Weedy Rice Infestation in Malaysia: What Do We Know and Where Do We Go?
Abstract
:1. Introduction
2. Weedy Rice Issues in Malaysia
3. Adaptive Traits of Weedy Rice
4. Weedy Rice Research in Malaysia
| Category | Methods | Findings | Area/Samples Coverage | References |
|---|---|---|---|---|
| Ecology | Field samplings, field surveys | More than 19,900 ha of rice farms were damaged by weedy rice. | Peninsular Malaysia | [25] |
| New biotypes of weedy rice (NBWR) with shorter plant height were first reported. Most farm blocks showed clump or under-dispersed spatial distribution. | Selangor | [2] | ||
| The relative abundant indices of annual weeds were more dominant than perennial weeds. | Kedah | [72] | ||
| The Lloyd patchiness index revealed that more than 50% of surveyed field blocks in five states displayed uniform distribution. | Peninsular Malaysia | [42] | ||
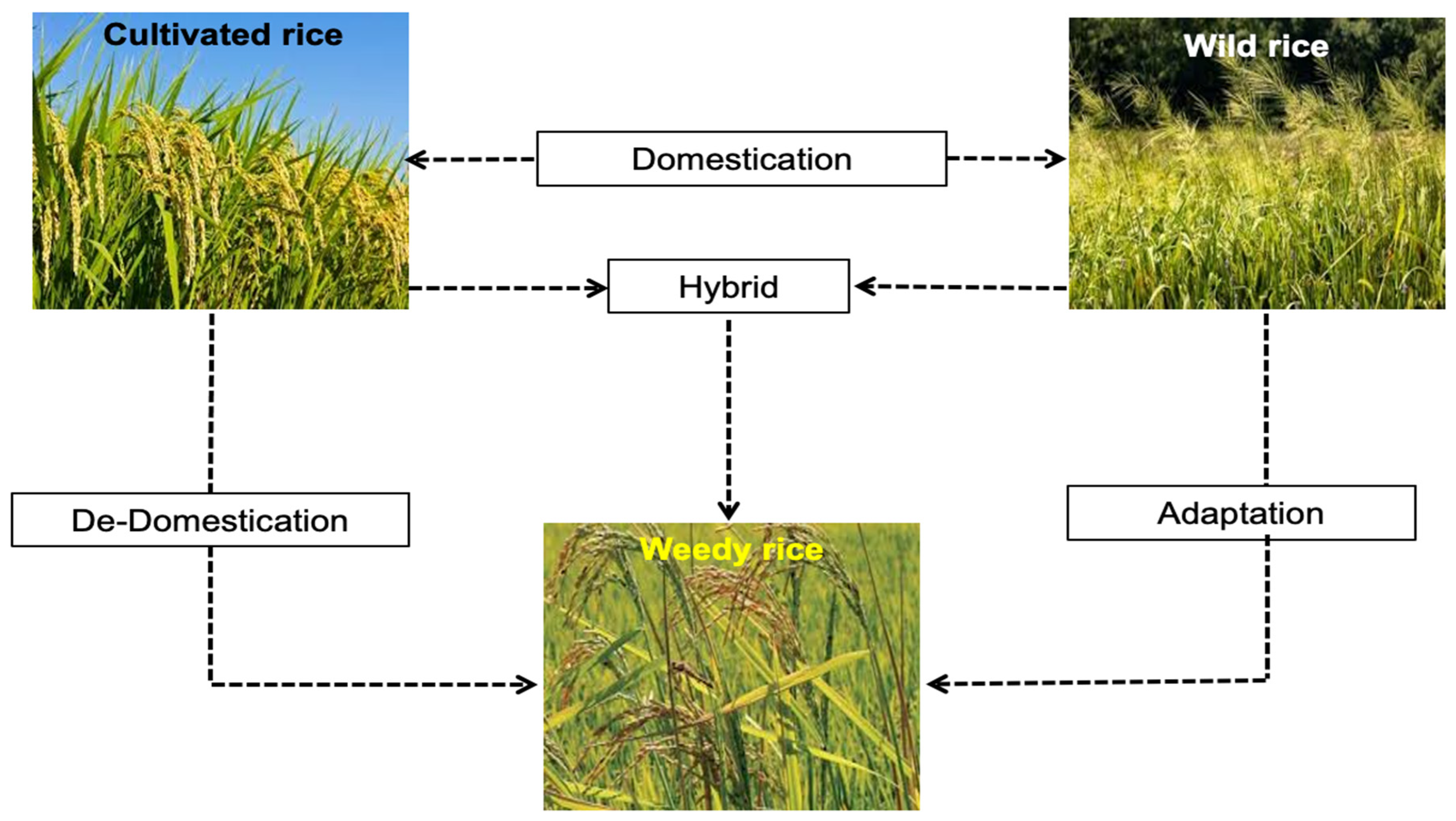
| Genomic | PCR, SSR | The high genetic diversity showed that Malaysian weedy rice had diverse origins: de-domestication from cultivated rice, adaptation from wild rice, and hybridization between cultivated and wild rice or between cultivated rice and weedy rice. | Peninsular Malaysia | [16,20,53,71,73,74] |
| Sabah weedy rice was shaped by accidentally introducing Peninsular Malaysia’s weedy rice strains. Cultivated rice was highly diverse compared to Peninsular, which supported the progenitor of Sabah cultivar-like weedy rice. | Sabah | [6,33] | ||
| All weedy rice populations showed cross-resistance to the IMI herbicides imazapic and imazapyr due to a Ser-653-Asn mutation in the AHAS gene resulting from an herbicide-insensitive AHAS enzyme. | Kedah, Perlis | [75] | ||
| The multiplex PCR assays employed a standard agarose-based gel electrophoresis system to simultaneously disclose at least two major grain quality (amylose content and fragrance) and biotic stress (blast, sheath blight, and bacterial leaf blight) genes in rice. | Not applicable | [8] | ||
| Morphology and Physiology | Greenhouse and field experiment | Heterogenous plant height, hull color, pericarp color, awn, and maturity period. Most weedy rice had seed shattering. Greater in growth development, tiller number, LAI | Peninsular Malaysia | [1,7,42,43,44,73,76,77] |
| The rice and weed dry matter, rice plant height, chlorophyll content, leaf area, number of tillers, filled grain, 1000 grain weight and grain yield were reduced with an increased crop–weed competition period. | Selangor, Pahang | [53,61] | ||
| The germination rate was found to be related to the degree of dormancy, but it had no influence on the range of cardinal temperatures. | Peninsular Malaysia | [78] | ||
| A high degree of seed dormancy retained their viability for more than 200 days once imbibed. | Selangor | [70] | ||
| Weed Management | Field samplings and surveys | Sites that practiced integrated weed management with regular surveillance and monitoring had less weedy rice infestation and higher rice yields and vice versa with control. Major weed populations are F. miliacea, L. Hyssopifolia and O. sativa. | Perlis, Kedah | [79] |
| Most farmers ignored the technology and deliberately disregarded stewardship guidelines. Their perceptions of the weedy rice issue varied from region to region, leading to differing methods for controlling weedy rice. Farmers were more likely to use herbicides than mechanical to control weedy rice. | Peninsular Malaysia | [80,81] | ||
| Greenhouse and field experiment | Weedy rice populations were effectively controlled, resulting in an average 15% yield increment under the Clearfield® Production System (CPS) in direct seeded fields. The critical period for weed control ranged from approximately 12–16 to 53–60 DAS. | Perak, Penang | [28,37] | |
| Weedy rice was found to be more resistant to OnDuty® (premix of imazapic and imazapyr) than the susceptible weedy rice. | Perlis, Kedah Penang | [39] | ||
| Weedy rice in Malaysia developed various degrees of resistance toward IMI herbicide. | Selangor | [66] | ||
