Automatic Detection of Rice Blast Fungus Spores by Deep Learning-Based Object Detection: Models, Benchmarks and Quantitative Analysis
Abstract
1. Introduction
- To construct a microscopic imageset of M. oryzae conidia captured in different magnification and different scenes, including the images of M. oryzae conidia mixed with other impurities;
- To train a variety of object detection algorithms based on deep learning to detect conidia of M. oryzae in different scenes, where different evaluation criteria were used to compare the performance of various object detection algorithms;
- To explore whether using the pre-trained model obtained from the self-built single class of spore data for training is helpful for the detection of M. oryzae conidia and other spores.
2. Materials and Methods
2.1. Dataset Construction
2.2. Image Pre-Processing and Labelling
2.3. Object Detection Frameworks and Backbone Networks
2.3.1. Faster R-CNN
2.3.2. Cascade R-CNN
2.3.3. YOLOv3 and DarkNet-53
2.3.4. ResNet
2.3.5. MobileNet
2.4. Transfer Learning
2.5. Experimental Scheme Design
2.6. Training Tools and Parameter Settings
2.7. Evaluating Indicators
3. Results and Discussion
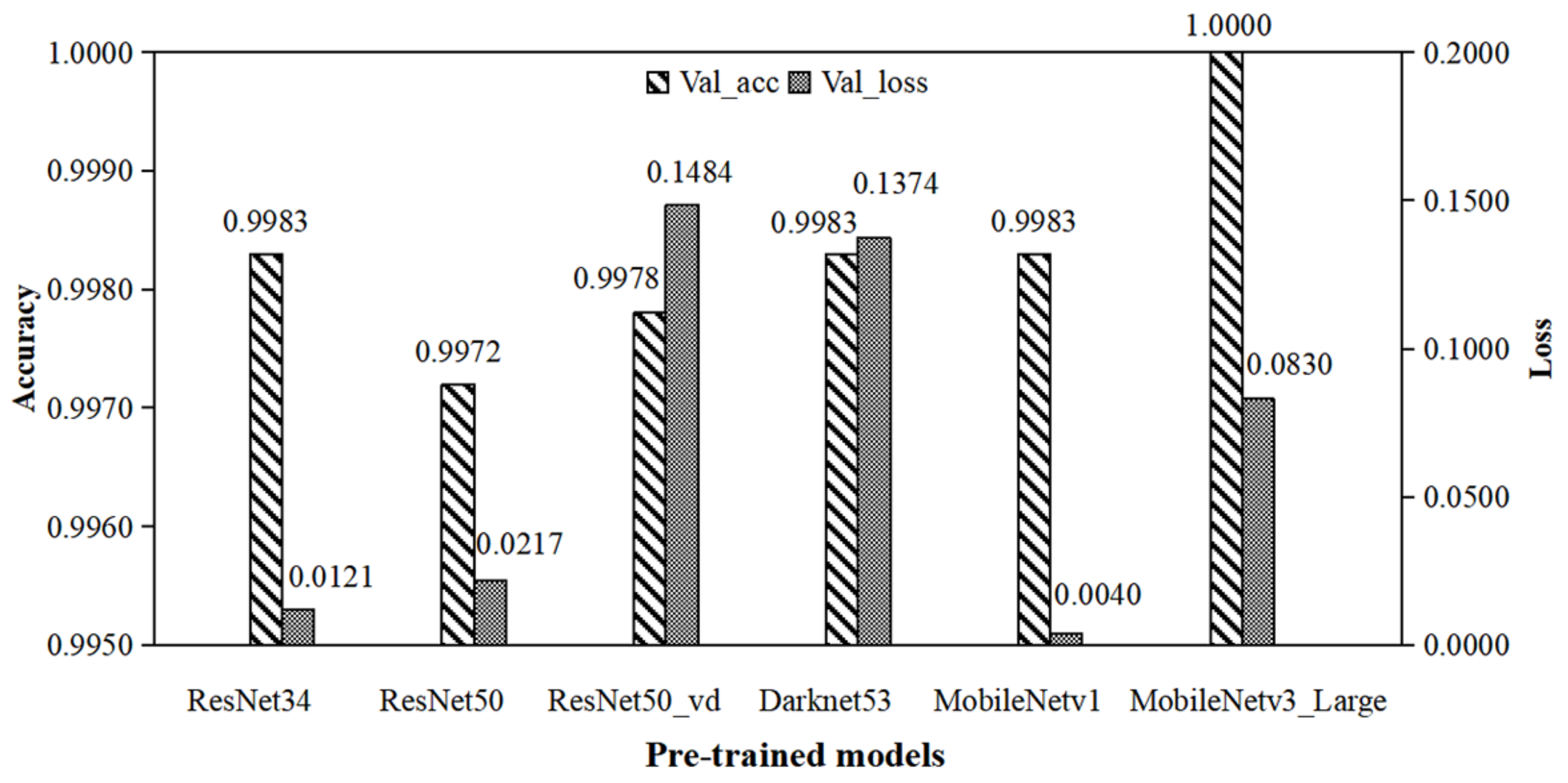
3.1. Influence of Model Pre-Training
3.2. Performance Comparison of Different Object Detection Algorithms
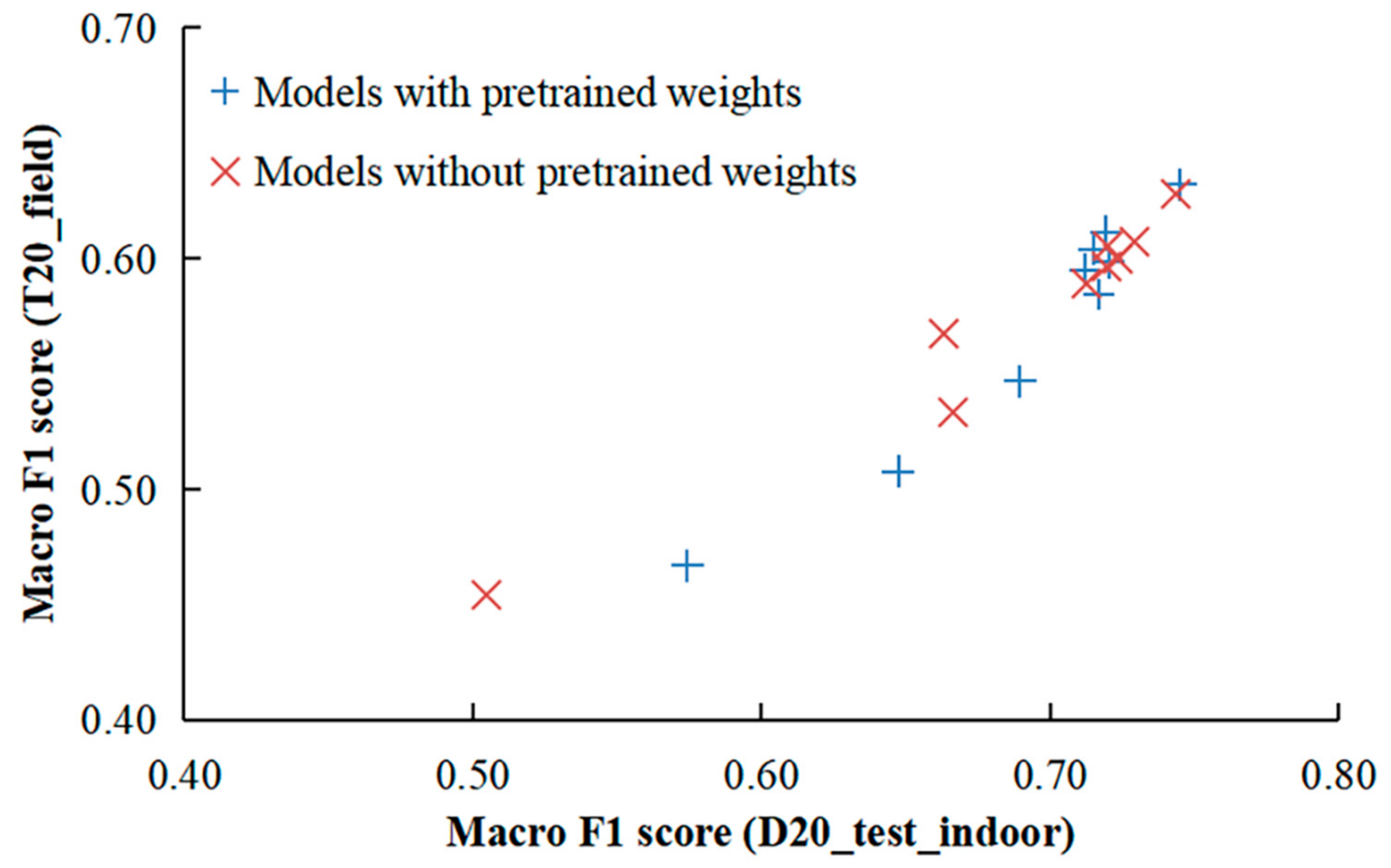
3.3. Analysis of Model Performance on Two Types of Test Sets
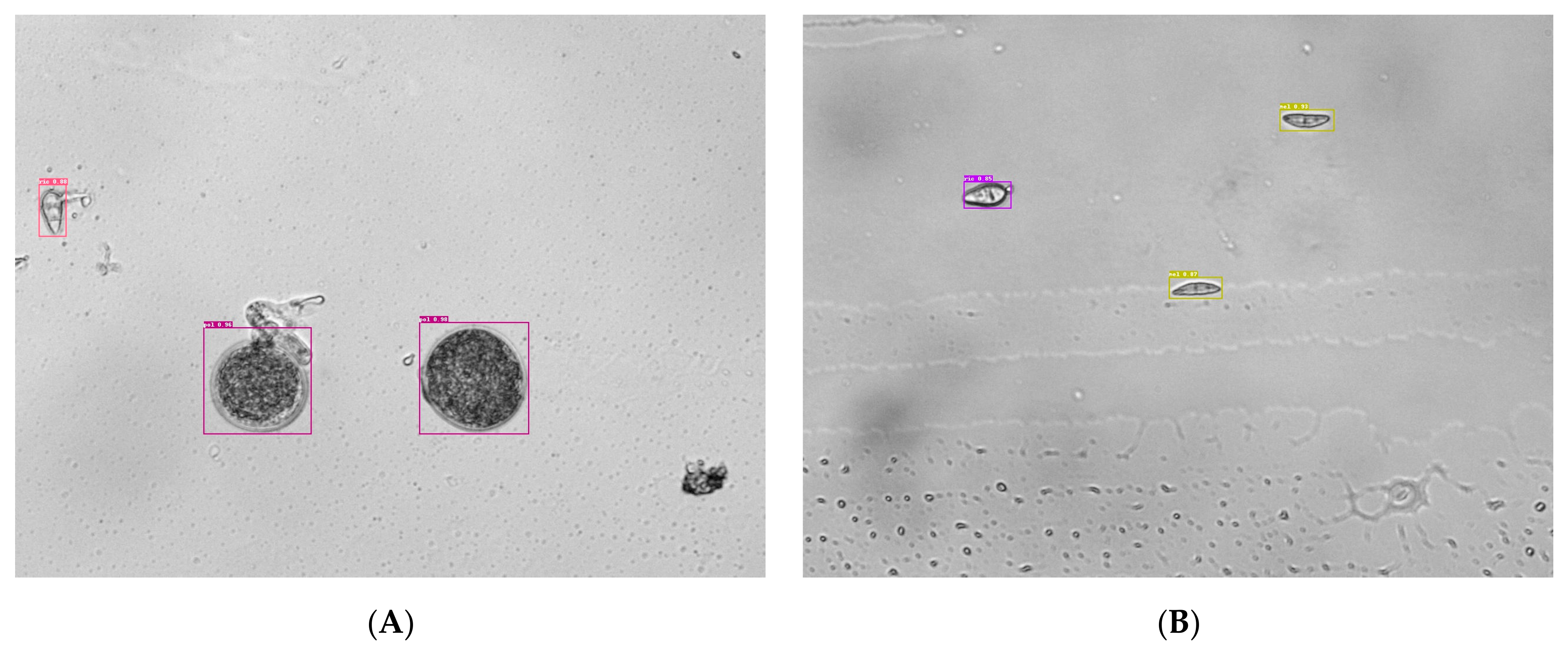
3.4. Detection Performance Comparison on M. oryzae Conidia
3.5. Performance Comparison with Previous Studies
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/faostat/ (accessed on 1 December 2023).
