Fruit Stalk Recognition and Picking Point Localization of New Plums Based on Improved DeepLabv3+
Abstract
:1. Introduction
2. Dataset Construction and Labeling
3. The Construction of the New Plum Fruit Stalk Segmentation Model and Picking Point Localization
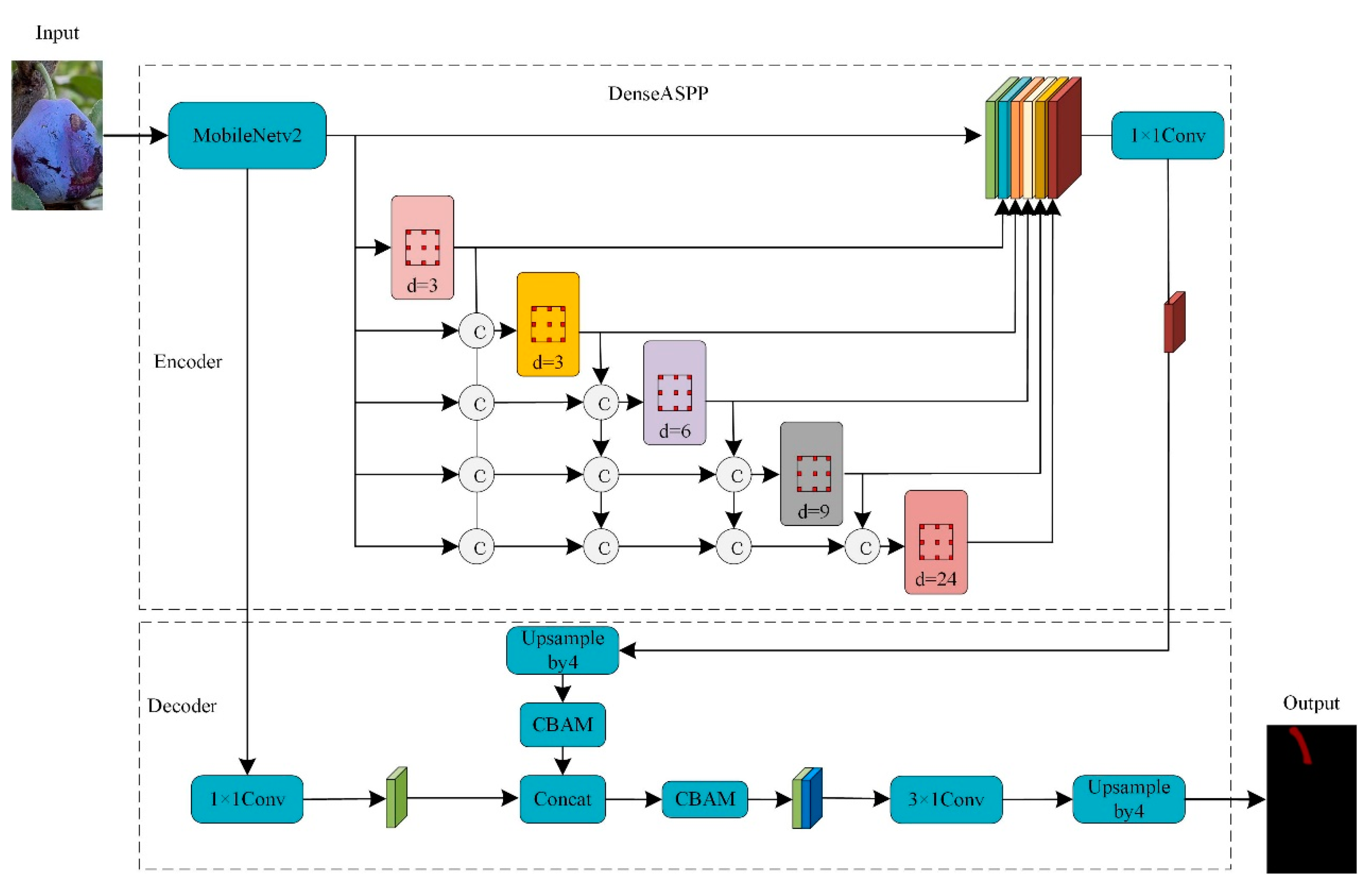
3.1. DeepLabv3+ Modeling
3.2. Improvements to the DeepLabv3+ Model
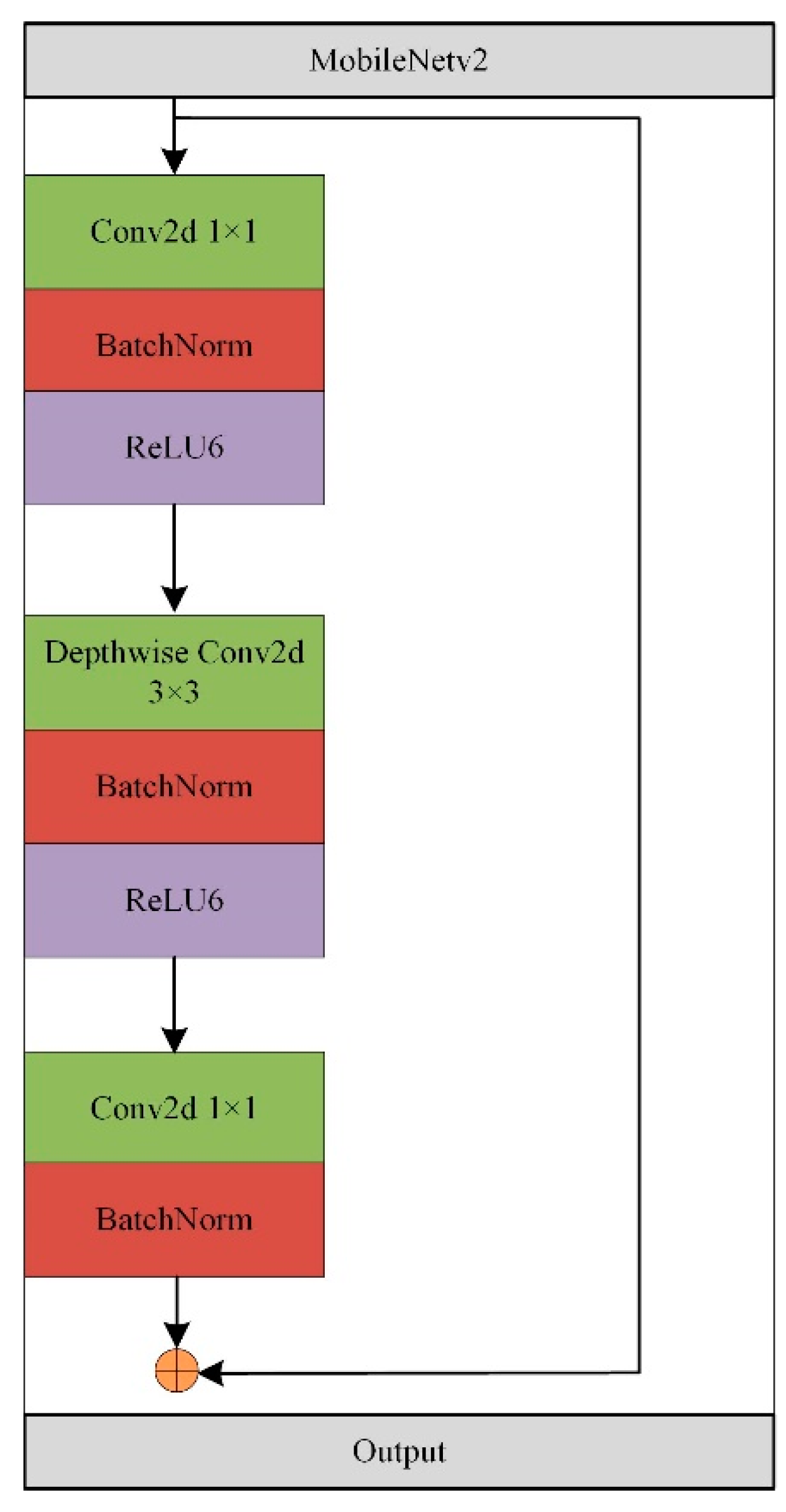
3.2.1. Lightweight Feature Extraction Module
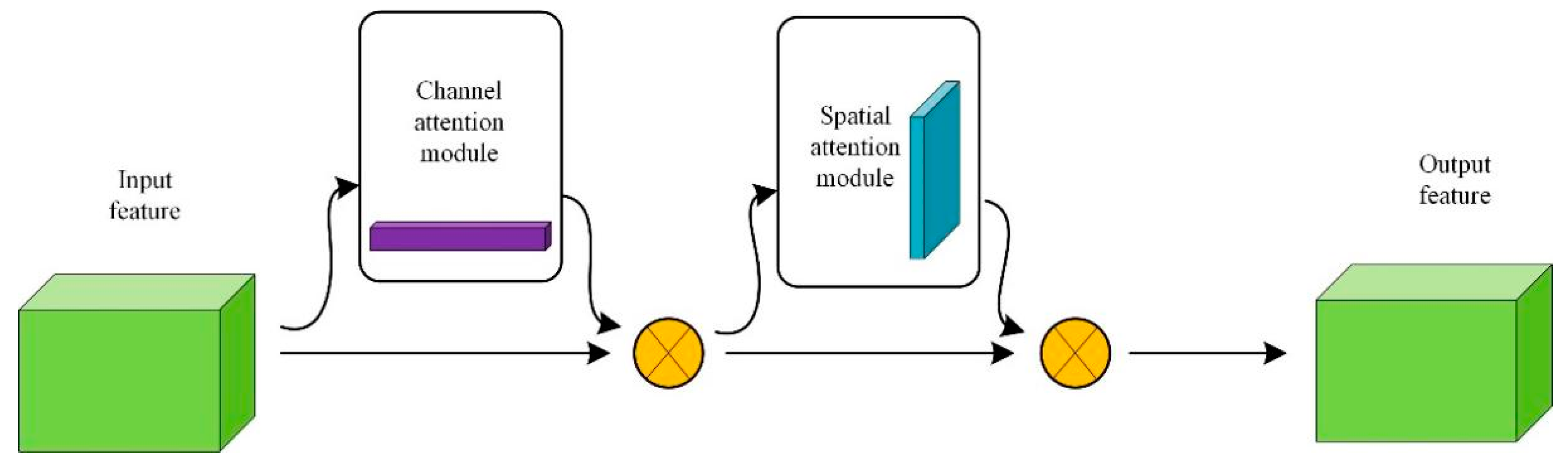
3.2.2. CBAM Attention Mechanism
3.2.3. Pyramid Pooling Module for Dense Void Spaces
3.3. Picking Point Location
4. Tests and Analysis
4.1. Experimental Environment
4.2. Model Evaluation Indicators
4.3. Comparative Analysis of Test Results
4.3.1. Comparative Tests of Attention Mechanisms
4.3.2. Comparison of Ablation Tests
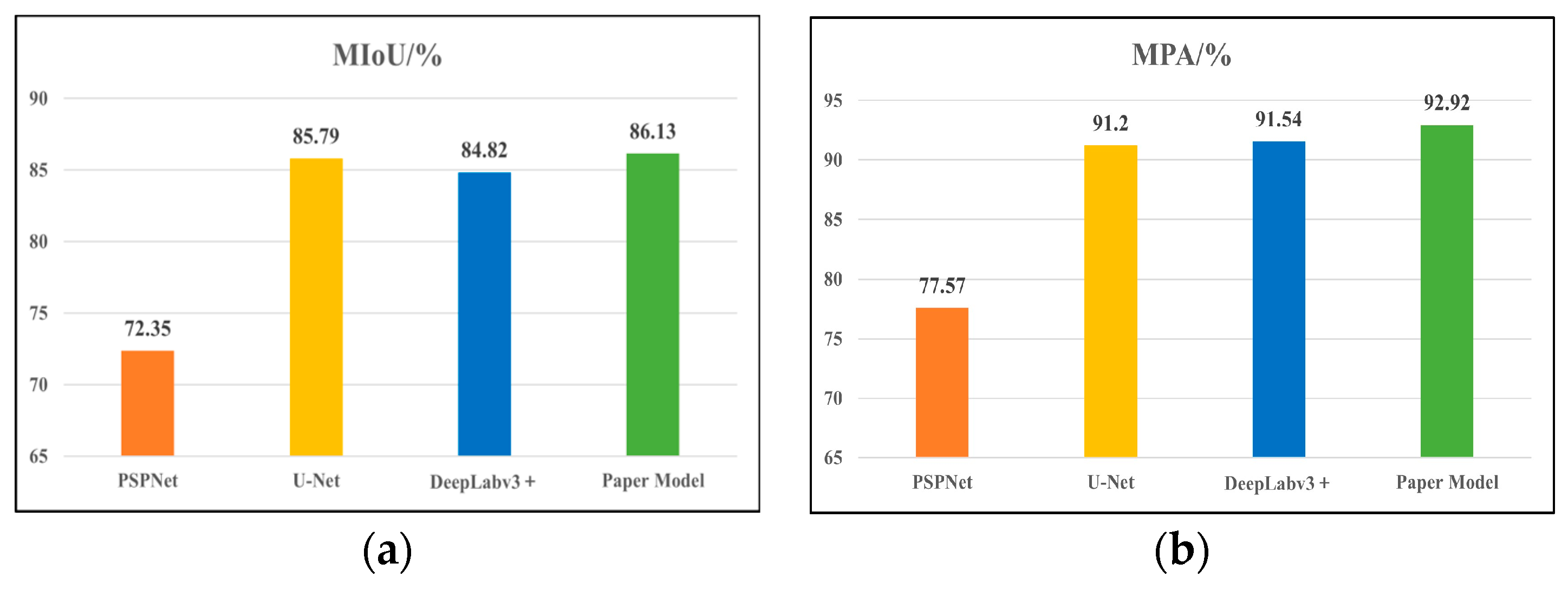
4.3.3. Comparison Experiments of Different Segmentation Models