| Metabolomic | Field sampling, NMR | Weedy rice and cultivated rice can be clearly distinguished based on metabolome profiles. It was found that the metabolite profiles of weedy rice from the east coast are different from the west coast area of Peninsular Malaysia. | Peninsular Malaysia | [82] |
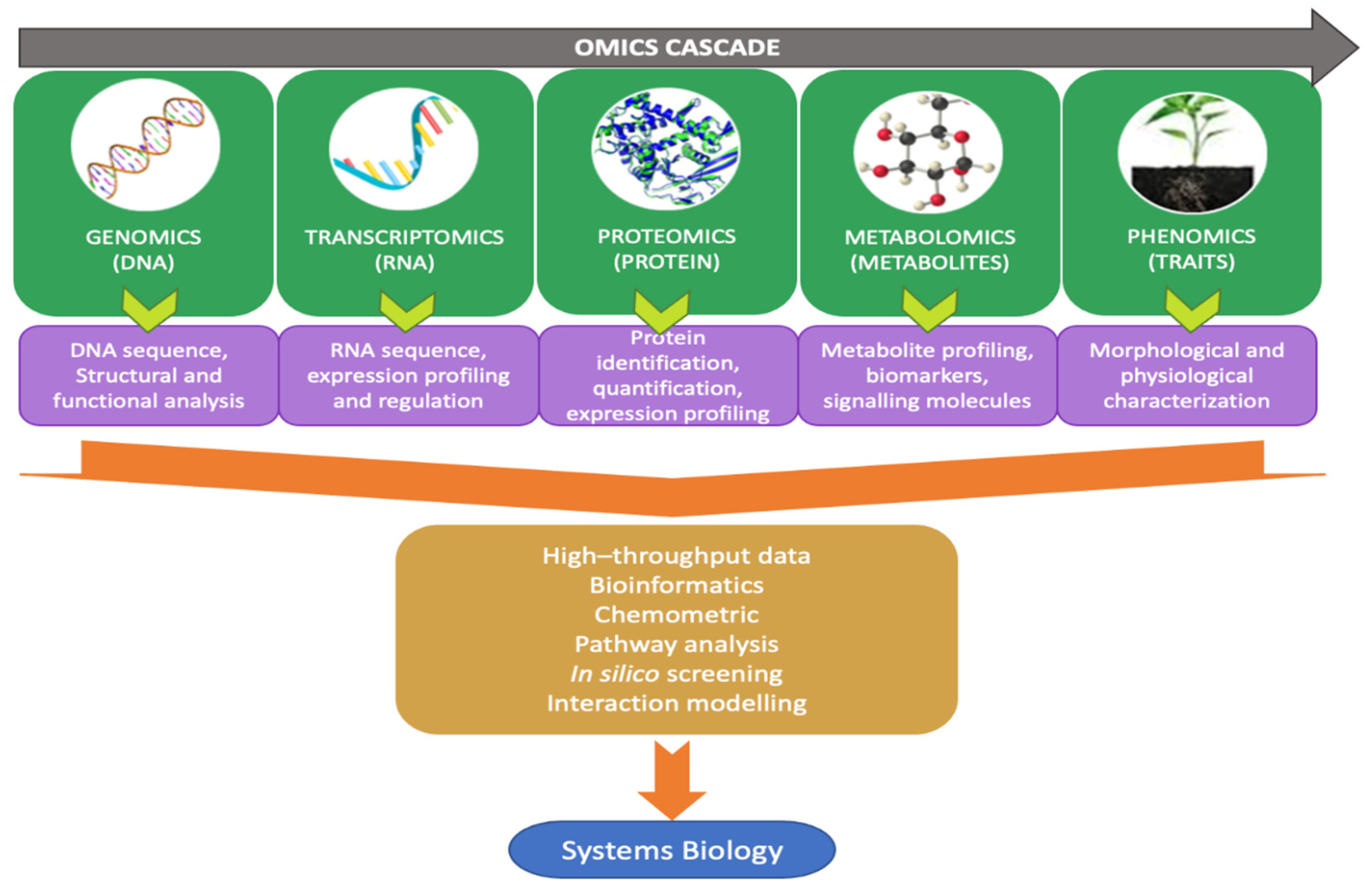
5. Omics Study as Knowledge-Based Weed Management Strategies for the Future
| Omics Study | Field of Research | References |
|---|---|---|
| Genomics | Evolution: origin, genetic variation, adaptation | [6,11,18,74,89,97,98,99,100,101,102,103,104,105] |
| Genomics | Phenotypic variation | [106] |
| Genomics | Herbicide resistance | [53] |
| Genomics | Anthocyanin nutrition | [107] |
| Genomics | Abiotic stress: temperature | [108] |
| Transcriptomics | Evolution: origin, genetic variation, adaptation | [109] |
| Transcriptomics | Phenotypic variation, hormone level | [110] |
| Transcriptomics | Abiotic stress: cold stress | [65,94,111,112] |
| Transcriptomics | Abiotic stress: drought | [113,114] |
| Transcriptomics | Abiotic: nutrient stress | [95] |
| Transcriptomics | Abiotic stress: temperature | [93] |
| Proteomics | Abiotic stress: drought | [114] |
| Proteomics | Biotic stress: insects | [92] |
| Metabolomics | Chemical profiles: accession, geographical origin | [82] |
6. Genomics Aspect of Weedy Rice
| Gene (s) | Important Morphological Traits with Response to Abiotic and Biotic Stress | References |
|---|---|---|
| sh4, qSH1 | Seed shattering | [116,124,125] |
| Rc | Pigmented pericarp | [54,74,107] |
| Bh4, Phr1 | Hull color (black) | [119,122] |
| An-1, LABA1 | Awn length and awn barb | [126,127] |
| Sdr4, DOG1, qSD7-1, CYP707A, NCED | Dormancy and germination | [51,128] |
| sd-1 | Plant height | [89,129] |
| Prog1 | Erect leave and erect panicles | [89,99,130] |
| OsLG1 | Closed panicle | [99,131] |
| Hd1 | Early flowering | [132] |
| AtCYP78A7 | Drought tolerance | [113] |
| HKT, NHX1, SOS1 | Salinity stress | [115,133] |
| RAB16, OVP1, APX1 | Cold tolerance | [111] |
| HSF2a, HSFA7 | Heat tolerance | [134] |
| ADH, PDC, OsB12D1 | Flooding tolerance | [135] |
| AHAS, ALS, EPSP | Herbicide resistance (imazapic–imazapyr, glyphosate) | [35,136] |
| Pi-ta | Blast resistant (Magnaporthe oryza B.) | [137] |
7. The Potential Use of Metabolomics in Weedy Rice Study
| Organ Parts | Category | Experimental Methodology | Metabolomics Acquisition Methods | Bioinformatics Tools | Statistical Analysis | Findings | Source of Samples | References |
|---|---|---|---|---|---|---|---|---|
| Grains | Natural variation | Field | LC-MS, GC-MS | R, SIMCA-P, Cytoscape, MeV, MetATT | PCA, Heatmap, LASSO, Metabolic pathway, Two-way ANOVA, ASCA, Pearson correlation | Japonica and indica subspecies were significantly different in relative abundances of metabolites and metabolic association networks. | China | [151] |
| Grains | Natural variation | Laboratory | SPME-GC-MS, NMR, LC-ESI-MS, GC-TOF-MS | MetAlign, AMIX, SIMCA-P, TagFinder | PCA, PLS-DA, HCA | Cultivars of jasmine and basmati showed different metabolic profiles. Storage grains had a significant effect on the metabolome in both cultivars. | Iran, Pakistan, Malaysia, Thailand, Indonesia, Philipines, Cambodia, Australia, Japan | [154] |
| Grains | Natural variation | Laboratory | NMR | Chenomx, SIMCA-P, SPSS | PCA, PLS-DA, ANOVA, Tukey’s | The PLS model demonstrated that α-linolenic acid, ϒ-oryzanol, α-tocopherol, ϒ-aminobutyric acid, 3-hydroxybutyric acid, fumaric acid, fatty acids, threonine, tryptophan, and vanillic acid were significantly correlated with black germinated rice. | Thailand | [155] |
| Grains | Natural variation | Greenhouse | GC-MS | MetaMiner, AMDIS, PageMan, MapMan | t-test, HCA, Heatmap, metabolic pathway | Increment in hexose phosphates, tricarboxylic acid cycle intermediates, and ϒ-aminobutyric acid after one hour of water imbibition. Later enrichment in carbohydrate, amino acid, and cell wall metabolism. | None | [161] |
| Grains | Natural variation | Field | HPLC | Chrompare, XLSTAT | PCA, HCA | Significant differences in the contents of the anthocyanins, cyanidin-3-glucoside and peonidin-3- glucoside of red, black and non-colored indica and japonica rice subspecies. | China | [162] |
