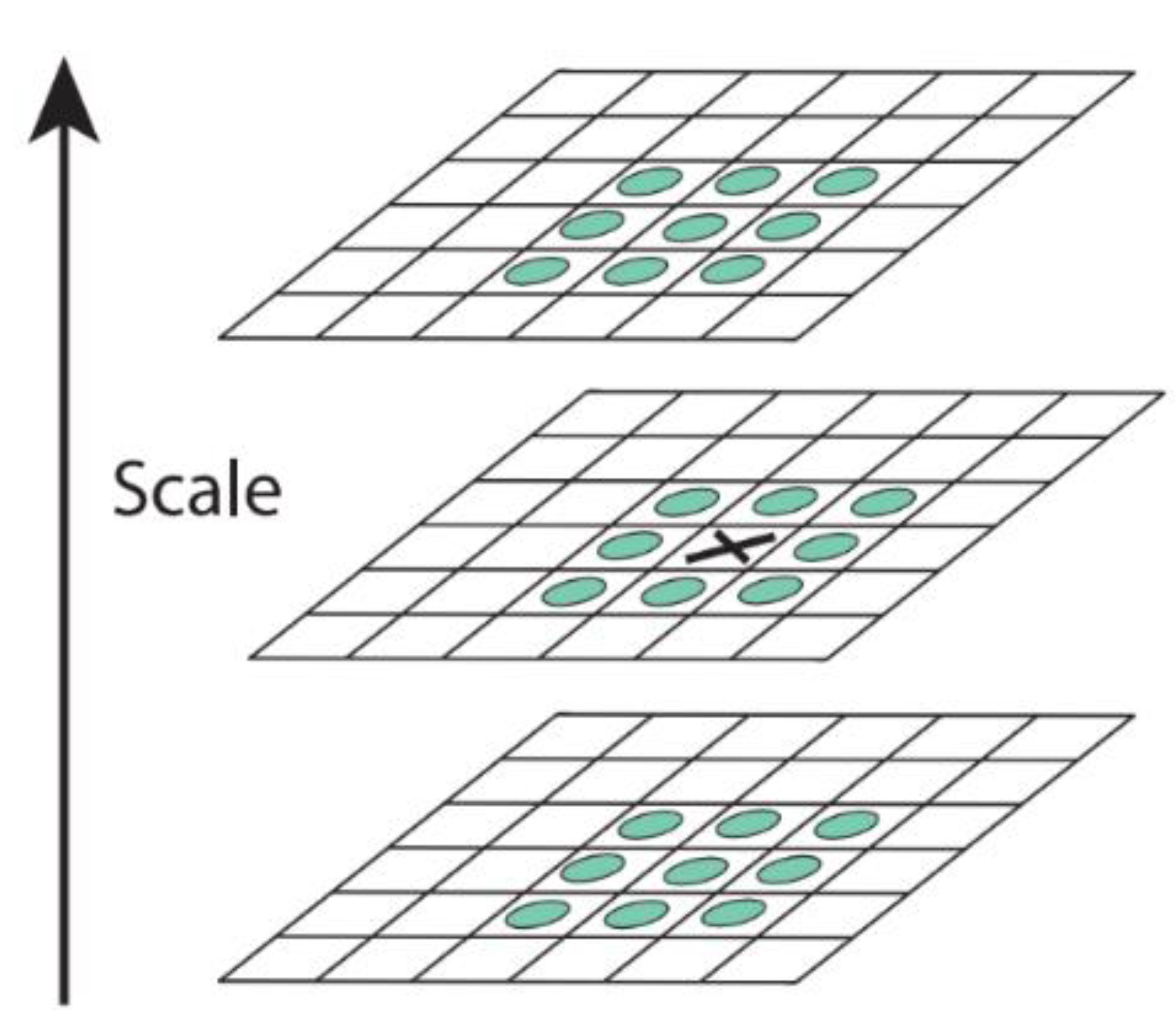
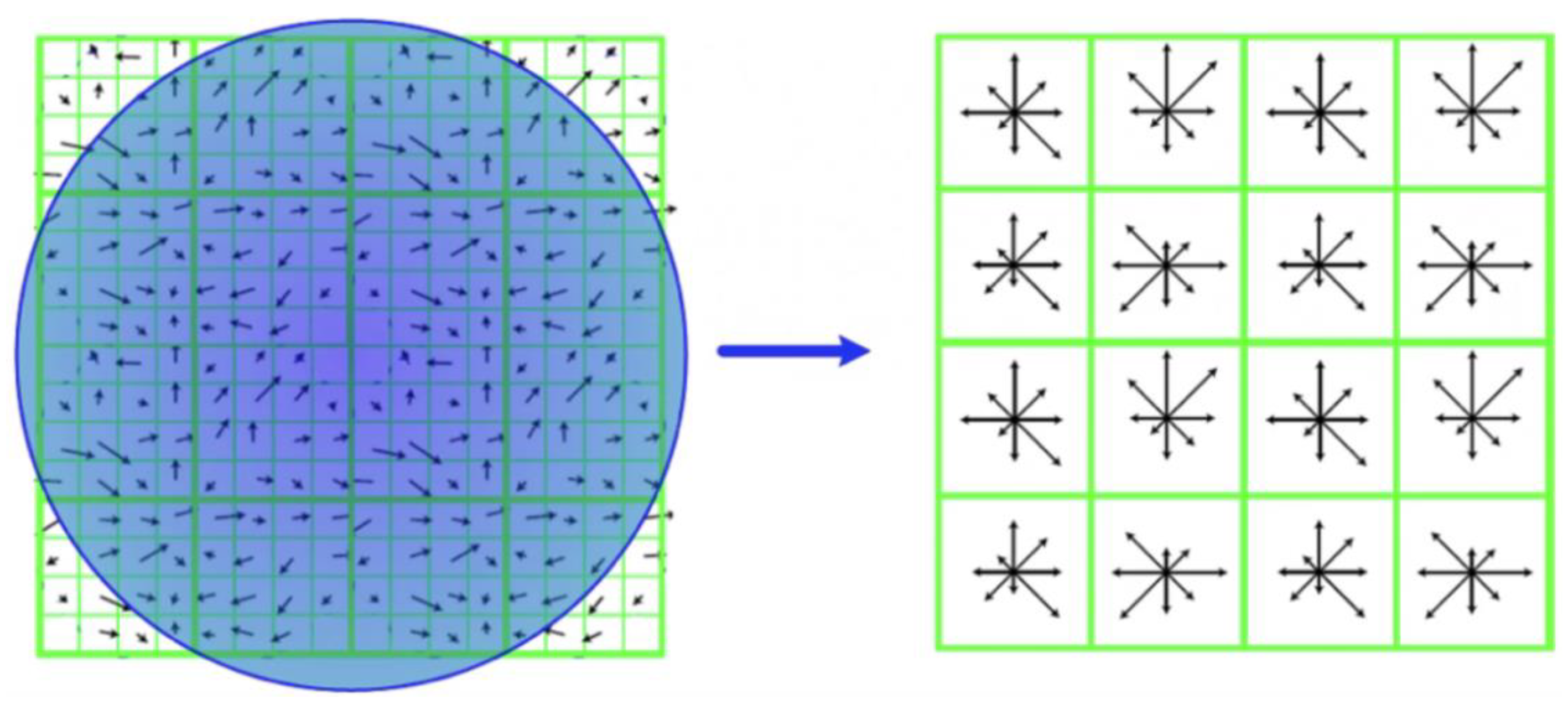
2.4.3. Scale-Invariant Feature
- (1)
- Constructing the scale space
- (2)
- Detecting the key points in the scale space
- (3)
- Locating the key points
- (4)
- Generating the feature description
2.5. Traditional Algorithm Classification
2.5.1. K Nearest Neighbor
2.5.2. Support Vector Machine
2.5.3. Random Forest
2.5.4. Data Set Parameter Setting and Training
2.6. Deep Learning Methods for Classification
2.6.1. Data Augmentation