- Deng, R.L.; Tao, M.; Xing, H.; Yang, X.L.; Liu, C.; Liao, K.F.; Qi, L. Automatic diagnosis of rice diseases using deep learning. Front. Plant Sci. 2021, 12, e701038. [Google Scholar] [CrossRef]
- Yang, N.; Hu, J.Q.; Zhou, X.; Wang, A.Y.; Yu, J.J.; Tao, X.Y.; Tang, J. A rapid detection method of early spore viability based on AC impedance measurement. J. Food Process Eng. 2020, 43, e13520. [Google Scholar] [CrossRef]
- Fernandez, J.; Orth, K. Rise of a cereal killer: The biology of Magnaporthe oryzae biotrophic growth. Trends Microbiol. 2018, 26, 582–597. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Yao, Z.F.; He, D.J. Automatic detection and counting of urediniospores of Puccinia striiformis f. sp. tritici using spore traps and image processing. Sci. Rep. 2018, 8, e13647. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.; Macher, J. Automated spore measurements using microscopy, image analysis, and peak recognition of near-monodisperse aerosols. Aerosol Sci. Technol. 2012, 46, 862–873. [Google Scholar] [CrossRef]
- Zhao, Z.Q.; Zheng, P.; Xu, S.T.; Wu, X.D. Object detection with deep learning: A review. IEEE Trans. Neural Netw. Learn. Syst. 2019, 30, 3212–3232. [Google Scholar] [CrossRef] [PubMed]
- Lowe, D.G. Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Dalal, N.; Triggs, B. Histograms of oriented gradients for human detection. In Proceedings of the 2005 IEEE Conference on Computer Vision and Pattern Recognition, San Diego, CA, USA, 20–25 June 2005; IEEE: Piscateville, NJ, USA, 2005; pp. 886–893. [Google Scholar] [CrossRef]
- Wu, X.W.; Sahoo, D.; Hoi, S.C.H. Recent advances in deep learning for object detection. Neurocomputing 2020, 396, 39–64. [Google Scholar] [CrossRef]
- Yang, G.F.; Yang, Y.; He, Z.K.; Zhang, X.Y.; He, Y. A rapid, low-cost deep learning system to classify strawberry disease based on cloud service. J. Integr. Agric. 2022, 21, 460–473. [Google Scholar]
- Xiao, Y.Z.; Tian, Z.Q.; Yu, J.C.; Zhang, Y.S.; Liu, S.; Du, S.Y.; Lan, X.G. A review of object detection based on deep learning. Multimed. Tools Appl. 2020, 79, 23729–23791. [Google Scholar] [CrossRef]
- Liu, L.; Ouyang, W.L.; Wang, X.G.; Fieguth, P.; Chen, J.; Liu, X.W.; Pietikainen, M. Deep learning for generic object detection: A survey. Int. J. Comput. Vis. 2020, 128, 261–318. [Google Scholar] [CrossRef]
- Fu, L.; Feng, Y.; Wu, J.; Liu, Z.; Gao, F.; Majeed, Y.; Al-Mallahi, A.; Zhang, Q.; Li, R.; Cui, Y. Fast and accurate detection of kiwifruit in orchard using improved YOLOv3-tiny model. Precis. Agric. 2021, 22, 754–776. [Google Scholar] [CrossRef]