| Grains | Natural variation | Field, genebank | LC-MS, NMR | None | None | Hyperin, isoquercitrin, quercetin, gentiobioside were isolated from the grains of sugary rice. | Korea | [163] |
| Grains | Natural variation | Field, genebank | HPLC | Statistix 8.0 | ANOVA | Brown and white rice contained lower quantities of phytochemicals compared to black and red rice. | France | [164] |
| Grains | Natural variation | Field, genebank | LC-MS, GC-MS | Chroma TOF, MassLynx, MetAlign, SIMCA-P, MeV, Statistica 18 | PCA, PLS-DA, box-whisker, ANOVA, Duncan’s test, Pearson’s correlations | Antioxidant compounds (cyanidin-3-glucoside, peonidin-3-glucoside, proanthocyanidin dimers, proanthocyanidin trimers, and catechin) mostly found in black and red rice seeds. | Korea | [165] |
| Grains | Natural variation | Greenhouse | NMR | Mestrenova | OPLS-DA | Metabolites of valine, threonine, alanine, glutamate, galactinol, β-glucose, α-glucose, raffinose, and fumaric acid influenced the separation of red rice and black rice. | Indonesia | [166] |
| Grains | Natural variation | Field | LC-MS, GC-MS | MeV, SIMCA-P, Cytoscape, SPSS, R | PCA, Heatmap, ANOVA, Metabolic pathway, t-test, Pearson’s correlation | The chalky endosperm had lower levels of metabolites compared to the translucent upper part. | China | [167] |
| Grains | Natural variation | Field, genebank | GC-MS | TraceFinder | Heatmap | Identified 66 metabolites in the rice samples and cultivar Jaya showed the highest number of metabolites. | India | [168] |
| Grains | Natural variation | Field, genebank | HR-MAS NMR | MATLAB, SIMCA-P, SAS | PCA, OPLS-DA, ANOVA, t-test | Waxy rice cultivars accumulated lipids and had high levels of glutamate, aspartate, asparagine, alanine, and sucrose compared to nonwaxy rice cultivars. | Korea | [169] |
| Cooked grains | Natural variation | Laboratory | LC-MS | MarkerLynx, SIMCA-P | PLS-DA, Kruskal-wallis test | Chemical diversity among the varieties clustered according to subspecies classifications: indica, japonica, and aus. | Philippines | [170] |
| Rice bran | Natural variation | Field, genebank | LC-MS | Metabolon, SIMCA-P | Metabolic pathway, PCA | The 71 rice bran compounds of significant variation by cultivar included 21 amino acids, 7 carbohydrates, 2 metabolites from cofactors and vitamins, 33 lipids, 6 nucleotides, and 2 secondary metabolites. Tryptophan, α-ketoglutarate, ϒ-tocopherol/β-tocopherol, and ϒ-tocotrienol were among the metabolites. | Cambodia, India, Kenya, Mali, Nepal, Nicaragua and USA | [156] |
| Grains and seedlings | Natural variation | Field, growth chamber | HPLC | Analyst 1.5 | Heatmap, metabolic pathway | 24 candidate genes were associated with various metabolic quantitative trait loci by data mining. | China | [171] |
| Seedlings | Natural variation | Laboratory | GC-MS | ChemStation, XLSTAT | PCA, MANOVA | Thai black and purple rice contained higher levels of metabolites than the red and colorless samples. | Thailand | [172] |
| Seedlings | Natural variation | Laboratory | GS-MS, LC-MS | SIMCA-P, SPSS | PCA, PLS-DA, metabolic pathway | 25 metabolites, including acidic compounds, amino acids, sugars, lipids, and secondary metabolites were identified as the components that contributed to the variations in the germinated brown rice group. | Korea | [173] |
| Seedlings | Natural variation | Laboratory | LC-ESI-MS | R | OPLS-DA | Phenylpropanoid biosynthesis and glutathione metabolism were continuously enriched during the seed germination and young seedling growth stages. | China | [174] |
| Leaves | Abiotic stress (Plant–drought–flood) | Field | LC-MS | XCMS, Msconverter, Excel, SIMCA-P, R, SPSS | PCA, PLS-DA, OPLS-DA, t-test, Heatmap, Euclidean distance, ANOVA | 102 different metabolites were identified from the rice spike between T1 (abrupt drought–flood alternation) and control treatment, 104 different metabolites were identified between T1 and CK1 (drought) treatment, and 116 different metabolites were identified between T1 and CK2 (flood) treatment. | China | [96] |
