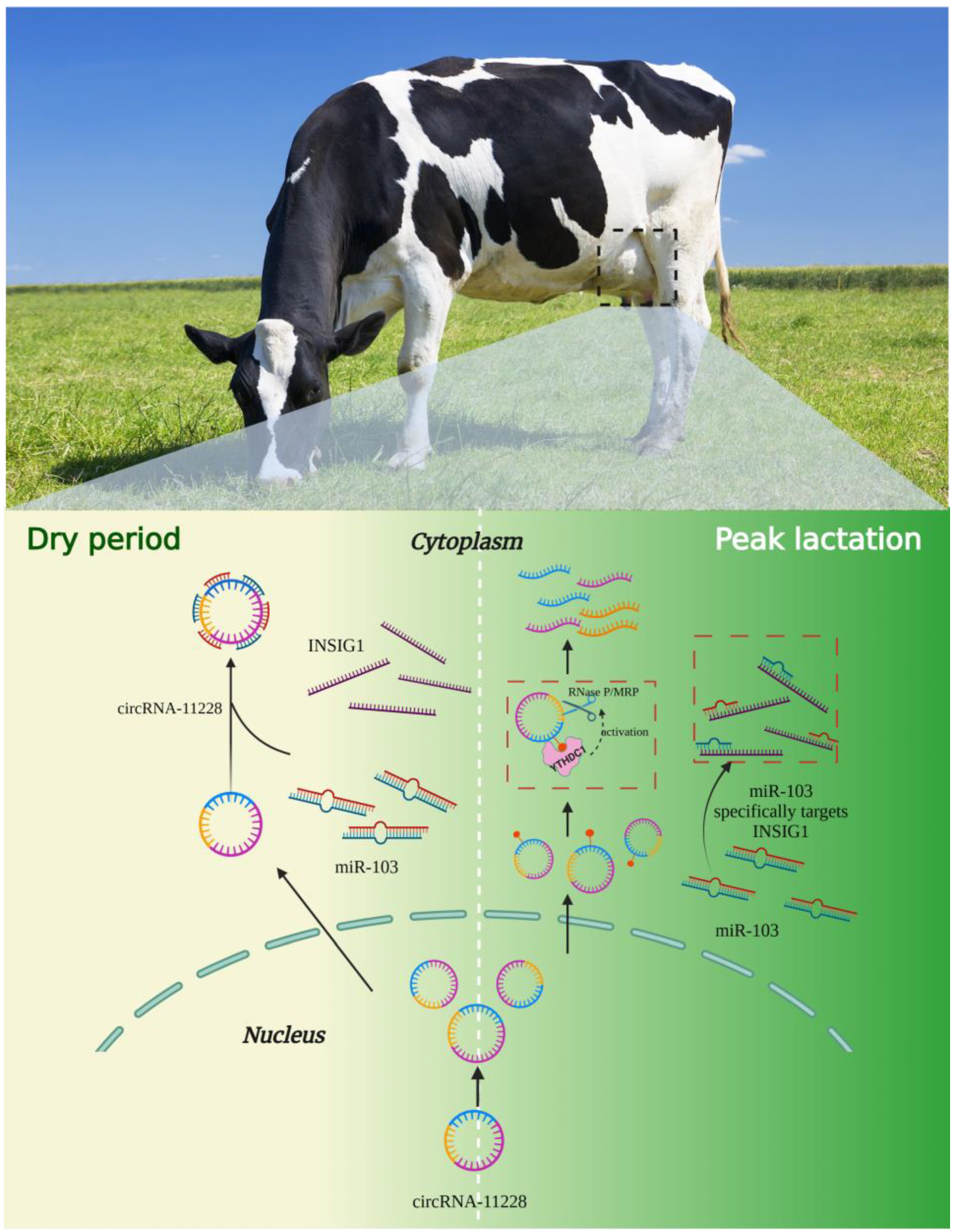
The Molecular Mechanism of circRNA-11228/miR-103/INSIG1 Pathway Regulating Milk Fat Synthesis in Bovine Mammary Epithelial Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture of BMECs (Bovine Mammary Epithelial Cells)
2.2. Triglycerides and Cholesterol Analysis
2.3. Oil Red O Staining
2.4. EdU Cell Proliferation Detection
2.5. Double Luciferase Report
2.6. Quantitative Real-Time PCR (qRT-PCR)
2.7. Western Blot
2.8. Data Analysis
3. Results
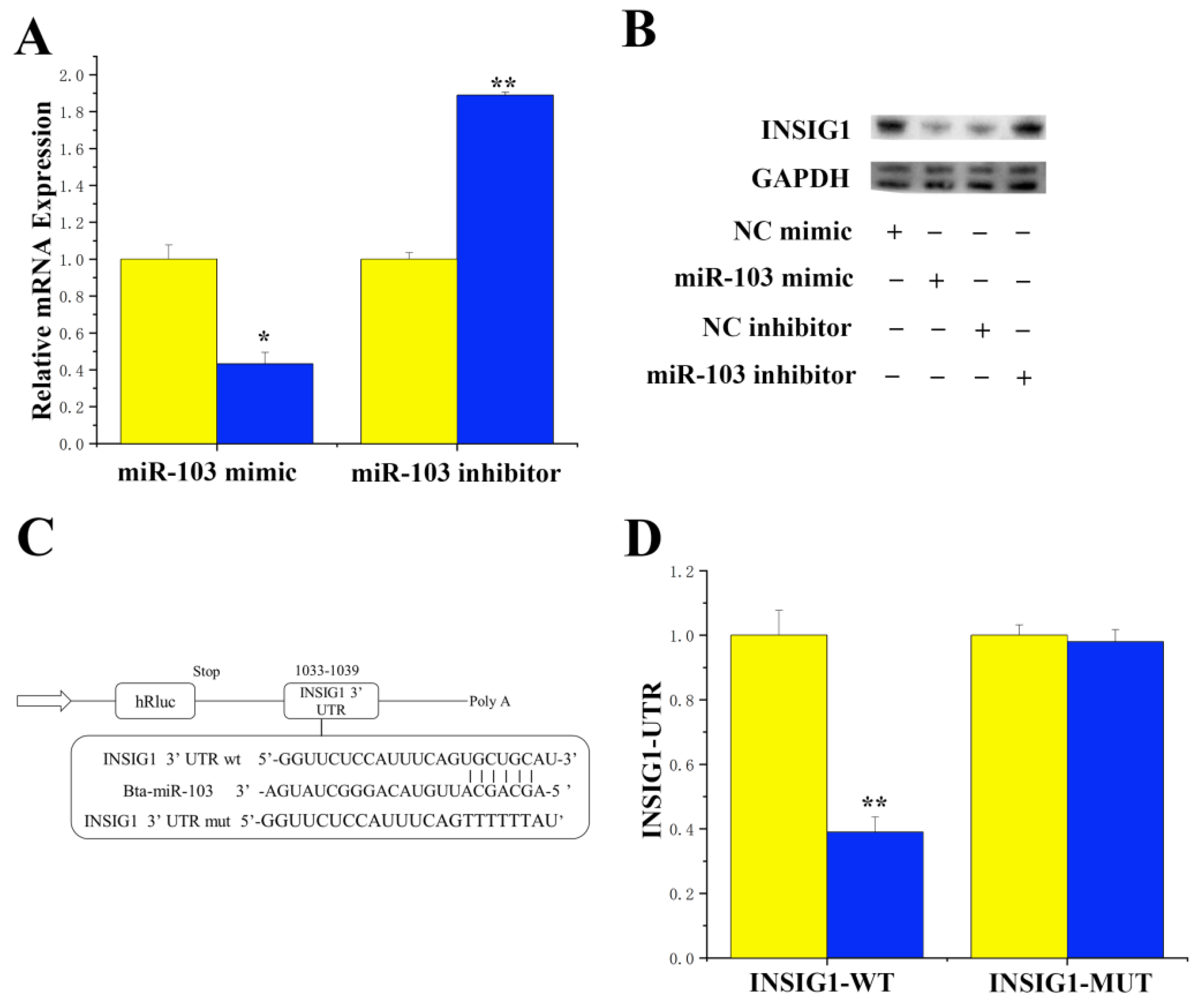
3.1. Targeting INSIG1 with miR-103 Specificity
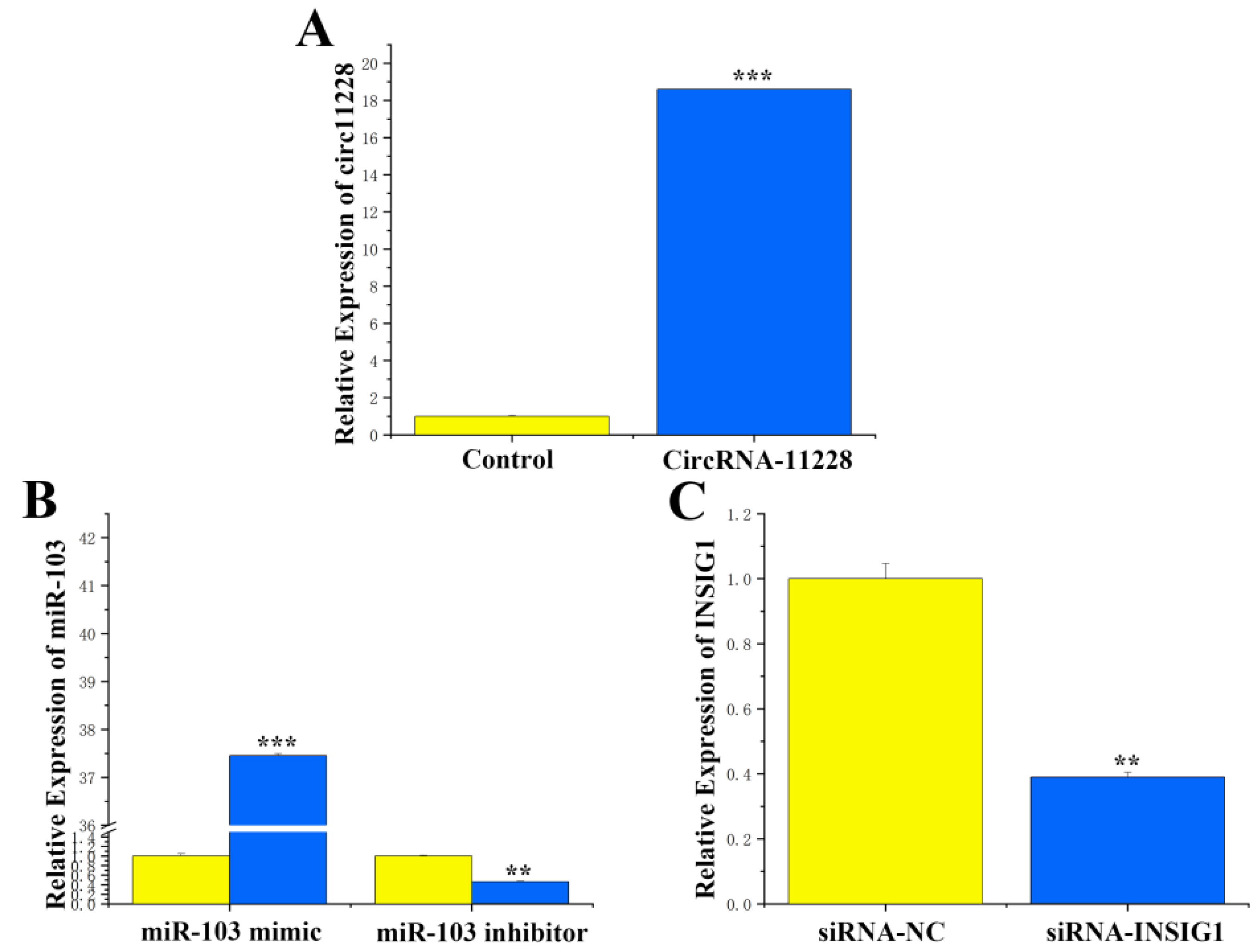
3.2. CircRNA-11228 Competitive Binding to miR-103
3.3. Transfection Efficiency of circRNA-11228, miR-103, and siRNA INSIG1
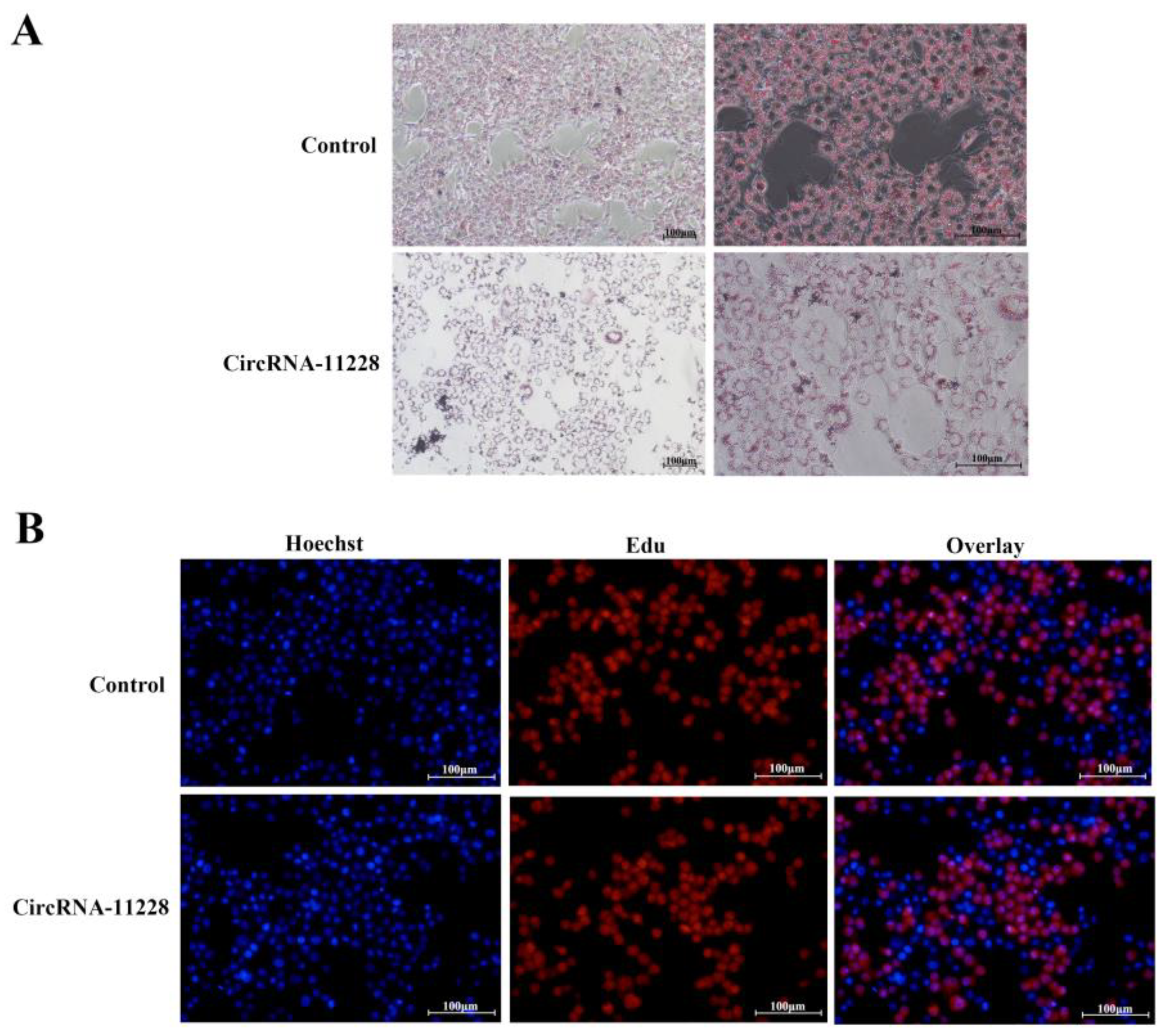
3.4. Functional Validation of circRNA-11228 in BMECs
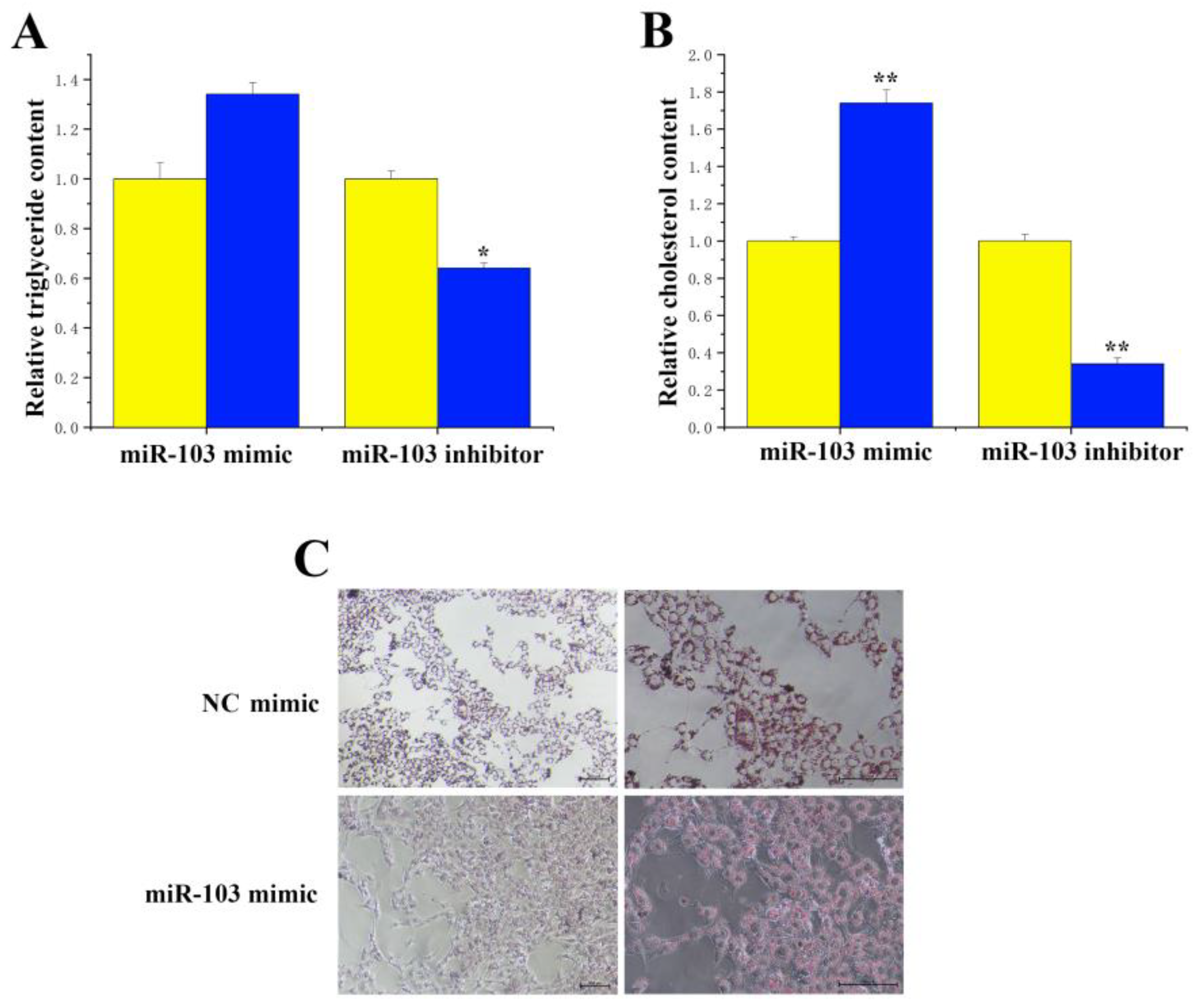
3.5. Functional Validation of miR-103 in BMECs
3.6. Functional Validation of INSIG1 in BMECs
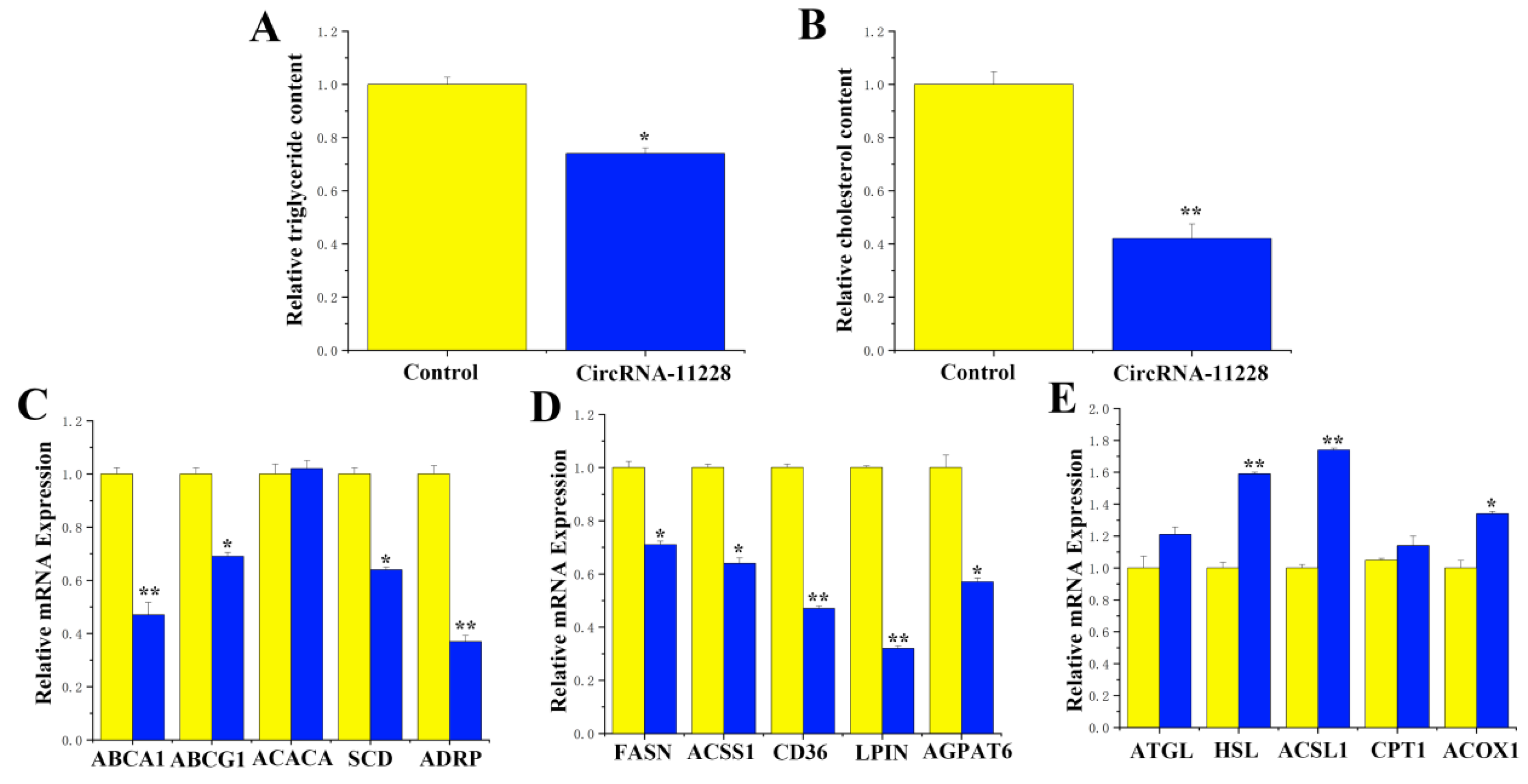
3.7. CircRNA-11228 Regulates Cholesterol Metabolism in BMECs by Adsorbing miR-133a
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Beermann, J.; Piccoli, M.T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef] [PubMed]
- Abe, B.T.; Wesselhoeft, R.A.; Chen, R.; Anderson, D.G.; Chang, H.Y. Circular RNA migration in agarose gel electrophoresis. Mol. Cell 2022, 82, 1768–1777.e1763. [Google Scholar] [CrossRef] [PubMed]