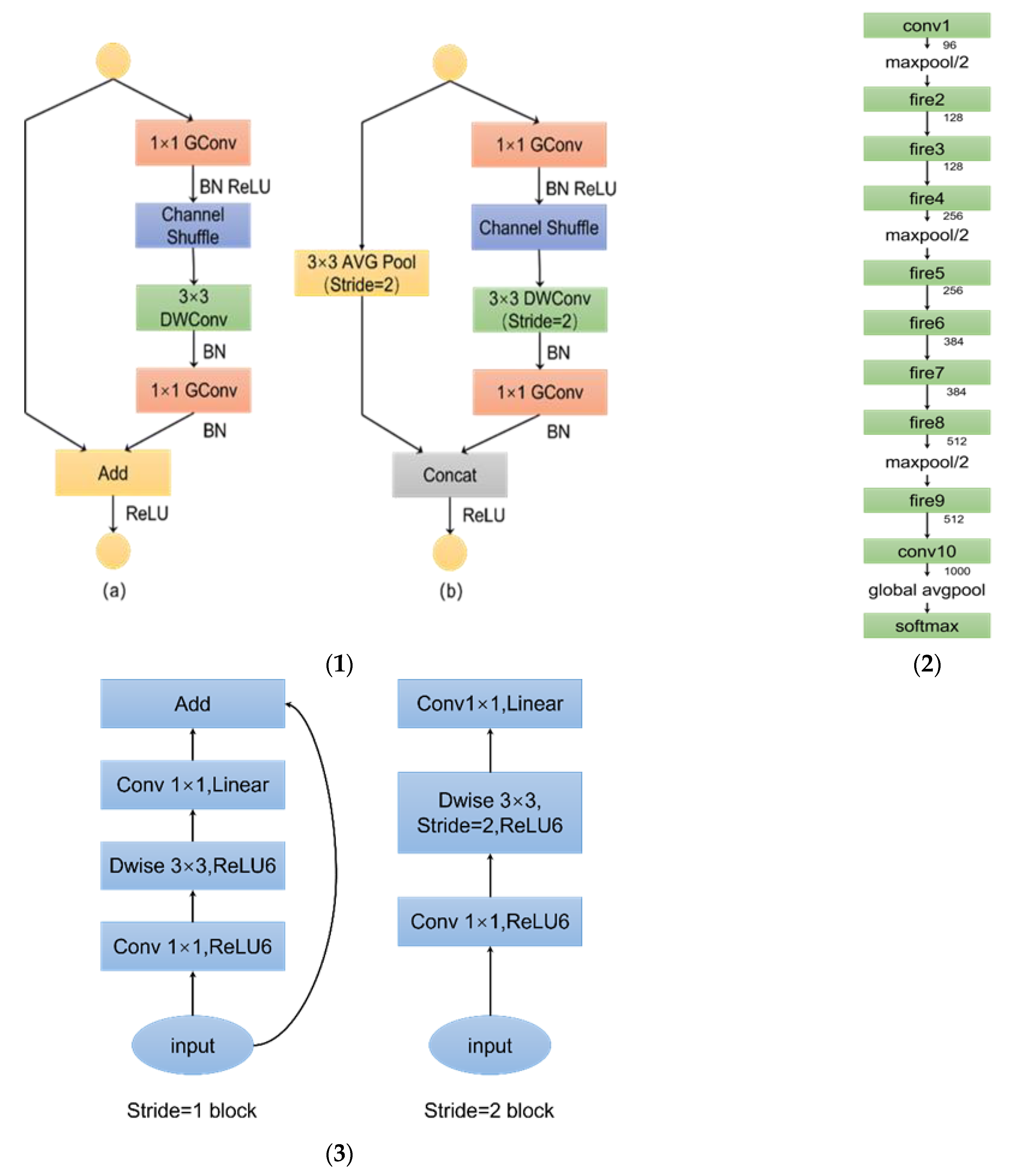
2.6.2. Convolutional Networks Recognition Algorithm Selection
- (1)
- Use 1 × 1 Convolutional substitution 3 × 3 convolution: the parameters are reduced to 1/9 of the original.
- (2)
- Reduce number of input channels: this section is implemented using squeeze layers.
- (3)
- Delay down-sampling operation to provide a larger activation map for convolutional layer: a larger activation map preserves more information and provides higher classification accuracy.