| Leaves | Abiotic stress (Plant–drought, heat) | Field | LCMS | R, ggplot2 | PCA, Venn diagram, heatmap | Decrement of some metabolites in stressed plants at specific development stages and organs: glycerophosphoglycerol, isocitric acid, ribitol, A116014 (Flag leaves/flowering), A214004, dehydroascorbic acid, dimer, glyceric acid, glycine, malic acid, phosphoric acid (flag leaves/early grain-filling), A147011, A180002, arbutin, aspartic acid, erythronic acid, galactonic acid, phosphoric acid (flowering spikelets). | Philippines | [158] |
| Leaves | Abiotic Stress (Plant–flood) | Greenhouse | GC-MS, NMR | AMDIS, MarkerLynx XS, MestReNova, Topspin, Minitab 15, SPSS | PCA, t-test | Metabolites of S-methyl methionine and the dipeptide alanylglycine were only detected by ¹H NMR. | USA | [175,176] |
| Leaves | Natural variation | Chamber | GS-MS, LC-MS | Vx Capture, MassLynx DataBridge, MetAlign, SIMCA-P, STATISTICA, Cytoscape | PCA, PLS-DA, OPLS-DA, Heatmap, ANOVA, Pearson’s correlation, metabolic pathway | Antioxidant activities of rice leaves were high in blue, white, and green light, followed by red and shade light of LED. | Korea | [177] |
| Leaves | Abiotic stress (Plant–drought) | Chamber | GC-MS | Chroma TOF, R | PCA, HCA, Heatmap Pearson correlation | Metabolite levels were mainly negatively correlated with performance parameters under drought stress. | Philippines, Vietnam | [178] |
| Leaves | Biotic Stress (Plant–insect) | Greenhouse | NMR | Chenomx, SIMCA-P, SPSS | PCA, Tukey’s test, ANOVA | The concentration of 10 metabolites was significantly altered between the infestation by planthopper and the control groups. | Thailand | [179] |
| Leaves | Abiotic stress (Plant–cold) | Greenhouse | GC-MS, CE-MS, LC-MS | None | PCA, Heatmap, Pathway | The accumulation of glucose, fructose, and sucrose involved in starch degradation, sucrose metabolism, and the glyoxylate cycle were upregulated in rice plants exposed to cold or dehydration. | Japan | [180] |
| Leaves | Biotic Stress (Plant–insect) | Greenhouse | GC-MS, LC-MS | XCMS, AMDIS, PeakView, MArkView, MetaboAnalyst, MetPA, SPSS | PCA, t-test, metabolic pathway, ANOVA | Alteration of metabolites in pathway analysis in both resistant cultivars compared to the control. Cyanoamino acids and lipid metabolism were induced in IR36, while changes in thiamine, taurine and hypotaurine metabolism in IR56. | Philippines | [181] |
| Leaves and roots | Abiotic stress (Plant–salinity) | Greenhouse | GC-MS | SAS, R | Heatmap, HCA, ANOVA | Sugars and amino acids increased significantly in the leaves and roots of both genotypes under salt stress, while organic acids increased in roots and decreased in leaves. | None | [182] |
| Leaves and roots | Abiotic Stress (Plant–drought) | Greenhouse | GC-MS | Chroma TOF, R | PCA, Two-way ANOVA, t-test | Higher accumulation of N-rich metabolites in shoots of the tolerant varieties, whereas in roots, the aus-type varieties showed a reduction in metabolites representative of glycolysis and the TCA cycle. | Philippines | [183] |
| Roots | Abiotic stress (Plant–salinity) | Chamber | NMR | Chenomx, MATLAB, MathWorks, SIMCA-P | PCA, PLS-DA, OPLS-DA | Salt-responsive metabolic markers of rice roots were identified, sucrose, allantoin and glutamate, whereas the levels of glutamine and alanine decreased. | Korea | [115] |
| Roots | Biotic Stress (Plant–fungus) | Cultures | GC-MS | AMDIS, SIMCA-P | PCA, OPLS-DA, Heatmap | Levels of metabolites of the shikimate and lignin biosynthesis pathways increased in the M. oryzae-challenged rice roots (Mo-roots) and reduced in H. oryzae-challenged rice roots (Ho-roots). Control showed a reduction in sucrose and maltose in both Ho-roots and Mo-roots. | China | [184] |
8. Study of Weedy Rice in Malaysia: Where Do We Go?
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Anuar, N.S.; Mazlan, N.; Ariff, E.E.; Juraimi, A.S.; Yusop, M.R. A comparative study of vegetative and reproductive growth of local weedy and Clearfield® rice varieties in Malaysia. ISSAAS J. 2014, 20, 41–51. [Google Scholar]