- Amaya, L.; Grigoryan, L.; Li, Z.; Lee, A.; Wender, P.A.; Pulendran, B.; Chang, H.Y. Circular RNA vaccine induces potent T cell responses. Proc. Natl. Acad. Sci. USA 2023, 120, e2302191120. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Tang, Z.; Huang, X.; Chen, W.; Zhou, J.; Liu, H.; Liu, C.; Kong, N.; Tao, W. Emerging mRNA technologies: Delivery strategies and biomedical applications. Chem. Soc. Rev. 2022, 51, 3828–3845. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Y.; Zhou, S.; Dain, L.; Mei, L.; Zhu, G. Circular RNA: An emerging frontier in RNA therapeutic targets, RNA therapeutics, and mRNA vaccines. J. Control Release 2022, 348, 84–94. [Google Scholar] [CrossRef]
- Hill, M.; Tran, N. miRNA:miRNA Interactions: A Novel Mode of miRNA Regulation and Its Effect on Disease. Adv. Exp. Med. Biol. 2022, 1385, 241–257. [Google Scholar]
- Smith, E.M.; Zhang, Y.; Baye, T.M.; Gawrieh, S.; Cole, R.; Blangero, J.; Carless, M.A.; Curran, J.E.; Dyer, T.D.; Abraham, L.J.; et al. INSIG1 influences obesity-related hypertriglyceridemia in humans. J. Lipid Res. 2010, 51, 701–708. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Wang, Z.; Xia, Y.; Shao, F.; Xia, W.; Wei, Y.; Li, X.; Qian, X.; Lee, J.H.; Du, L.; et al. The gluconeogenic enzyme PCK1 phosphorylates INSIG1/2 for lipogenesis. Nature 2020, 580, 530–535. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Tao, J.; Yu, X.; Wu, Y.; Chen, Y.; You, K.; Zhang, J.; Getachew, A.; Pan, T.; Zhuang, Y.; et al. Hypomorphic ASGR1 modulates lipid homeostasis via INSIG1-mediated SREBP signaling suppression. JCI Insight 2021, 6, e147038. [Google Scholar] [CrossRef]