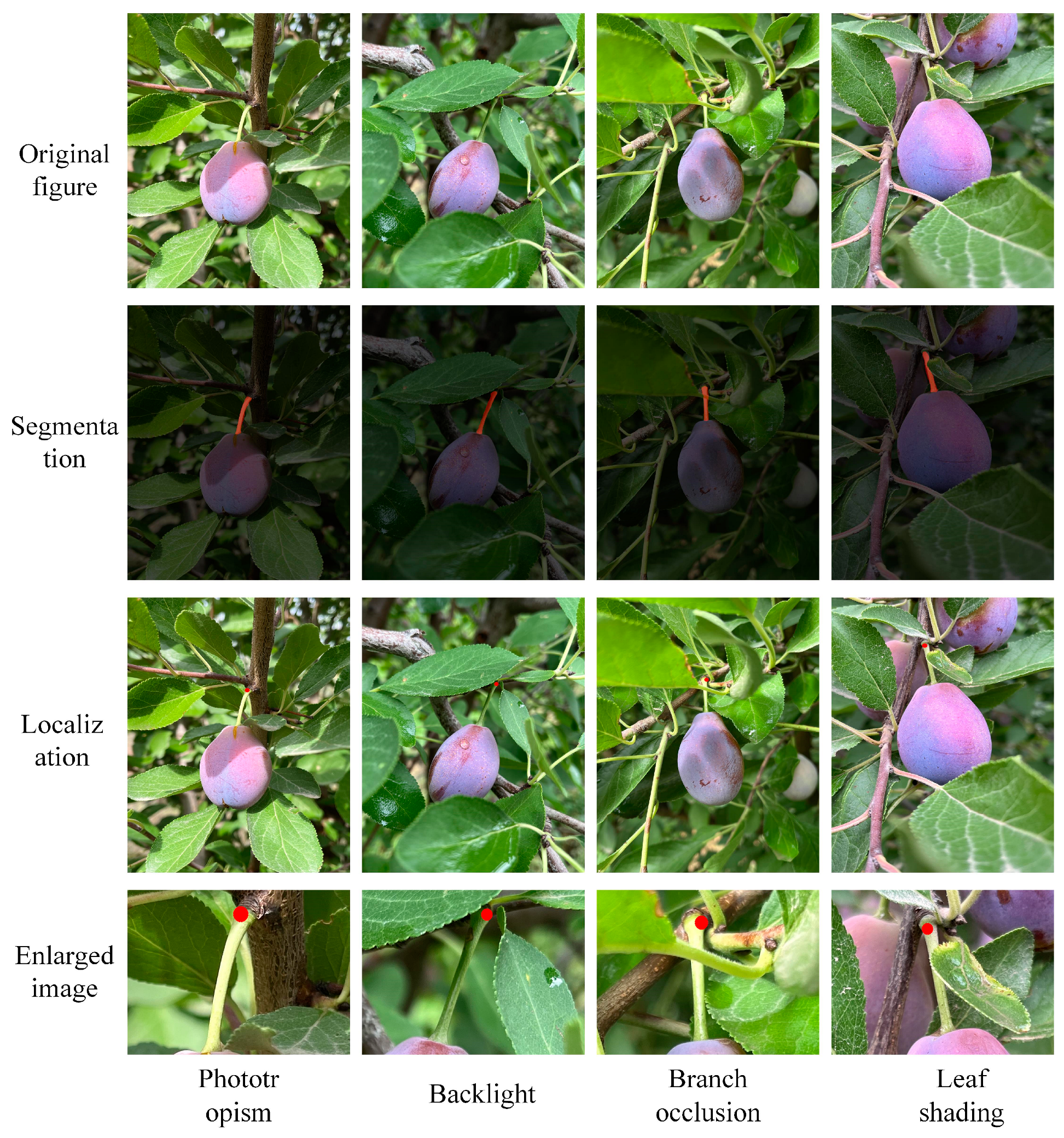
4.3.4. Positioning Tests
4.3.5. Field Segmentation and Localization Experiments in Orchards
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Feng, Q.; Wang, X.; Wang, G.; Li, Z. Design and test of tomatoes harvesting robot. In Proceedings of the IEEE International Conference on Information and Automation, Lijiang, China, 8–10 August 2015; pp. 949–952. [Google Scholar]
- Yao, Z.; Zhao, C.; Zhang, T. Agricultural machinery automatic navigation technology. iScience 2023, 27, 108714. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.M.; Harnal, S.; Gautam, V.; Tiwari, R.; Upadhyay, S. Weed density estimation in soya bean crop using deep convolutional neural networks in smart agriculture. J. Plant Dis. Prot. 2022, 129, 593–604. [Google Scholar] [CrossRef]