- Baki, B.B.; Shakirin, M.M. Spatio-temporal distribution pattern of new biotypes of weedy rice (Oryza sativa L.) in Selangor north-west project, Malaysia. Korean J. Weed Sci. 2010, 30, 68–83. [Google Scholar] [CrossRef]
- Azmi, M.; Karim, S.M.R. Weedy Rice-Biology, Ecology and Management; Malaysian Agricultural Research and Development Institute (MARDI): Kuala Lumpur, Malaysia, 2008. [Google Scholar]
- Rathore, M.; Singh, R.; Kumar, B.; Chauhan, B.S. Characterization of functional traits diversity among Indian cultivated and weedy rice populations. Sci. Rep. 2016, 6, 24176. [Google Scholar] [CrossRef] [PubMed]
- Mispan, M.S.; Bzoor, M.; Mahmod, I.F.; MD-Akhir, A.H.; Zulrushdi, A. Managing weedy rice (Oryza sativa L.) in Malaysia: Challenges and ways forward. J. Res. Weed Sci. 2019, 2, 149–167. [Google Scholar]
- Neik, T.X.; Chai, J.Y.; Tan, S.Y.; Sudo, M.P.S.; Cui, Y.; Jayaraj, J.; Teo, S.S.; Olsen, K.M.; Song, B.K. When west meets east: The origins and spread of weedy rice between continental and island Southeast Asia. G3 2019, 9, 2941–2950. [Google Scholar] [CrossRef] [PubMed]
- Sudianto, E.; Neik, T.X.; Tam, S.M.; Chuah, T.S.; Idris, A.A.; Olsen, K.M.; Song, B.K. Morphology of Malaysian weedy rice (Oryza sativa): Diversity, origin and implications for weed management. Weed Sci. 2016, 64, 501–512. [Google Scholar] [CrossRef]
- Mohd Hanafiah, N.; Cheng, A.; Lim, P.E.; Sethuraman, G.; Mohd Zain, N.A.; Baisakh, N.; Mispan, M.S. Novel PCR-Based Multiplex Assays for Detecting Major Quality and Biotic Stress in Commercial and Weedy Rice. Life 2022, 12, 1542. [Google Scholar] [CrossRef] [PubMed]
- Li, L.F.; Olsen, K.M. Population genomics of weedy crop relatives: Insights from weedy rice. In Population Genomics; Rajora, O.P., Ed.; Springer: Cham, Switzerland, 2020; pp. 1–25. [Google Scholar]
- Shrestha, S.; Stallworth, S.; Tseng, T.M. Weedy rice: Competitive ability, evolution, and diversity. In Integrated view of Population Genetics; Rafael, T.M., Magnolia, A.C., Eds.; Intech Open: London, UK, 2018; pp. 27–40. [Google Scholar]
- De Leon, T.B.; Karn, E.; Al-Khatib, K.; Espino, L.; Blank, T.; Andaya, C.B.; Andaya, V.C.; Brim-DeForest, W. Genetic variation and possible origins of weedy rice found in California. Ecol. Evol. 2019, 9, 5835–5848. [Google Scholar] [CrossRef] [PubMed]
- Vigueira, P.A.; Olsen, K.M.; Wagner, C.R.; Chittick, Z.B.; Vigueira, C.C. Weedy rice from South Korea arose from two distinct de-domestication events. Front. Agron. 2020, 2, 602612. [Google Scholar] [CrossRef]
- Choi, J.Y.; Platts, A.E.; Fuller, D.Q.; Wing, R.A.; Purugganan, M.D. The rice paradox: Multiple origins but single domestication in Asian rice. Mol. Biol. Evol. 2017, 34, 969–979. [Google Scholar] [CrossRef]
- Kim, H.; Jung, J.; Singh, N.; Greenberg, A.; Doyle, J.J.; Tyagi, W.; Chung, J.W.; Kimball, J.; Hamilton, R.S.; McCouch, S.R. Population dynamics among six major groups of the Oryza rufipogon species complex, wild relative of cultivated Asian rice. Rice 2016, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Zhu, W.; Rong, J.; Xu, X.; Chen, J.; Lu, B.R. Evidences of introgression from cultivated rice to Oryza rufipogon (Poaceae) populations based on SSR fingerprinting: Implications for wild rice differentiation and conservation. Evol. Ecol. 2016, 20, 501–522. [Google Scholar] [CrossRef]
- Song, B.K.; Chuah, T.S.; Tam, S.M.; Olsen, K.M. Malaysian weedy rice shows its true stripes: Wild Oryza and elite rice cultivars shape agricultural weed evolution in Southeast Asia. Mol. Ecol. 2014, 23, 5003–5017. [Google Scholar] [CrossRef]
- Londo, J.P.; Schaal, B.A. Origins and population genetics of weedy red rice in the USA. Mol. Ecol. 2007, 16, 4523–4535. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Dai, W.; Wu, C.; Song, X.; Qiang, S. Genetic diversity and origin of Japonica- and Indica-like rice biotypes of weedy rice in the Guangdong and Liaoning provinces of China. Gen. Res. Crop Evol. 2012, 59, 399–410. [Google Scholar] [CrossRef]