- Parvathi, S.; Selvi, S.T. Detection of maturity stages of coconuts in complex background using Faster R-CNN model. Biosyst. Eng. 2021, 202, 119–132. [Google Scholar] [CrossRef]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. In Proceedings of the 2014 IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 23–28 June 2014; IEEE: Piscateville, NJ, USA, 2014; pp. 580–587. [Google Scholar] [CrossRef]
- He, K.M.; Zhang, X.Y.; Ren, S.Q.; Sun, J. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 37, 1904–1916. [Google Scholar] [CrossRef]
- Redmon, J.; Divvala, S.; Girshick, R.; Farhadi, A. You only look once: Unified, real-time object detection. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; IEEE: Piscateville, NJ, USA, 2016; pp. 779–788. [Google Scholar] [CrossRef]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.Y.; Berg, A.C. SSD: Single shot multibox detector. In Proceedings of the 14th European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; IEEE: Piscateville, NJ, USA, 2016; pp. 21–37. [Google Scholar] [CrossRef]
- Pham, M.T.; Courtrai, L.; Friguet, C.; Lefevre, S.; Baussard, A. YOLO-Fine: One-stage detector of small objects under various backgrounds in remote sensing images. Remote Sens. 2020, 12, 2501. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, J.; Tang, F.; Zhang, H.; Cui, Z.; Zhou, H. An automatic detector for fungal spores in microscopic images based on deep learning. Appl. Eng. Agric. 2021, 37, 85–94. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar] [CrossRef]
- Ren, S.Q.; He, K.M.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 1137–1149. [Google Scholar] [CrossRef]
- Kubera, E.; Kubik-Komar, A.; Kurasinski, P.; Piotrowska-Weryszko, K.; Skrzypiec, M. Detection and recognition of pollen grains in multilabel microscopic images. Sensors 2022, 22, 2690. [Google Scholar] [CrossRef]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. In Proceedings of the 2017 IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; IEEE: Piscateville, NJ, USA, 2017; pp. 2999–3007. [Google Scholar] [CrossRef]
- Shakarami, A.; Menhaj, M.B.; Mahdavi-Hormat, A.; Tarrah, H. A fast and yet efficient YOLOv3 for blood cell detection. Biomed. Signal Process. Control 2021, 66, e102495. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. YOLOv3: An Incremental Improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar] [CrossRef]
- Yang, N.; Qian, Y.; EL-Mesery, H.S.; Zhang, R.B.; Wang, A.Y.; Tang, J. Rapid detection of rice disease using microscopy image identification based on the synergistic judgment of texture and shape features and decision tree-confusion matrix method. J. Sci. Food. Agric. 2019, 99, 6589–6600. [Google Scholar] [CrossRef]