- Ionica, M.E.; Nour, V.; Trandafir, I.; Cosmulescu, S.; Botu, M. Physical and chemical properties of some European plum cultivars (Prunus domestica L.). Not. Bot. Horti Agrobot. Cluj-Napoca 2013, 41, 499–503. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, J.; Yang, K.; Wang, L.; Su, F.; Chen, X. Semantic segmentation of high-resolution remote sensing images based on a class feature attention mechanism fused with Deeplabv3+. Comput. Geosci. 2022, 158, 104969. [Google Scholar] [CrossRef]
- Kaur, P.; Harnal, S.; Tiwari, R.; Upadhyay, S.; Bhatia, S.; Mashat, A.; Alabdali, A.M. Recognition of leaf disease using hybrid convolutional neural network by applying feature reduction. Sensors 2022, 22, 575. [Google Scholar] [CrossRef] [PubMed]
- Bac, C.W.; Hemming, J.; Van Henten, E.J. Stem localization of sweet-pepper plants using the support wire as a visual cue. Comput. Electron. Agric. 2014, 105, 111–120. [Google Scholar] [CrossRef]
- Xiong, J.; Lin, R.; Liu, Z.; He, Z.; Tang, L.; Yang, Z.; Zou, X. The recognition of litchi clusters and the calculation of picking point in a nocturnal natural environment. Biosyst. Eng. 2018, 166, 44–57. [Google Scholar] [CrossRef]
- Ji, C.; Zhang, J.; Yuan, T.; Li, W. Research on key technology of truss tomato harvesting robot in greenhouse. Appl. Mech. Mater. 2014, 442, 480–486. [Google Scholar] [CrossRef]
- Luo, L.; Tang, Y.; Lu, Q.; Chen, X.; Zhang, P.; Zou, X. A vision methodology for harvesting robot to detect cutting points on peduncles of double overlapping grape clusters in a vineyard. Comput. Ind. 2018, 99, 130–139. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, K.; Liu, H.; Yang, L.; Zhang, D. Real-time visual localization of the picking points for a ridge-planting strawberry harvesting robot. IEEE Access 2020, 8, 116556–116568. [Google Scholar] [CrossRef]