- (1)
- Cross-entropy loss function
- (2)
- Adam optimization algorithm
3. Results and Discussion
3.1. Lettuce Images and Image Features
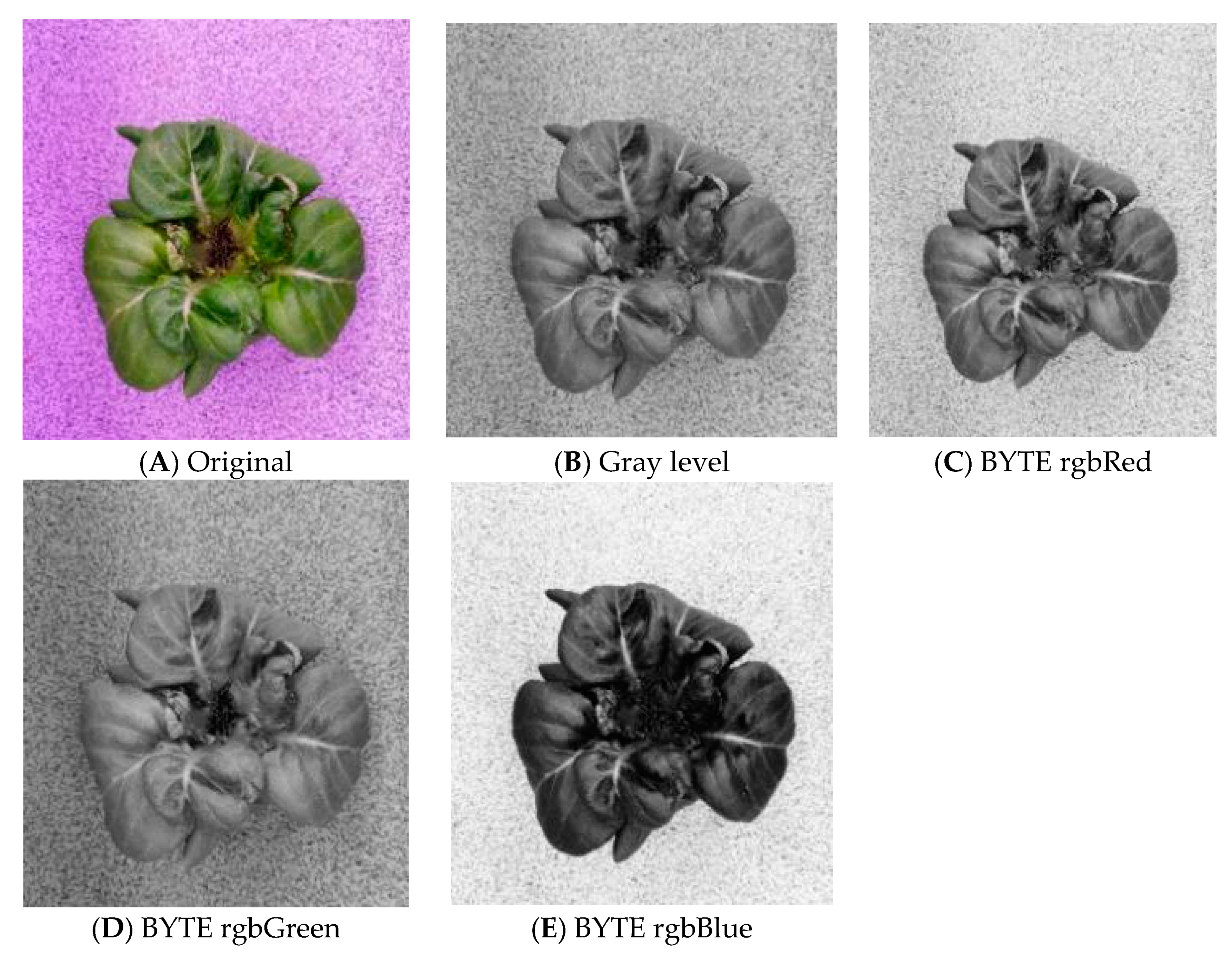
3.1.1. Channel Separation
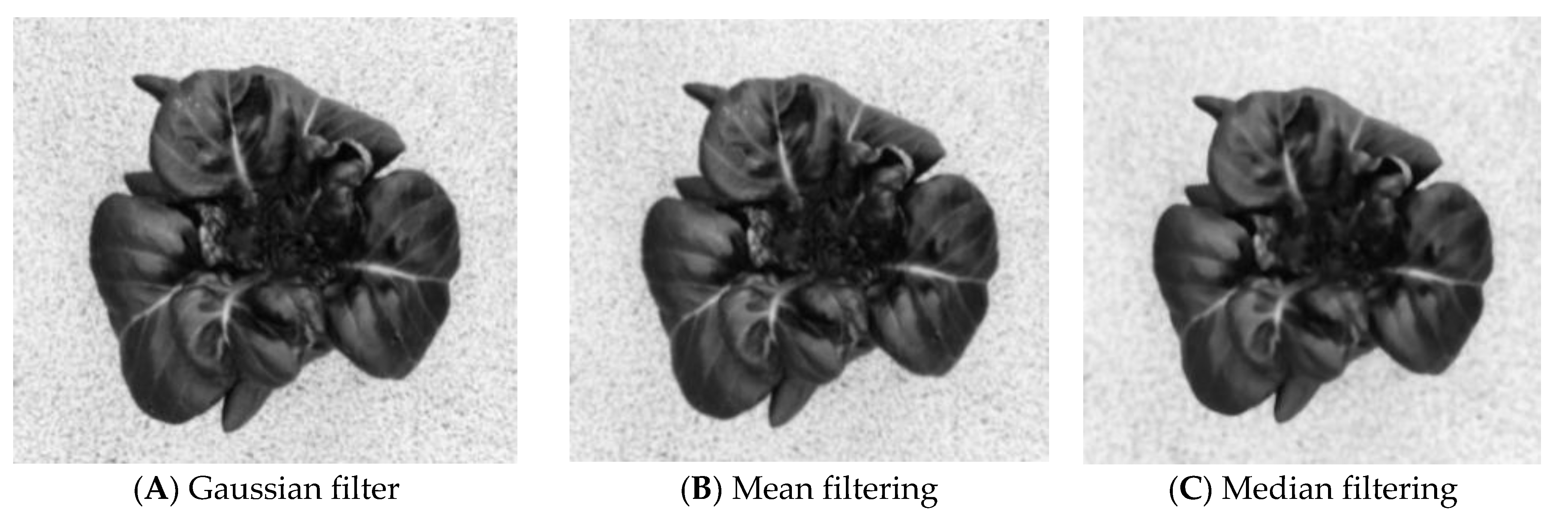
3.1.2. Image Filtering
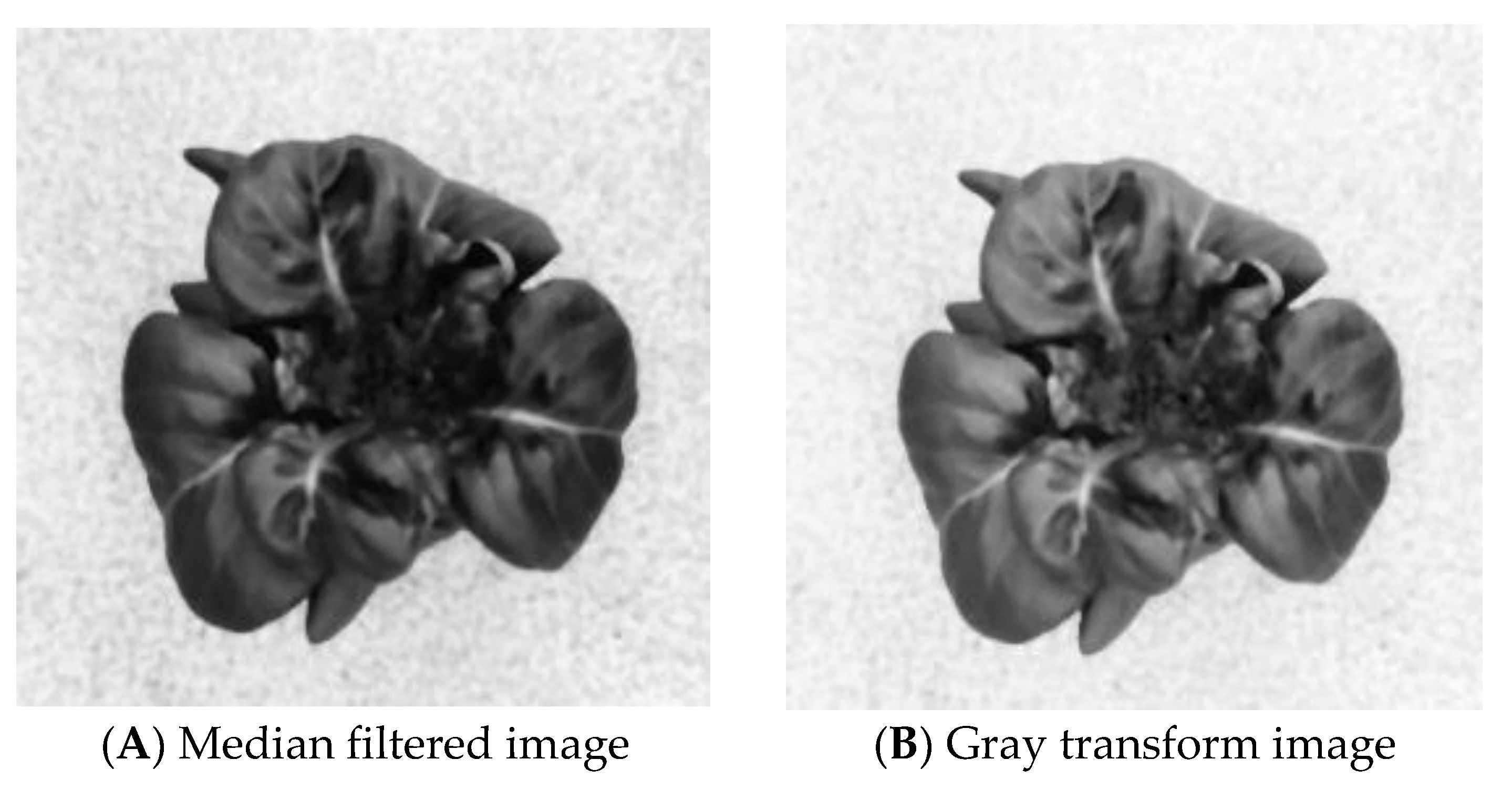
3.1.3. Gray Level Transformation
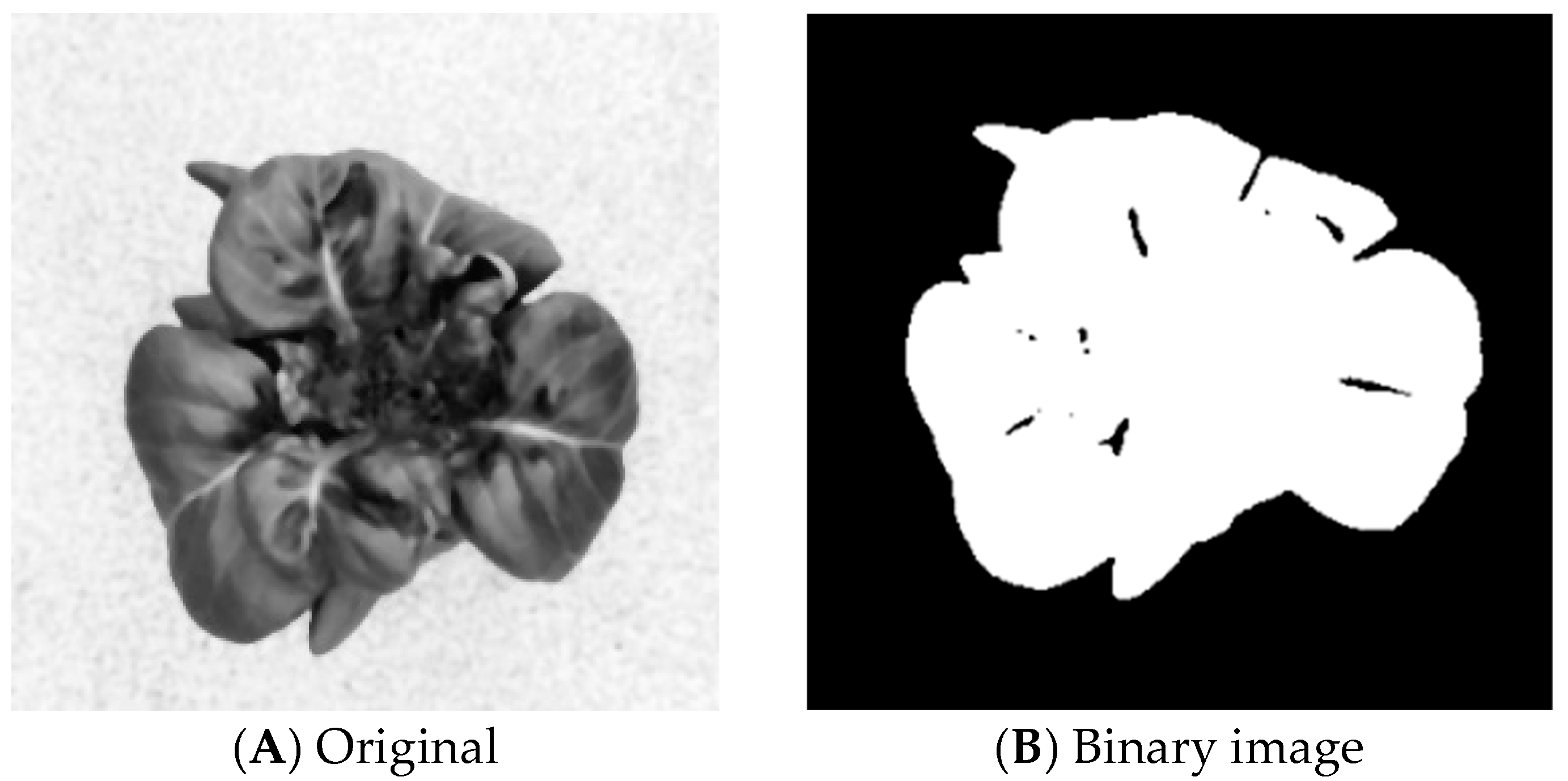
3.1.4. Threshold Segmentation
3.1.5. Foreground Cut and Scale
3.2. Evaluation Index of Machine Learning Algorithm
3.3. Classification Methods with Different Feature Extraction
3.4. Classification Methods with Different Feature Extraction
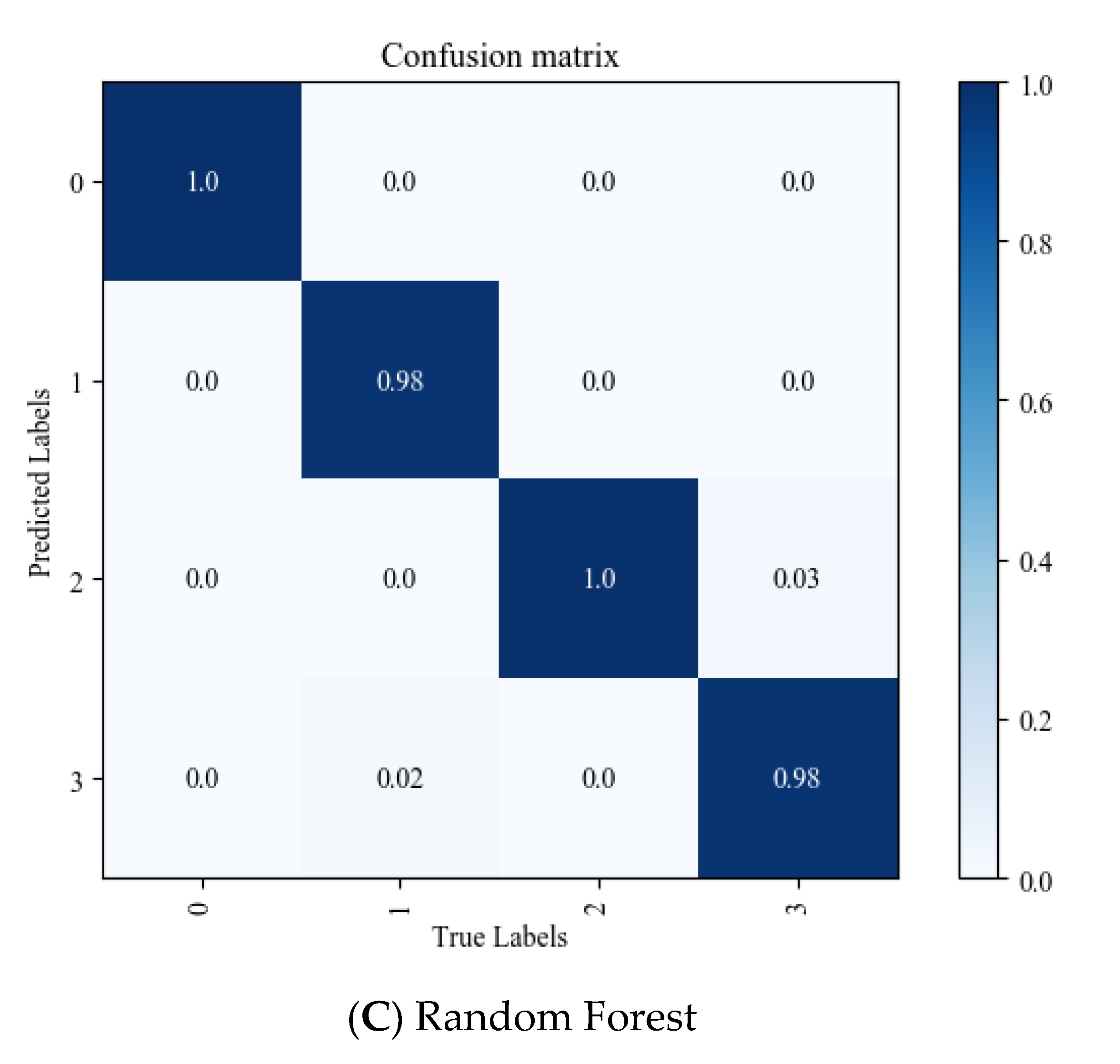
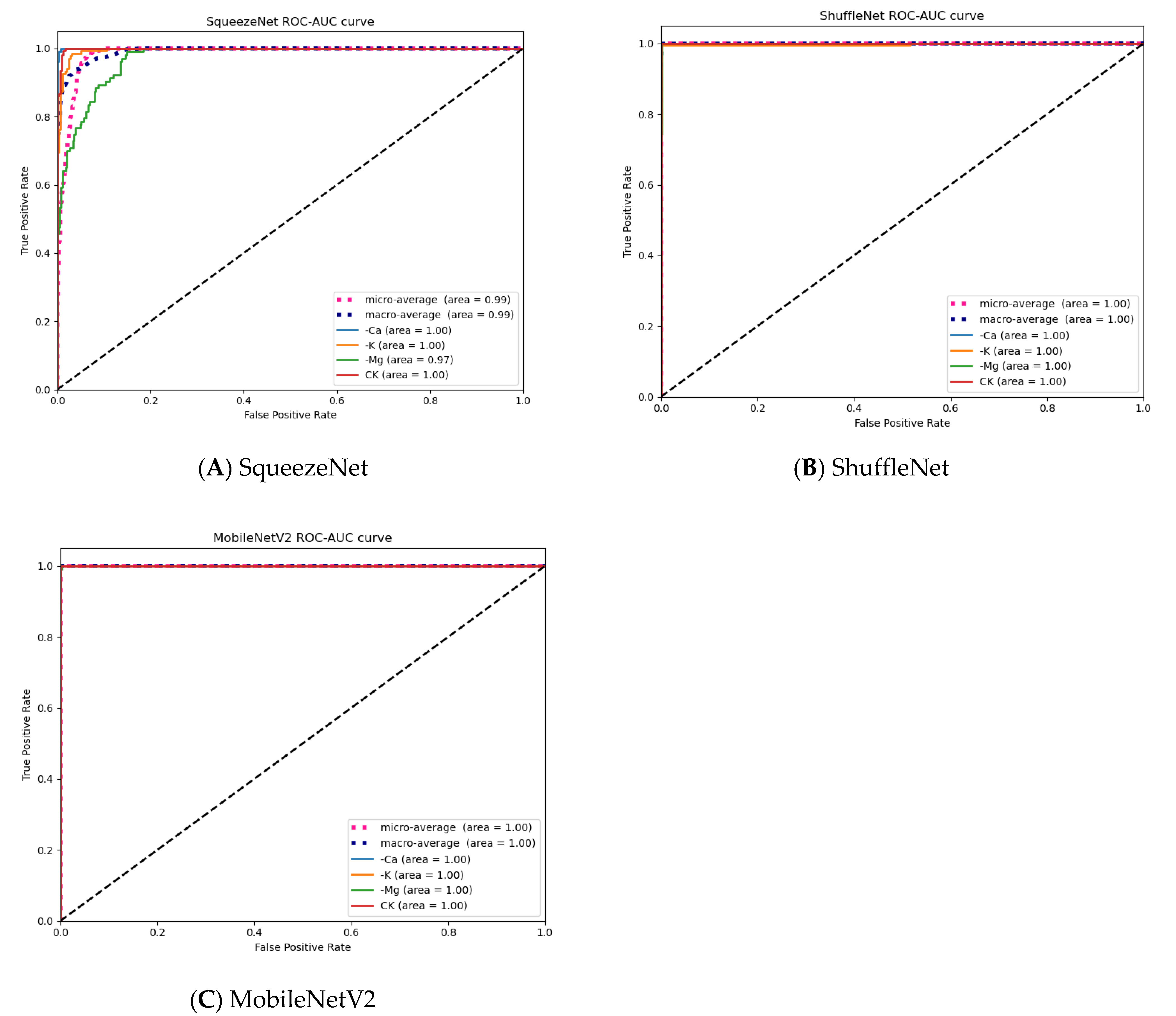
3.5. Visualization Evaluation
3.6. Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Guo, D. Diagnosis of Several Pathological Diseases and Physiological Diseases of Citrus Using Hyperspectral Imaging. Master’s Thesis, Southwest University, Chongqing, China, 2016. [Google Scholar]
- Kim, W.-S.; Lee, D.-H.; Kim, Y.-J. Machine vision-based automatic disease symptom detection of onion downy mildew. Comput. Electron. Agric. 2020, 168, 105099. [Google Scholar] [CrossRef]
- Varshney, D.; Babukhanwala, B.; Khan, J.; Saxena, D.; Singh, A. Machine Learning Techniques for Plant Disease Detection. In Proceedings of the 2021 5th International Conference on Trends in Electronics and Informatics (ICOEI), Tirunelveli, India, 3–5 June 2021. [Google Scholar]
- Fallas-Corrales, R.; van der Zee, S.E.A.T.M. Chapter 42—Diagnosis and management of nutrient constraints in papaya. In Fruit Crops; Srivastava, A.K., Hu, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 607–628. [Google Scholar]
- Taha, M.F.; Abdalla, A.; ElMasry, G.; Gouda, M.; Zhou, L.; Zhao, N.; Liang, N.; Niu, Z.; Hassanein, A.; Al-Rejaie, S.; et al. Using Deep Convolutional Neural Network for Image-Based Diagnosis of Nutrient Deficiencies in Plants Grown in Aquaponics. Chemosensors 2022, 10, 45. [Google Scholar] [CrossRef]
- Maione, C.; Barbosa, R.M. Recent applications of multivariate data analysis methods in the authentication of rice and the most analyzed parameters: A review. Crit. Rev. Food Sci. Nutr. 2019, 59, 1868–1879. [Google Scholar] [CrossRef]
- Takahashi, K.; Kim, K.; Ogata, T.; Sugano, S. Tool-body assimilation model considering grasping motion through deep learning. Robot. Auton. Syst. 2017, 91, 115–127. [Google Scholar] [CrossRef]
- Zhang, B.; He, X.; Ouyang, F.; Gu, D.; Dong, Y.; Zhang, L.; Mo, X.; Huang, W.; Tian, J.; Zhang, S. Radiomic machine-learning classifiers for prognostic biomarkers of advanced nasopharyngeal carcinoma. Cancer Lett. 2017, 403, 21–27. [Google Scholar] [CrossRef]
- Zhao, Y.; Li, J.; Yu, L. A deep learning ensemble approach for crude oil price forecasting. Energy Econ. 2017, 66, 9–16. [Google Scholar] [CrossRef]