- Kanapeckas, K.L.; Vigueira, C.C.; Ortiz, A.; Gettler, K.A.; Burgos, N.R.; Fischer, A.J.; Lawton-Rauh, A.L. Escape to ferality: The endoferal origin of weedy rice from crop rice through de-domestication. PLoS ONE 2016, 11, e0162676. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, M.Z.; Watanabe, H.; Vaughan, D.A.; Okuno, K. Padi angin: Its probable origin and diversity revealed through DNA polymorphism. In Proceedings of the 1st National Congress on Genetics, Kuala Lumpur, Malaysia, 7–8 November 1994; pp. 164–167. [Google Scholar]
- Wahab, A.H.; Suhaimi, O. Padi angin: Characteristics, adverse effects and methods of its eradication. Teknol. Padi 1991, 8, 21–31. [Google Scholar]
- Mohammed Zuki, I.; Kamarudin, D. Status and control of padi angin in the Muda area. In Padi Angin Workshop; Malaysian Agricultural Research and Development Institute (MARDI): Penang, Malaysia, 1994; p. 12. [Google Scholar]
- Azmi, M.; Abdullah, M.Z.; Mislamah, B.; Baki, B.B. Management of weedy rice (Oryza sativa L.): The Malaysian experience. In Wild and Weedy Rice in Rice Ecosystems in Asia—A Review; Baki, B.B., Chin, D.V., Mortimer, M., Eds.; IRRI: Manila, Philippines, 2000; pp. 25–34. [Google Scholar]
- Karim, R.S.M.; Zainal, M.; Mansor, M.; Azmi, M. Weedy rice: A cancerous threat for rice growers in Malaysia. In Proceedings of the National Rice Conference, Perak, Malaysia, 28–30 June 2010; Malaysian Agricultural Research and Development Institute (MARDI): Serdang, Malaysia, 2010; pp. 327–329. [Google Scholar]
- Baki, B.B.; Bakar, M.A.; Man, A.B. Weedy rice (Oryza sativa L.) in Peninsular Malaysia. In Wild and Weedy Rice in Rice Ecosystems in Asia—A Review; Baki, B.B., Chin, D.V., Mortimer, M., Eds.; IRRI: Manila, Philippines, 2000; pp. 25–34. [Google Scholar]
- Baki, B.B. Invasive weed species in Malaysia: Species, impacts and management. Malays. J. Sci. 2004, 23, 1–42. [Google Scholar]
- Azmi, M.; Chin, D.V.; Vongsaroj, P.; Johnson, D.E. Emerging issues in weed management of direct-seeded rice in Malaysia, Vietnam, and Thailand. In Rice Is Life: Scientific Perspectives for the 21st Century; International Rice Research Institute: Los Baños, Philippines, 2005; pp. 196–198. [Google Scholar]
- Azmi, M.; Baki, B.B. Weed flora landscapes of the MUDA rice granary in the new millennium: A descriptive analysis. J. Tropic. Agric. Food Sci. 2007, 35, 319–331. [Google Scholar]
- Azmi, M.; Muhamad, H.; Johnson, D.E. Impact of Weedy Rice Infestation on Rice Yield and Influence of Crop Establishment Technique. In Proceedings of the 20th Asian-Pacific Weed Science Society Conference, Ho Chi Minh City, Vietnam, 7–11 November 2005. [Google Scholar]
- Karim, R.S.M.; Azmi, M.; Ismail, S. Weed problems and their management in rice fields of Malaysia: An overview. Weed Biol. Manag. 2004, 4, 177–186. [Google Scholar] [CrossRef]
- Chauhan, B.S. Strategies to manage weedy rice in Asia. Crop Prot. 2013, 48, 51–56. [Google Scholar] [CrossRef]
- Kadir, A.; Kamariah, O. Bridging the technology gap through effective expansion programs. In Proceedings of the International Rice Conference, Alor Setar, Malaysia, 13–16 October 2003; pp. 208–216. [Google Scholar]
- San Sudo, M.P.; Yesudasan, R.; Neik, T.X.; Masilamany, D.; Jayaraj, J.; Teo, S.S.; Rahman, S.; Song, B.K. The details are in the genome-wide SNPs: Fine scale evolution of the Malaysian weedy rice. Plant Sci. 2021, 310, 110985. [Google Scholar] [CrossRef] [PubMed]
- Olajumoke, B.; Juraimi, A.S.; Uddin, M.; Husni, M.H.; Alam, M. Competitive ability of cultivated rice against weedy rice biotypes: A review. Chil. J. Agric. Res. 2016, 76, 243–252. [Google Scholar] [CrossRef]
- Ruzmi, R.; Ahmad-Hamdani, M.S.; Abidin, M.Z.Z.; Roma-Burgos, N. Evolution of imidazolinone-resistant weedy rice in Malaysia: The current status. Weed Sci. 2021, 69, 598–608. [Google Scholar] [CrossRef]
- Mispan, M.S.; Jalaluddin, A.; Majrashi, A.A.; Baki, B.B. Weed Science in Malaysia: An Analysis. In Weed Science in the Asian Pacific Region; Rao, V.S., Yaduraju, N.T., Chandrasena, N.R., Hassan, G., Sharma, A.R., Eds.; Indian Society of Weed Science: Jabalpur, India, 2015; pp. 197–212. [Google Scholar]
- Azmi, M.; Azlan, S.; Yim, K.M.; George, T.V.; Chew, S.E. Control of weedy rice in direct seeded rice using the Clearfield® production system in Malaysia. Pak. J. Weed Sci. Res. 2012, 18, 49–53. [Google Scholar]