- Cheng, C.; Ru, P.; Geng, F.; Liu, J.; Yoo, J.Y.; Wu, X.; Cheng, X.; Euthine, V.; Hu, P.; Guo, J.Y.; et al. Glucose-Mediated N-glycosylation of SCAP Is Essential for SREBP-1 Activation and Tumor Growth. Cancer Cell 2015, 28, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Azzu, V.; Vacca, M.; Kamzolas, I.; Hall, Z.; Leslie, J.; Carobbio, S.; Virtue, S.; Davies, S.E.; Lukasik, A.; Dale, M.; et al. Suppression of insulin-induced gene 1 (INSIG1) function promotes hepatic lipid remodelling and restrains NASH progression. Mol. Metab. 2021, 48, 101210. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Qiu, L.; Teng, X.; Zhang, Y.; Miao, Y. Effect of INSIG1 on the milk fat synthesis of buffalo mammary epithelial cells. J. Dairy Res. 2020, 87, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Jiao, H.; Gao, W.; Huang, L.; Shi, C.; Zhang, F.; Wu, J.; Luo, J. Fatty Acid Desaturation Is Suppressed in Mir-26a/b Knockout Goat Mammary Epithelial Cells by Upregulating INSIG1. Int. J. Mol. Sci. 2023, 24, 10028. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.X.; Rothenberg, M.E. MicroRNA. J. Allergy Clin. Immunol. 2018, 141, 1202–1207. [Google Scholar] [CrossRef] [PubMed]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Xia, C.; Cen, P.; Li, S.; Yu, L.; Zhu, J.; Jin, J. MiR-103-3p promotes hepatic steatosis to aggravate nonalcoholic fatty liver disease by targeting of ACOX1. Mol. Biol. Rep. 2022, 49, 7297–7305. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, Z.; Zhang, Z.; Liu, G.; Sun, S.; Sun, C. miR-103 promotes 3T3-L1 cell adipogenesis through AKT/mTOR signal pathway with its target being MEF2D. Biol. Chem. 2015, 396, 235–244. [Google Scholar] [CrossRef]