- Trang, K.; TonThat, L.; Thao, N.G.M. Plant Leaf Disease Identification by Deep Convolutional Autoencoder as a Feature Extraction Approach. In Proceedings of the 2020 17th International Conference on Electrical Engineering/Electronics, Computer, Telecommunications and Information Technology (ECTI-CON), Phuket, Thailand, 24–27 June 2020; pp. 522–526. [Google Scholar]
- Govardhan, M.; Veena, M.B. Diagnosis of Tomato Plant Diseases using Random Forest. In Proceedings of the 2019 Global Conference for Advancement in Technology (GCAT), Bangalore, India, 18–20 October 2019; pp. 1–5. [Google Scholar]
- Balakrishna, K.; Rao, M. Tomato Plant Leaves Disease Classification Using KNN and PNN. Int. J. Comput. Vis. Image Process. 2019, 9, 51–63. [Google Scholar] [CrossRef]
- Paul, S.G.; Biswas, A.A.; Saha, A.; Zulfiker, M.S.; Ritu, N.A.; Zahan, I.; Rahman, M.; Islam, M.A. A real-time application-based convolutional neural network approach for tomato leaf disease classification. Array 2023, 19, 100313. [Google Scholar] [CrossRef]
- Qin, F.; Liu, D.X.; Sun, B.D.; Ruan, L.; Ma, Z.; Wang, H. Image recognition of four different alfalfa leaf diseases based on deep learning and support vector machine. J. China Agric. Univ. 2017, 22, 123–133. [Google Scholar]
- Syed-Ab-Rahman, S.F.; Hesamian, M.H.; Prasad, M. Citrus disease detection and classification using end-to-end anchor-based deep learning model. Appl. Intell. 2022, 52, 927–938. [Google Scholar] [CrossRef]
- Agarwal, M.; Gupta, S.K.; Biswas, K.K. Development of Efficient CNN model for Tomato crop disease identification. Sustain. Comput. Inform. Syst. 2020, 28, 100407. [Google Scholar] [CrossRef]
- Chen, J.; Chen, J.; Zhang, D.; Sun, Y.; Nanehkaran, Y.A. Using deep transfer learning for image-based plant disease identification. Comput. Electron. Agric. 2020, 173, 105393. [Google Scholar] [CrossRef]
- Francis, M.; Deisy, C. Disease Detection and Classification in Agricultural Plants Using Convolutional Neural Networks—A Visual Understanding. In Proceedings of the 2019 6th International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 7–8 March 2019; pp. 1063–1068. [Google Scholar]
- Militante, S.V.; Gerardo, B.D.; Dionisio, N.V. Plant Leaf Detection and Disease Recognition using Deep Learning. In Proceedings of the 2019 IEEE Eurasia Conference on IOT, Communication and Engineering (ECICE), Yunlin, Taiwan, 3–6 October 2019; pp. 579–582. [Google Scholar]
- Azimi, S.; Kaur, T.; Gandhi, T.K. A deep learning approach to measure stress level in plants due to Nitrogen deficiency. Measurement 2021, 173, 108650. [Google Scholar] [CrossRef]
- Ghosal, S.; Blystone, D.; Singh, A.K.; Ganapathysubramanian, B.; Singh, A.; Sarkar, S. An explainable deep machine vision framework for plant stress phenotyping. Proc. Natl. Acad. Sci. USA 2018, 115, 4613–4618. [Google Scholar] [CrossRef]