- Bzour, M.I.; Zuki, F.M.; Mispan, M.S. Introduction of imidazolinone herbicide and Clearfield rice between weedy rice—Control efficiency and environmental concerns. Environ. Rev. 2018, 26, 181–198. [Google Scholar] [CrossRef]
- Dilipkumar, M.; Burgos, N.R.; Chuah, T.S.; Ismail, S. Cross-resistance to imazapic and imazapyr in a weedy rice (Oryza sativa) biotype found in Malaysia. Planta Daninha 2018, 36, e018182239. [Google Scholar] [CrossRef]
- Dilipkumar, M.; Ahmad-Hamdani, M.S.; Rahim, H.; Chuah, T.S.; Burgos, N.R. Survey on weedy rice (Oryza spp.) management practice and adoption of Clearfield® rice technology in Peninsular Malaysia. Weed Sci. 2021, 69, 558–564. [Google Scholar] [CrossRef]
- Wedger, M.J.; Roma-Burgos, N.; Olsen, K.M. Genomic revolution of US weedy rice in response to 21st century agricultural technologies. Commun. Biol. 2022, 5, 885. [Google Scholar] [CrossRef]
- Mahmod, I.F.; Saiman, M.Z.; Mohamed, Z.; Ishak, M.N.; Mispan, M.S. Morphological variation, distribution and relationship of weedy rice (Oryza sativa L.) in Peninsular Malaysia. Weed Biol. Manag. 2021, 21, 86–99. [Google Scholar] [CrossRef]
- Zainuddin, H.; Azmi, M.; Othman, A.S. Morphological study of the relationships between weedy rice accessions (Oryza sativa complex) and commercial rice varieties in Penang’s rice granary area. Trop. Life Sci. Res. 2010, 21, 47–62. [Google Scholar]
- Ahmed, Q.N.; Roy, R.; Kamaruzaman, M.; Othman, A.S. Vegetative and reproductive growth of weedy rice in Selangor, Malaysia: A comparative study with commercial rice varieties. Malays. Appl. Biol. 2012, 41, 29–35. [Google Scholar]
- Ratnasekera, D. Weedy rice: A threat to rice production in Sri Lanka. J. Univ. Ruhuna 2015, 3, 2–13. [Google Scholar] [CrossRef]
- Mahmod, I.F.; Barakbah, S.S.; Osman, N.; Omar, O. Physiological response of local rice varieties to aerobic condition. Int. J. Agric. Biol. 2014, 16, 738–744. [Google Scholar]
- Stallworth, S.; Shrestha, S.; Schumaker, B.; Roma-Burgos, N.; Tseng, T.M. screening diverse weedy rice (Oryza sativa ssp.) mini germplasm for tolerance to heat and complete submergence stress during seedling stage. Front. Agron. 2021, 3, 642335. [Google Scholar] [CrossRef]
- Svizzero, S. Agronomic practices and biotechnological methods dealing with the occurrence, dispersion, proliferation and adaptation of weedy rice (Oryza sativa f. spontanea). Int. J. Pest Manag. 2021, 70, 62–75. [Google Scholar] [CrossRef]
- Xu, R.; Sun, C. What happened during domestication of wild to cultivated rice. Crop J. 2021, 9, 564–576. [Google Scholar] [CrossRef]
- Abraham, C.T.; Jose, N. Weedy rice invasion in rice fields of India and management options. J. Crop Weed. 2014, 10, 365–374. [Google Scholar]
- Zhang, Z.; Dai, W.; Song, X.; Qiang, S. A model of the relationship between weedy rice seed-bank dynamics and rice-crop infestation and damage in Jiangsu Province, China. Pest Manag. Sci. 2014, 70, 716–724. [Google Scholar] [CrossRef]
- Gu, X.Y.; Kianian, S.F.; Foley, M.E. Seed dormancy imposed by covering tissues interrelates to shattering and seed morphological characteristics in weedy rice. Crop Sci. 2005, 45, 948–955. [Google Scholar] [CrossRef]
- Engku, A.K.; Norida, M.; Juraimi, A.S.; Rafii, M.Y.; Abdullah, S.N.A.; Alam, M.A. Gene flow from Clearfield® rice to weedy rice under field conditions. Plant Soil Environ. 2016, 62, 16–22. [Google Scholar] [CrossRef]
- Chen, L.J.; Lee, D.S.; Song, Z.P.; Suh, H.S.; Lu, B.R. Gene flow from cultivated rice (Oryza sativa) to its weedy and wild relatives. Ann. Bot. 2004, 93, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Mispan, M.S.; Zhang, L.; Feng, J.; Gu, X.Y. Quantitative trait locus and haplotype analyses of wild and crop-mimic traits in U.S. weedy rice. G3 2013, 3, 1049–1059. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.R.; Yang, X.; Ellstrand, N.C. Fitness correlates of crop transgene flow into weedy populations: A case study of weedy rice in China and other examples. Evol. Appl. 2016, 9, 857–870. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.K.; Li, M.J.; Li, J.Y.; Li, X.J.; Yang, X.H.; Tong, Y.P.; Zhang, A.M.; Li, B.; Lin, J.X.; Kuang, T.Y.; et al. Dynamic changes in flag leaf angle contribute to high photosynthetic capacity. Chin. Sci. Bull. 2009, 54, 3045–3052. [Google Scholar] [CrossRef]