- Wang, Z.; Shen, W.; Zhu, M.; Xu, M.; Qiu, M.; Zhang, D.; Chen, S. MiR-199-3p Suppressed Inflammatory Response by Targeting MECP2 to Alleviate TRX-Induced PHN in Mice. Cell Transplant. 2022, 31, 9636897221108192. [Google Scholar] [CrossRef]
- Chen, Z.; Lu, Q.; Liang, Y.; Cui, X.; Wang, X.; Mao, Y.; Yang, Z. Circ11103 Interacts with miR-128/PPARGC1A to Regulate Milk Fat Metabolism in Dairy Cows. J. Agric. Food Chem. 2021, 69, 4490–4500. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Lu, Q.; Zhang, X.; Zhang, Z.; Cao, X.; Wang, K.; Lu, X.; Yang, Z.; Loor, J.J.; Jiao, P. Circ007071 Inhibits Unsaturated Fatty Acid Synthesis by Interacting with miR-103-5p to Enhance PPARγ Expression in the Dairy Goat Mammary Gland. J. Agric. Food Chem. 2022, 70, 13719–13729. [Google Scholar] [CrossRef] [PubMed]
- Sveinbjornsson, G.; Ulfarsson, M.O.; Thorolfsdottir, R.B.; Jonsson, B.A.; Einarsson, E.; Gunnlaugsson, G.; Rognvaldsson, S.; Arnar, D.O.; Baldvinsson, M.; Bjarnason, R.G.; et al. Multiomics study of nonalcoholic fatty liver disease. Nat. Genet. 2022, 54, 1652–1663. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, D.; Zhang, M.; Sun, J.; Li, W.; Jiang, R.; Han, R.; Wang, Y.; Tian, Y.; Kang, X.; et al. miRNA-223 targets the GPAM gene and regulates the differentiation of intramuscular adipocytes. Gene 2019, 685, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Hu, Z.; Cui, A.; Liu, Z.; Ma, F.; Xue, Y.; Liu, Y.; Zhang, F.; Zhao, Z.; Yu, Y.; et al. Post-translational regulation of lipogenesis via AMPK-dependent phosphorylation of insulin-induced gene. Nat. Commun. 