- Peng, H.; Xue, C.; Shao, Y.; Chen, K.; Xiong, J.; Xie, H.; Zhang, L. Semantic segmentation of litchi branches using DeepLabV3+ model. IEEE Access 2020, 8, 164546–164555. [Google Scholar] [CrossRef]
- Ning, Z.; Lou, L.; Liao, J.; Wen, H.; Wei, H.; Lu, Q. Recognition and the optimal picking point location of grape stems based on deep learning. Trans. Chin. Soc. Agric. Eng. 2021, 37, 222–229. [Google Scholar]
- Rong, Q.; Hu, C.; Hu, X.; Xu, M. Picking point recognition for ripe tomatoes using semantic segmentation and morphological processing. Comput. Electron. Agric. 2023, 210, 107923. [Google Scholar] [CrossRef]
- Yan, C.; Chen, Z.; Li, Z.; Liu, R.; Li, Y.; Xiao, H.; Xie, B. Tea sprout picking point identification based on improved DeepLabV3+. Agriculture 2022, 12, 1594. [Google Scholar] [CrossRef]
- Wu, L.; Su, L.; Jia, G.; Ma, Y.; Li, B.; He, S. Image Segmentation of Potato Roots Using an Improved DeepLabv3+ Network. Trans. Chin. Soc. Agric. Eng. 2023, 39, 134–144. [Google Scholar]
- Chen, L.C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Zhu, Y.; Liu, S.; Wu, X.; Gao, L.; Xu, Y. Multi-class segmentation of navel orange surface defects based on improved DeepLabv3+. J. Agric. Eng. 2024, 55. [Google Scholar] [CrossRef]
- Chollet, F. Xception: Deep learning with depthwise separable convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, 16–21 June 2012; pp. 1251–1258. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted residuals and linear bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision, Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Yang, M.; Yu, K.; Zhang, C.; Li, Z.; Yang, K. Denseaspp for semantic segmentation in street scenes. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 3684–3692. [Google Scholar]
- DB65/T 4475-2021; Quality Grading of Prunus Domestica. Market Supervision Administration of Xinjiang Uygur Autonomous Region: Urumqi, China, 2021.