- Tran, D.; Cédric, C. Early Diagnosis of Iron Deficiency in Commercial Tomato Crop Using Electrical Signals. Front. Sustain. Food Syst. 2021, 5, 631529. [Google Scholar] [CrossRef]
- Xu, Z.; Guo, X.; Zhu, A.; He, X.; Zhao, X.; Han, Y.; Subedi, R. Using Deep Convolutional Neural Networks for Image-Based Diagnosis of Nutrient Deficiencies in Rice. Comput. Intell. Neurosci. 2020, 2020, 7307252. [Google Scholar] [CrossRef]
- Yi, J.; Krusenbaum, L.; Unger, P.; Hüging, H.; Seidel, S.J.; Schaaf, G.; Gall, J. Deep Learning for Non-Invasive Diagnosis of Nutrient Deficiencies in Sugar Beet Using RGB Images. Sensors 2020, 20, 5893. [Google Scholar] [CrossRef]
- Han, X. Research on Recognition Method of Tomato Leaf Deficiency Based on Convolutionneural Network. Master’s Thesis, Northwest A&F University, Xianyang, China, 2022. [Google Scholar]
- Rizkiana, A.; Nugroho, A.P.; Salma, N.M.; Afif, S.; Masithoh, R.E.; Sutiarso, L.; Okayasu, T. Plant growth prediction model for lettuce (Lactuca sativa.) in plant factories using artificial neural network. IOP Conf. Ser. Earth Environ. Sci. 2021, 733, 012027. [Google Scholar] [CrossRef]
- Hao, X.; Jia, J.; Gao, W.; Guo, X.; Zhang, W.; Zheng, L.; Wang, M. MFC-CNN: An automatic grading scheme for light stress levels of lettuce (Lactuca sativa L.) leaves. Comput. Electron. Agric. 2020, 179, 105847. [Google Scholar] [CrossRef]
- Sai, K.; Sood, N.; Saini, I. Classification of various nutrient deficiencies in tomato plants through electrophysiological signal decomposition and sample space reduction. Plant Physiol. Biochem. 2022, 186, 266–278. [Google Scholar] [CrossRef]
- Sathyavani, R.; JaganMohan, K.; Kalaavathi, B. Classification of nutrient deficiencies in rice crop using denseNet-BC. Mater. Today Proc. 2022, 56, 1783–1789. [Google Scholar] [CrossRef]
- Hamdani, H.; Septiarini, A.; Sunyoto, A.; Suyanto, S.; Utaminingrum, F. Detection of oil palm leaf disease based on color histogram and supervised classifier. Optik 2021, 245, 167753. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, X.; Lin, M.; Sun, J. ShuffleNet: An Extremely Efficient Convolutional Neural Network for Mobile Devices. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 6848–6856. [Google Scholar]
- Grigorescu, S.E.; Petkov, N.; Kruizinga, P. Comparison of texture features based on Gabor filters. IEEE Trans. Image Process. 2002, 11, 1160–1167. [Google Scholar] [CrossRef]
- Manjunath, B.S.; Ma, W.Y. Texture features for browsing and retrieval of image data. IEEE Trans. Pattern Anal. Mach. Intell. 1996, 18, 837–842. [Google Scholar] [CrossRef]
- Wu, C.-M.; Chen, Y.-C.; Hsieh, K.-S. Texture features for classification of ultrasonic liver images. IEEE Trans. Med. Imaging 1992, 11, 141–152. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Ojala, T.; Pietikainen, M.; Harwood, D. Performance evaluation of texture measures with classification based on Kullback discrimination of distributions. In Proceedings of the 12th International Conference on Pattern Recognition, Jerusalem, Israel, 9–13 October 1994; Volume 581, pp. 582–585. [Google Scholar]