2019, 10, 623. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Xu, J.; Ma, Q.; Liu, Z.; Sudhahar, V.; Cao, Y.; Wang, L.; Zeng, X.; Zhou, Y.; Zhang, M.; et al. PRKAA1/AMPKα1-driven glycolysis in endothelial cells exposed to disturbed flow protects against atherosclerosis. Nat. Commun. 2018, 9, 4667. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Ma, Q.; Xu, J.; Liu, Z.; Mao, X.; Zhou, Y.; Cai, Y.; Da, Q.; Hong, M.; Weintraub, N.L.; et al. Endothelial AMPKα1/PRKAA1 exacerbates inflammation in HFD-fed mice. Br. J. Pharmacol. 2022, 179, 1661–1678. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Luo, J.; Gao, W.; Song, N.; Tian, H.; Zhu, L.; Jiang, Q.; Loor, J.J. CRISPR/Cas9-Induced Knockout of miR-24 Reduces Cholesterol and Monounsaturated Fatty Acid Content in Primary Goat Mammary Epithelial Cells. Foods 2022, 11, 2012. [Google Scholar] [CrossRef] [PubMed]
- Ng, R.; Wu, H.; Xiao, H.; Chen, X.; Willenbring, H.; Steer, C.J.; Song, G. Inhibition of microRNA-24 expression in liver prevents hepatic lipid accumulation and hyperlipidemia. Hepatology 2014, 60, 554–564. [Google Scholar] [CrossRef]
- Chen, C.; Wang, S.; Zhang, M.; Chen, B.; You, C.; Xie, D.; Liu, Y.; Monroig, Ó.; Tocher, D.R.; Waiho, K.; et al. miR-24 is involved in vertebrate LC-PUFA biosynthesis as demonstrated in marine teleost Siganus canaliculatus. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 619–628. [Google Scholar] [CrossRef]