- Zhang, T.Y.; Suen, C.Y. A fast parallel algorithm for thinning digital patterns. Commun. ACM 1984, 27, 236–239. [Google Scholar] [CrossRef]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Wang, Q.; Wu, B.; Zhu, P.; Li, P.; Zuo, W.; Hu, Q. ECA-Net: Efficient channel attention for deep convolutional neural networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020; pp. 11534–11542. [Google Scholar]
- Liu, Y.; Shao, Z.; Teng, Y.; Hoffmann, N. NAM: Normalization-based attention module. arXiv 2021, arXiv:2111.12419. [Google Scholar]
- Zhao, H.; Shi, J.; Qi, X.; Wang, X.; Jia, J. Pyramid scene parsing network. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2881–2890. [Google Scholar]
- Yang, X.; Li, X.; Ye, Y.; Lau, R.Y.; Zhang, X.; Huang, X. Road detection and centerline extraction via deep recurrent convolutional neural network U-Net. IEEE Trans. Geosci. Remote Sens. 2019, 57, 7209–7220. [Google Scholar] [CrossRef]







| Network Models | Backbone Network | Attention Module | MIoU/% | MPA/% |
|---|---|---|---|---|
| DeepLabv3+ | Xception | NONE | 84.82 | 91.54 |
| DeepLabv3+ | MobileNetv2 | ECA | 85.51 | 91.84 |
| DeepLabv3+ | MobileNetv2 | NAM | 85.65 | 91.87 |
| DeepLabv3+ | MobileNetv2 | SE | 85.95 | 92.57 |
| DeepLabv3+ | MobileNetv2 | CBAM | 86.13 | 92.92 |
| Test | MobileNetv2 | CBAM | DenseASPP | MIoU/% | MPA/% |
|---|---|---|---|---|---|
| 1 | × | × | × | 84.82 | 91.54 |
| 2 | √ | × | × | 84.64 | 90.79 |
| 3 | √ | √ | × | 84.96 | 91.99 |
| 4 | √ | √ | √ | 86.13 | 92.92 |
| Network Models | Backbone Network | MIoU/% | MPA/% | Model Size/MB |
|---|---|---|---|---|
| PSPNet | Resnet50 | 72.35 | 77.57 | 178 |
| U-Net | Resnet50 | 85.79 | 91.20 | 167 |
| DeepLabv3+ | Xception | 84.82 | 91.54 | 209 |
| Paper Model | MobileNetv2 | 86.13 | 92.92 | 59.6 |
| Serial Number | Test Environment | Pixel Coordinate Point |
|---|---|---|
| Sample 1 | Phototropism | (1011, 918) |
| Sample 2 | Backlight | (1303, 889) |
| Sample 3 | Branch occlusion | (1049, 862) |
| Sample 4 | Leaf shading | (719, 450) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Dong, G.; Fan, X.; Xu, Y.; Liu, T.; Zhou, J.; Jiang, H. Fruit Stalk Recognition and Picking Point Localization of New Plums Based on Improved DeepLabv3+. Agriculture 2024, 14, 2120. https://doi.org/10.3390/agriculture14122120
Chen X, Dong G, Fan X, Xu Y, Liu T, Zhou J, Jiang H. Fruit Stalk Recognition and Picking Point Localization of New Plums Based on Improved DeepLabv3+. Agriculture. 2024; 14(12):2120. https://doi.org/10.3390/agriculture14122120
Chicago/Turabian StyleChen, Xiaokang, Genggeng Dong, Xiangpeng Fan, Yan Xu, Tongshe Liu, Jianping Zhou, and Hong Jiang. 2024. "Fruit Stalk Recognition and Picking Point Localization of New Plums Based on Improved DeepLabv3+" Agriculture 14, no. 12: 2120. https://doi.org/10.3390/agriculture14122120
APA StyleChen, X., Dong, G., Fan, X., Xu, Y., Liu, T., Zhou, J., & Jiang, H. (2024). Fruit Stalk Recognition and Picking Point Localization of New Plums Based on Improved DeepLabv3+. Agriculture, 14(12), 2120. https://doi.org/10.3390/agriculture14122120