- Ojala, T.; Pietikäinen, M.; Harwood, D. A comparative study of texture measures with classification based on featured distributions. Pattern Recognit. 1996, 29, 51–59. [Google Scholar] [CrossRef]
- Ojala, T.; Valkealahti, K.; Oja, E.; Pietikäinen, M. Texture discrimination with multidimensional distributions of signed gray-level differences. Pattern Recognit. 2001, 34, 727–739. [Google Scholar] [CrossRef]
- Ojala, T.; Pietikainen, M.; Maenpaa, T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 971–987. [Google Scholar] [CrossRef]
- Pietikäinen, M. Image Analysis with Local Binary Patterns. In Proceedings of the Image Analysis: 14th Scandinavian Conference, SCIA 2005, Joensuu, Finland, 19–22 June 2005; Springer: Berlin/Heidelberg, Germany, 2005; pp. 115–118. [Google Scholar]
- Stricker, M.; Orengo, M. Similarity of Color Images. In Proceedings of the Is&T/Spie’s Symposium on Electronic Imaging: Science and Technology, San Jose, CA, USA, 5–10 February 1995; Volume 2420. [Google Scholar] [CrossRef]
- Swain, M.; Ballard, D. Color Indexing. Int. J. Comput. Vis. 1991, 7, 11–32. [Google Scholar] [CrossRef]
- Iandola, F.; Han, S.; Moskewicz, M.; Ashraf, K.; Dally, W.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5MB model size. arXiv 2016, arXiv:1602.07360. [Google Scholar]
- Peng, D.; Deng, A. Improved research on uniform feature extraction based on SIFTalgorithm. Geomat. Spat. Inf. Technol. 2021, 44, 46–50, 56. [Google Scholar]
- Lowe, D.G. Distinctive Image Features from Scale-Invariant Keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Ding, G.; Qiao, Y.; Yi, W.; Du, L. Improved SIFT extraction and matching based on spectral image space. Trans. Beijing Inst. Technol. 2022, 42, 192–199. [Google Scholar] [CrossRef]
- Abdullah, N.E.; Rahim, A.A.; Hashim, H.; Kamal, M.M. Classification of Rubber Tree Leaf Diseases Using Multilayer Perceptron Neural Network. In Proceedings of the 2007 5th Student Conference on Research and Development, Selangor, Malaysia, 11–12 December 2007; pp. 1–6. [Google Scholar]
- Tan, L.; Lu, J.; Jiang, H. Tomato Leaf Diseases Classification Based on Leaf Images: A Comparison between Classical Machine Learning and Deep Learning Methods. AgriEngineering 2021, 3, 542–558. [Google Scholar] [CrossRef]
- Yang, J.; Qiao, P.; Li, Y.; Wang, N. A review of machine-learning classification and algorithms. Stat. Decis. 2019, 35, 36–40. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Kingma, D.; Ba, J. Adam: A Method for Stochastic Optimization. In Proceedings of the 3rd International Conference for Learning Representations, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Lu, J.; Tan, L.; Jiang, H. Review on Convolutional Neural Network (CNN) Applied to Plant Leaf Disease Classification. Agriculture 2021, 11, 707. [Google Scholar] [CrossRef]