- Rodriguez, R.E.; Schommer, C.; Palatnik, J.F. Control of cell proliferation by microRNAs in plants. Curr. Opin. Plant Biol. 2016, 34, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Gong, W.; Wang, Q.; Wang, P.; Shi, T.; Mahmut, A.; Qin, J.; Yao, Y.; Yan, W.; Chen, D.; et al. Atrophic skeletal muscle fibre-derived small extracellular vesicle miR-690 inhibits satellite cell differentiation during ageing. J. Cachexia Sarcopenia Muscle 2022, 13, 3163–3180. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhang, C.; Zhang, J.; Jiao, Z.; Dong, N.; Wang, G.; Wang, Z.; Wang, L. Localized injection of miRNA-21-enriched extracellular vesicles effectively restores cardiac function after myocardial infarction. Theranostics 2019, 9, 2346–2360. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, H.; Zeng, D.; Xiong, J.; Luo, J.; Chen, X.; Chen, T.; Xi, Q.; Sun, J.; Ren, X.; et al. The novel importance of miR-143 in obesity regulation. Int. J. Obes. 2023, 47, 100–108. [Google Scholar] [CrossRef]
- Bork, S.; Horn, P.; Castoldi, M.; Hellwig, I.; Ho, A.D.; Wagner, W. Adipogenic differentiation of human mesenchymal stromal cells is down-regulated by microRNA-369-5p and up-regulated by microRNA-371. J. Cell Physiol. 2011, 226, 2226–2234. [Google Scholar] [CrossRef] [PubMed]
- Benito-Vicente, A.; Uribe, K.B.; Rotllan, N.; Ramírez, C.M.; Jebari-Benslaiman, S.; Goedeke, L.; Canfrán-Duque, A.; Galicia-García, U.; Saenz De Urturi, D.; Aspichueta, P.; et al. miR-27b Modulates Insulin Signaling in Hepatocytes by Regulating Insulin Receptor Expression. Int. J. Mol. Sci. 2020, 21, 8675. [Google Scholar] [CrossRef] [PubMed]