- Wang, Z.; Chu, G.; Wang, J.; Huang, X.; Gao, F.; Ding, X. Spores detection of rice blast by IKSVM based on HOG features. Trans. Chin. Soc. Agric. Mach. 2018, 49, 387–392, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Qi, L.; Jiang, Y.; Li, Z.; Ma, X.; Zheng, Z.; Wang, W. Automatic detection and counting method for spores of rice blast based on micro image processing. Trans. Chin. Soc. Agric. Eng. 2015, 31, 186–193, (In Chinese with English Abstract). [Google Scholar] [CrossRef]
- Lee, S.H.; Goeau, H.; Bonnet, P.; Joly, A. New perspectives on plant disease characterization based on deep learning. Comput. Electron. Agric. 2020, 170, e105220. [Google Scholar] [CrossRef]
- Chen, J.D.; Chen, J.X.; Zhang, D.F.; Sun, Y.D.; Nanehkaran, Y.A. Using deep transfer learning for image-based plant disease identification. Comput. Electron. Agric. 2020, 173, e105393. [Google Scholar] [CrossRef]
- Jiang, J.L.; Liu, H.Y.; Zhao, C.; He, C.; Ma, J.F.; Cheng, T.; Zhu, Y.; Cao, W.X.; Yao, X. Evaluation of diverse convolutional neural networks and training strategies for wheat leaf disease identification with field-acquired photographs. Remote Sens. 2022, 14, 3446. [Google Scholar] [CrossRef]
- Feng, Q.; Xu, P.; Ma, D.; Lan, G.; Wang, F.; Wang, D.; Yun, Y. Online recognition of peanut leaf diseases based on the data balance algorithm and deep transfer learning. Precis. Agric. 2023, 24, 560–586. [Google Scholar] [CrossRef]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollar, P.; Zitnick, C.L. Microsoft COCO: Common objects in context. In Proceedings of the 13th European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; IEEE: Piscateville, NJ, USA, 2014; pp. 740–755. [Google Scholar] [CrossRef]
- Girshick, R. Fast R-CNN. In Proceedings of the 2015 IEEE International Conference on Computer Vision, Santiago, Chile, 11–18 December 2015; IEEE: Piscateville, NJ, USA, 2015; pp. 1440–1448. [Google Scholar] [CrossRef]
- Cai, Z.W.; Vasconcelos, N. Cascade R-CNN: Delving into high quality object detection. In Proceedings of the 2018 IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; IEEE: Piscateville, NJ, USA, 2018; pp. 6154–6162. [Google Scholar] [CrossRef]
- He, K.M.; Zhang, X.Y.; Ren, S.Q.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 26 June–1 July 2016; IEEE: Piscateville, NJ, USA, 2016; pp. 770–778. [Google Scholar] [CrossRef]
- He, T.; Zhang, Z.; Zhang, H.; Zhang, Z.Y.; Xie, J.Y.; Li, M. Bag of tricks for image classification with convolutional neural networks. In Proceedings of the 2019 IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; IEEE: Piscateville, NJ, USA, 2019; pp. 558–567. [Google Scholar] [CrossRef]