- Tenaye, F.; Diriba, C.; Kuyu, M. Coffee Arabica Nutrient Deficiency Detection System Using Image Processing Techniques. Biomed. J. Sci. Tech. Res. 2022, 46, 37600–37606. [Google Scholar] [CrossRef]
- Guerrero, R.; Renteros, B.; Castañeda, R.; Villanueva, A.; Belupú, I. Detection of nutrient deficiencies in banana plants using deep learning. In Proceedings of the 2021 IEEE International Conference on Automation/XXIV Congress of the Chilean Association of Automatic Control (ICA-ACCA), Valparaíso, Chile, 22–26 March 2021; pp. 1–7. [Google Scholar]
- Luz, P.; Marin, M.; Devechio, F.; Romualdo, L.; Zuñiga, A.; Oliveira, M.; Herling, V.; Bruno, O. Boron Deficiency Precisely Identified on Growth Stage V4 of Maize Crop Using Texture Image Analysis. Commun. Soil Sci. Plant Anal. 2018, 49, 1–11. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, X.; Hou, Z.; Ning, J.; Zhang, Z. Discrimination of nitrogen fertilizer levels of tea plant (Camellia sinensis) based on hyperspectral imaging. J. Sci. Food Agric. 2018, 98, 4659–4664. [Google Scholar] [CrossRef]
- Bahtiar, A.R.; Pranowo; Santoso, A.J.; Juhariah, J. Deep Learning Detected Nutrient Deficiency in Chili Plant. In Proceedings of the 2020 8th International Conference on Information and Communication Technology (ICoICT), Yogyakarta, Indonesia, 24–26 June 2020; pp. 1–4. [Google Scholar]
- Ibrahim, S.; Hasan, N.; Sabri, N.; Abu Samah, K.A.F.; Rahimi Rusland, M. Palm leaf nutrient deficiency detection using convolutional neural network (CNN). Int. J. Nonlinear Anal. Appl. 2022, 13, 1949–1956. [Google Scholar] [CrossRef]
- Song, Y.; Liu, L.; Rao, Y.; Zhang, X.; Jin, X. FA-Net: A Fused Feature for Multi-Head Attention Recoding Network for Pear Leaf Nutritional Deficiency Diagnosis with Visual RGB-Image Depth and Shallow Features. Sensors 2023, 23, 4507. [Google Scholar] [CrossRef] [PubMed]
- Espejo-Garcia, B.; Malounas, I.; Mylonas, N.; Kasimati, A.; Fountas, S. Using EfficientNet and transfer learning for image-based diagnosis of nutrient deficiencies. Comput. Electron. Agric. 2022, 196, 106868. [Google Scholar] [CrossRef]







| Elements | Compound | All Nutrient Elements Group (CK)/(g/L) | Nutrient Deficiency Group/(g/L) | ||
|---|---|---|---|---|---|
| Potassium Depletion (-K) | Calcium Deficiency (-Ca) | Magnesium Deficiency (-Mg) | |||
| Macro elements | Ca(NO3)2·H2O | 0.70845 | 0.70845 | - | 0.70845 |
| KNO3 | 1.001 | - | 1.011 | 1.001 | |
| NH4H2PO4 | 0.4003 | 0.4003 | 0.4003 | 0.4003 | |
| MgSO4·7H2O | 0.49294 | 0.49294 | 0.49294 | - | |
| NaNO3 | - | 0.8499 | 0.50994 | - | |
| EDTA-2Na (×1000) | 3.73 | 3.73 | 3.73 | 3.73 | |