- Sakai, E.; Imaizumi, T.; Suzuki, R.; Taracena-Gándara, M.; Fujimoto, T.; Sakurai, F.; Mizuguchi, H. miR-27b targets MAIP1 to mediate lipid accumulation in cultured human and mouse hepatic cells. Commun. Biol. 2023, 6, 669. [Google Scholar] [CrossRef]
- Generoso, G.; Janovsky, C.; Bittencourt, M.S. Triglycerides and triglyceride-rich lipoproteins in the development and progression of atherosclerosis. Curr. Opin. Endocrinol. Diabetes Obes. 2019, 26, 109–116. [Google Scholar] [CrossRef]
- McKenzie, K.M.; Lee, C.M.; Mijatovic, J.; Haghighi, M.M.; Skilton, M.R. Medium-Chain Triglyceride Oil and Blood Lipids: A Systematic Review and Meta-Analysis of Randomized Trials. J. Nutr. 2021, 151, 2949–2956. [Google Scholar] [CrossRef]
- Peng, J.Y.; Cai, D.K.; Zeng, R.L.; Zhang, C.Y.; Li, G.C.; Chen, S.F.; Yuan, X.Q.; Peng, L. Upregulation of Superenhancer-Driven LncRNA FASRL by USF1 Promotes De Novo Fatty Acid Biosynthesis to Exacerbate Hepatocellular Carcinoma. Adv. Sci. 2022, 10, e2204711. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.G.; Zhu, S.T.; Cheng, H.M.; Zhang, X.; Cheng, G.; Thu, P.M.; Wang, S.P.; Li, H.J.; Ding, M.; Qiang, L.; et al. Discovery of a potent SCAP degrader that ameliorates HFD-induced obesity, hyperlipidemia and insulin resistance via an autophagy-independent lysosomal pathway. Autophagy 2021, 17, 1592–1613. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.X.; Chen, L.L. Circular RNAs: Characterization, cellular roles, and applications. Cell 2022, 185, 2016–2034. [Google Scholar] [CrossRef] [PubMed]