- Howard, A.G.; Menglong, Z.; Chen, B.; Kalenichenko, D.; Weijun, W.; Weyand, T.; Andreetto, M.; Adam, H. MobileNets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar] [CrossRef]
- Howard, A.; Sandler, M.; Chu, G.; Chen, L.; Chen, B.; Tan, M.X.; Wang, W.J.; Zhu, Y.K.; Pang, R.M.; Vasudevan, V.; et al. Searching for MobileNetV3. In Proceedings of the 2019 IEEE International Conference on Computer Vision, Seoul, Republic of Korea, 27 October–2 November 2019; IEEE: Piscateville, NJ, USA, 2019; pp. 1314–1324. [Google Scholar] [CrossRef]
- Fraiwan, M.; Faouri, E.; Khasawneh, N. Classification of corn diseases from leaf images using deep transfer learning. Plants 2022, 11, 2668. [Google Scholar] [CrossRef] [PubMed]
- Gogoi, M.; Kumar, V.; Begum, S.A.; Sharma, N.; Kant, S. Classification and detection of rice diseases using a 3-Stage CNN architecture with transfer learning approach. Agriculture 2023, 13, 1505. [Google Scholar] [CrossRef]
- Zhu, X.Z.; Hu, H.; Lin, S.; Dai, J.F. Deformable ConvNets v2: More deformable, better results. In Proceedings of the 2019 IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; IEEE: Piscateville, NJ, USA, 2019; pp. 9300–9308. [Google Scholar] [CrossRef]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
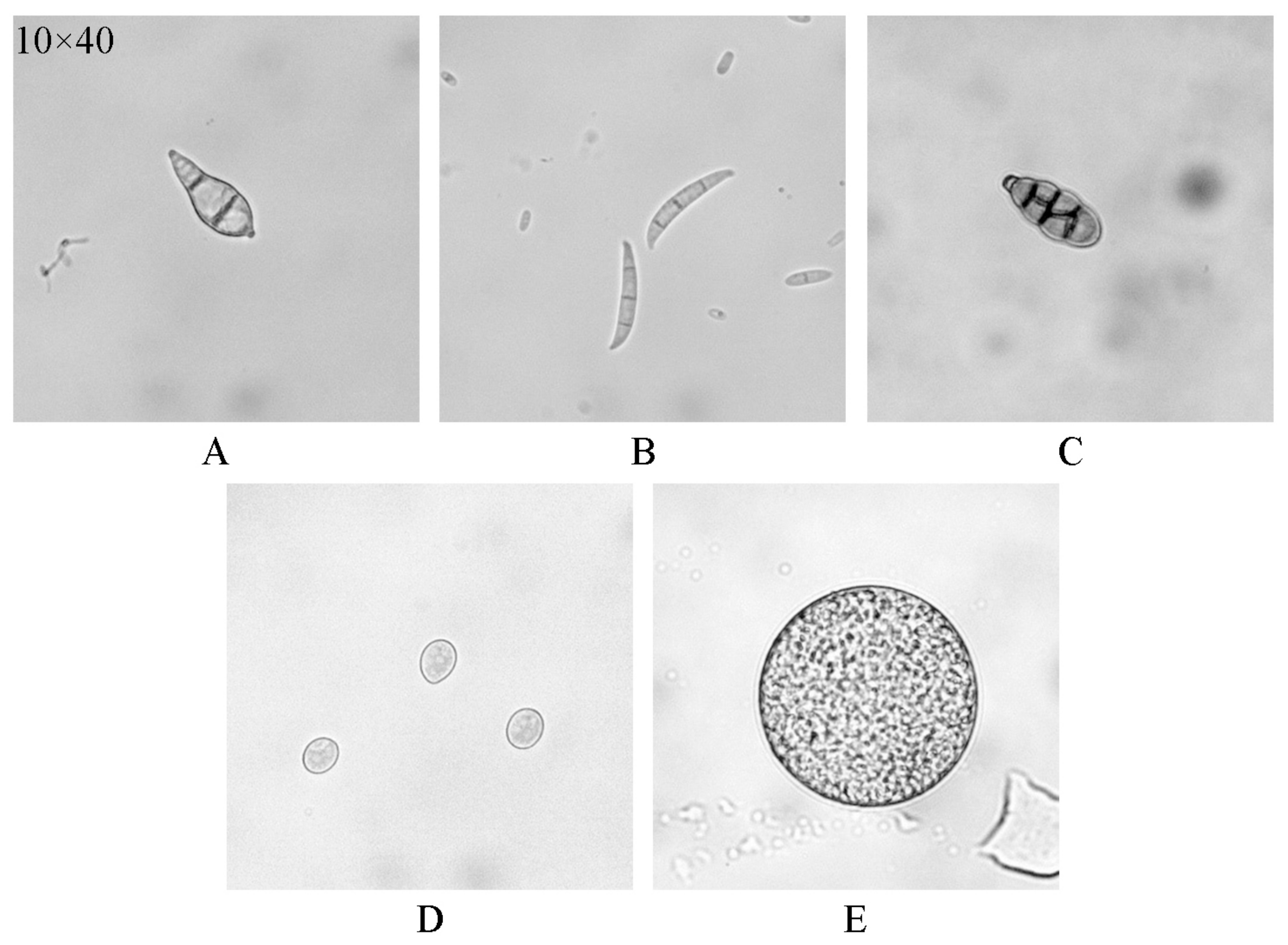





| (a) | ||||||
| ric 1 | mel 2 | bet 3 | str 4 | pol 5 | ||
| D40_Train | 1520 | 1425 | 1465 | 1441 | 1316 | |
| D40_Validation | 380 | 357 | 366 | 360 | 329 | |
| (b) | ||||||
| Datasets | ric | ric&mel | ric&bet | ric&str | ric&pol | pol |
| D20_Train | 200 | 200 | 200 | 200 | / | 100 |
| D20_Validation | 50 | 50 | 50 | 50 | / | 25 |
| D20_test_indoor | 50 | 50 | 50 | 50 | / | 25 |
| T20_field | 20 | 20 | 20 | 20 | 20 | / |
| Pre-Trained Models | Object Detection Algorithms | |
|---|---|---|
| Epochs | 100 | 100 |
| Initial learning rate | 0.1 | 0.001 |
| Milestone 1 | 30, 60, 90 | 50, 80 |
| Warmup steps 2 | / | 4000 |
| Batch_size | 32 | 4 |
| Pretrained weight | False | True/False |
| Input size | default | default |
| Models | Pre-Train 1 | FPS (f/s) | mAP(0.5) | mAP_ric(0.5:0.95) | ||
|---|---|---|---|---|---|---|
| D20_Test_ Indoor | T20_Field | D20_Test_ Indoor | T20_Field | |||
| Cascade_RCNN_ ResNet50 | True | 11.5 | 0.976 | 0.812 | 0.739 | 0.586 |
| False | 11.7 | 0.969 | 0.813 | 0.742 | 0.546 | |
| ave | 11.6 | 0.973 2 | 0.813 | 0.741 | 0.566 | |
| Cascade_RCNN_ ResNet 50_vd | True | 11.4 | 0.953 | 0.789 | 0.710 | 0.513 |
| False | 11.6 | 0.963 | 0.776 | 0.719 | 0.504 | |
| ave | 11.5 | 0.958 | 0.783 | 0.715 | 0.509 | |
| Faster_RCNN _ResNet34 | True | 20.1 | 0.967 | 0.804 | 0.694 | 0.451 |