| FeSO4·7H2O (×1000) | 2.78 | 2.78 | 2.78 | 2.78 | |
| Trace elements (×1000) | H3BO3 | 0.866 | 0.866 | 0.866 | 0.866 |
| ZnSO4·7H2O | 0.863 | 0.863 | 0.863 | 0.863 | |
| MnSO4·H2O | 0.848 | 0.848 | 0.848 | 0.848 | |
| CuSO4·5H2O | 0.175 | 0.175 | 0.175 | 0.175 | |
| CoCl2·6H2O | 0.024 | 0.024 | 0.024 | 0.024 | |
| (NH4)6MoO24·4H2O | 0.1441 | 0.1441 | 0.1441 | 0.1441 | |
| Parameters | Content |
|---|---|
| Operating system | Windows 10 |
| Learning framework | Pytorch1.5.1 |
| Programming language | Python3.7.3 |
| Hardware environment | 8 GB RAM |
| Hyperparameter | Value |
|---|---|
| Learning rate | 0.0001 |
| The momentum | 0.99 |
| Batch size | 256 |
| Epochs | 30 |
| Varieties | (Recall Percentage, F1-Score Percentage) of Feature Extraction Methods | |||||||
|---|---|---|---|---|---|---|---|---|
| LBP | GLCM | LBP + GLCM | color | Color + LBP | Color + GLCM | All | SIFT | |
| -Ca | (81, 87) | (68, 76) | (95, 96) | (92, 96) | (89, 94) | (92, 96) | (92, 96) | (92, 92) |
| -Mg | (68, 71) | (71, 76) | (81, 88) | (97, 97) | (97, 98) | (97, 98) | (97, 98) | (90, 95) |
| -K | (88, 84) | (79, 74) | (95, 89) | (99, 97) | (99, 94) | (99, 95) | (99, 96) | (99, 90) |
| CK | (85, 86) | (87, 89) | (87, 91) | (98, 98) | (93, 95) | (95, 96) | (96, 97) | (87, 93) |
| average | (81, 82) | (76, 79) | (90, 91) | (97, 97) | (95, 95) | (96, 96) | (96, 97) | (92, 93) |
| Classifiers | Evaluating Indicator | |||
|---|---|---|---|---|
| Accuracy | Precision | Recall | F1-Score | |
| KNN | 97.0% | 97.1% | 96.7% | 96.8% |
| SVM | 97.0% | 96.7% | 97.2% | 96.9% |
| Random Forest | 97.6% | 97.9% | 97.4% | 97.6% |
| Algorithms | Evaluating Indicator | ||||||
|---|---|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1-Score | Params/M | FLOPs/G | Time/ms | |
| ShuffleNet | 99.9% | 99.9% | 99.9% | 99.8% | 1.37 | 0.04 | 80.89 |
| SqueezeNet | 99.5% | 99.5% | 99.5% | 99.5% | 1.24 | 0.35 | 238.22 |
| MobileNetV2 | 99.8% | 99.5% | 99.5% | 99.5% | 3.50 | 0.31 | 97.12 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, J.; Peng, K.; Wang, Q.; Sun, C. Lettuce Plant Trace-Element-Deficiency Symptom Identification via Machine Vision Methods. Agriculture 2023, 13, 1614. https://doi.org/10.3390/agriculture13081614
Lu J, Peng K, Wang Q, Sun C. Lettuce Plant Trace-Element-Deficiency Symptom Identification via Machine Vision Methods. Agriculture. 2023; 13(8):1614. https://doi.org/10.3390/agriculture13081614
Chicago/Turabian StyleLu, Jinzhu, Kaiqian Peng, Qi Wang, and Cong Sun. 2023. "Lettuce Plant Trace-Element-Deficiency Symptom Identification via Machine Vision Methods" Agriculture 13, no. 8: 1614. https://doi.org/10.3390/agriculture13081614
APA StyleLu, J., Peng, K., Wang, Q., & Sun, C. (2023). Lettuce Plant Trace-Element-Deficiency Symptom Identification via Machine Vision Methods. Agriculture, 13(8), 1614. https://doi.org/10.3390/agriculture13081614