- Wen, S.Y.; Qadir, J.; Yang, B.B. Circular RNA translation: Novel protein isoforms and clinical significance. Trends Mol. Med. 2022, 28, 405–420. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yang, L.; Chen, L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef]
- Belousova, E.A.; Filipenko, M.L.; Kushlinskii, N.E. Circular RNA: New Regulatory Molecules. Bull. Exp. Biol. Med. 2018, 164, 803–815. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Wu, Y.; Wang, Y.; Yang, X.; Gao, R.; Lu, Q.; Lv, X.; Chen, Z. The Molecular Mechanism of circRNA-11228/miR-103/INSIG1 Pathway Regulating Milk Fat Synthesis in Bovine Mammary Epithelial Cells. Agriculture 2024, 14, 538. https://doi.org/10.3390/agriculture14040538
Li X, Wu Y, Wang Y, Yang X, Gao R, Lu Q, Lv X, Chen Z. The Molecular Mechanism of circRNA-11228/miR-103/INSIG1 Pathway Regulating Milk Fat Synthesis in Bovine Mammary Epithelial Cells. Agriculture. 2024; 14(4):538. https://doi.org/10.3390/agriculture14040538
Chicago/Turabian StyleLi, Xiaofen, Yanni Wu, Yuhao Wang, Xiaozhi Yang, Rui Gao, Qinyue Lu, Xiaoyang Lv, and Zhi Chen. 2024. "The Molecular Mechanism of circRNA-11228/miR-103/INSIG1 Pathway Regulating Milk Fat Synthesis in Bovine Mammary Epithelial Cells" Agriculture 14, no. 4: 538. https://doi.org/10.3390/agriculture14040538
APA StyleLi, X., Wu, Y., Wang, Y., Yang, X., Gao, R., Lu, Q., Lv, X., & Chen, Z. (2024). The Molecular Mechanism of circRNA-11228/miR-103/INSIG1 Pathway Regulating Milk Fat Synthesis in Bovine Mammary Epithelial Cells. Agriculture, 14(4), 538. https://doi.org/10.3390/agriculture14040538