| False | 19.2 | 0.969 | 0.801 | 0.696 | 0.469 | |
| ave | 19.6 | 0.968 | 0.803 | 0.695 | 0.460 | |
| Faster_RCNN _ResNet50_vd | True | 3.5 | 0.946 | 0.776 | 0.715 | 0.416 |
| False | 3.5 | 0.954 | 0.796 | 0.722 | 0.468 | |
| ave | 3.5 | 0.950 | 0.786 | 0.719 | 0.442 | |
| Yolov3_ResNet34 | True | 42.4 | 0.980 | 0.822 | 0.702 | 0.504 |
| False | 42.5 | 0.978 | 0.800 | 0.676 | 0.499 | |
| ave | 42.5 | 0.979 | 0.811 | 0.689 | 0.502 | |
| Yolov3_ResNet50_vd | True | 36.8 | 0.822 | 0.663 | 0.423 | 0.300 |
| False | 36.5 | 0.713 | 0.654 | 0.376 | 0.342 | |
| ave | 36.7 | 0.768 | 0.659 | 0.400 | 0.321 | |
| Yolov3_ DarkNet53 | True | 35.0 | 0.979 | 0.818 | 0.705 | 0.527 |
| False | 37.7 | 0.980 | 0.803 | 0.694 | 0.506 | |
| ave | 36.4 | 0.980 | 0.811 | 0.700 | 0.517 | |
| Yolov3_MobileNetv1 | True | 42.0 | 0.924 | 0.692 | 0.613 | 0.431 |
| False | 43.3 | 0.945 | 0.745 | 0.612 | 0.441 | |
| ave | 42.7 | 0.935 | 0.719 | 0.613 | 0.436 | |
| Yolov3_MobileNetv3_large | True | 42.5 | 0.966 | 0.727 | 0.650 | 0.476 |
| False | 42.6 | 0.939 | 0.774 | 0.623 | 0.477 | |
| ave | 42.6 | 0.953 | 0.751 | 0.637 | 0.477 | |
| Data Location | Detection Rate | Spore Classes | Time (s/f) | Test Images | |
|---|---|---|---|---|---|
| DT/ CM [28] 1 | Table 4 | 0.940 | Rice blast spores and rice smut spores | 18 | 500 |
| HOG/ IKSVM [29] 2 | Table 1 | 0.982 | Rice blast spores | 4.8 | 150 |
| FCM-Canny/ DT-GF-WA [30] 3 | Table 1 | 0.985 | Rice blast spores | / | 100 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, H.; Lai, Q.; Huang, Q.; Cai, D.; Huang, D.; Wu, B. Automatic Detection of Rice Blast Fungus Spores by Deep Learning-Based Object Detection: Models, Benchmarks and Quantitative Analysis. Agriculture 2024, 14, 290. https://doi.org/10.3390/agriculture14020290
Zhou H, Lai Q, Huang Q, Cai D, Huang D, Wu B. Automatic Detection of Rice Blast Fungus Spores by Deep Learning-Based Object Detection: Models, Benchmarks and Quantitative Analysis. Agriculture. 2024; 14(2):290. https://doi.org/10.3390/agriculture14020290
Chicago/Turabian StyleZhou, Huiru, Qiang Lai, Qiong Huang, Dingzhou Cai, Dong Huang, and Boming Wu. 2024. "Automatic Detection of Rice Blast Fungus Spores by Deep Learning-Based Object Detection: Models, Benchmarks and Quantitative Analysis" Agriculture 14, no. 2: 290. https://doi.org/10.3390/agriculture14020290
APA StyleZhou, H., Lai, Q., Huang, Q., Cai, D., Huang, D., & Wu, B. (2024). Automatic Detection of Rice Blast Fungus Spores by Deep Learning-Based Object Detection: Models, Benchmarks and Quantitative Analysis. Agriculture, 14(2), 290. https://doi.org/10.3390/agriculture14020290