- Fogliatto, S.; Ferrero, A.; Vidotto, F. How can weedy rice stand against abiotic stresses? A review. Agronomy 2020, 10, 1284. [Google Scholar] [CrossRef]
- Chauhan, B.S.; Johnson, D.E. The role of seed ecology in improving weed management strategies in the tropics. Adv. Agron. 2010, 105, 221–262. [Google Scholar]
- Dass, A.; Shekhawat, K.; Choudhary, A.K.; Sepat, S.; Rathore, S.S.; Mahajan, G.; Chauhan, B.S. Weed management in rice using crop competition-a review. Crop Prot. 2017, 95, 45–52. [Google Scholar] [CrossRef]
- Hakim, M.A.; Juraimi, A.S.; Hanafi, M.M.; Ismail, M.R.; Selamat, A.; Rafii, M.Y.; Latif, M.A. Biochemical and anatomical changes and yield reduction in rice (Oryza sativa L.) under varied salinity regimes. Biomed. Res. Int. 2014, 1, 208584. [Google Scholar]
- Cheng, A.; Mohd Hanafiah, N.; Harikrishna, J.A.; Eem, L.P.; Baisakh, N.; Mispan, M.S. A Reappraisal of Polyploidy Events in Grasses (Poaceae) in a Rapidly Changing World. Biology 2022, 11, 636. [Google Scholar] [CrossRef]
- Vaghefi, N.; Nasir Shamsudin, M.; Radam, A.; Rahim, K.A. Modelling the impact of climate change on rice production: An overview. J. Appl. Sci. 2013, 13, 5649–5660. [Google Scholar] [CrossRef]
- Ding, G.H.; Liu, X.L.; Ma, D.R.; Wang, X.X.; Yang, G.; Gao, M.C.; Gao, Q.; Sun, J.; Chen, W.F. Responses of weedy rice to drought stress at germination and seedling stages. In Applied Mechanics and Materials; Trans Tech Publications Ltd.: Bäch, Switzerland, 2013; Volume 316, pp. 451–459. [Google Scholar]
- Han, B.; Ma, X.; Cui, D.; Wang, Y.; Geng, L.; Cao, G.; Zhang, H.; Han, L. Comprehensive evaluation and analysis of the mechanism of cold tolerance based on the transcriptome of weedy rice seedlings. Rice 2020, 13, 12. [Google Scholar] [CrossRef] [PubMed]
- Bzour, M.I.; Zuki, F.M.; Mispan, M.S. Evaluation of the emergence of imidazolinone resistant weedy rice (Oryza sativa L.) in Malaysia. Appl. Ecol. Environ. Res. 2020, 18, 7189–7199. [Google Scholar] [CrossRef]
- Burgos, N.R.; Singh, V.; Tseng, T.M.; Black, H.; Young, N.D.; Huang, Z.; Hyma, K.E.; Gealy, D.R.; Caicedo, A.L. The impact of herbicide-resistant rice technology on phenotypic diversity and population structure of United States weedy rice. Plant Physiol. 2014, 166, 1208–1220. [Google Scholar] [CrossRef] [PubMed]
- Dilipkumar, M.; Chuah, T.S.; Goh, S.S.; Sahid, I. Weed management issues, challenges, and opportunities in Malaysia. Crop Prot. 2020, 134, 104347. [Google Scholar] [CrossRef]
- Ruzmi, R.; Ahmad-Hamdani, M.S.; Bakar, B.B. Prevalence of herbicide-resistant weed species in Malaysian rice fields: A review. Weed Biol. Manag. 2017, 17, 3–16. [Google Scholar] [CrossRef]
- Zulrushdi, A.Q.; Rejab, N.A.; Mahmod, I.F.; Mohamed, Z.; Ishak, M.N.; Mispan, M.S. Seed morphological traits associated with weedy rice escaped from imazapic+ imazapyr herbicide treatment. Phytoparasitica 2022, 50, 1123–1132. [Google Scholar] [CrossRef]
- Abdullah, M.Z.; Vaughan, A.D.; Watanabe, H.; Okuno, K. The origin of weedy rice in Peninsular Malaysia. MARDI Res. J. 1996, 24, 169–174. [Google Scholar]
- Hakim, M.A.; Juraimi, A.S.; Ismail, M.R.; Hanafi, M.M.; Selamat, A. Distribution of weed population in the costal rice growing area of Kedah in peninsular Malaysia. J. Agron. 2010, 9, 9–16. [Google Scholar] [CrossRef]
- Hussain, Z.; Man, A.; Othman, A.S. Association of commercial rice varieties with weedy rice accessions (Oryza sativa complex) in Pulau Pinang’s rice granary area. Trop. Life Sci. Res. 2011, 22, 1–11. [Google Scholar]
- Cui, Y.; Song, B.K.; Li, L.F.; Huang, Z.; Caicedo, A.L.; Jia, Y.; Olsen, K.M. Little white lies: Pericarp color provides insights into the origins and evolution of southeast Asian weedy rice. G3 2016, 6, 4105–4114. [Google Scholar] [CrossRef] [PubMed]
- Ruzmi, R.; Ahmad-Hamdani, M.S.; Mazlan, N. Ser-653-Asn substitution in the acetohydroxyacid synthase gene confers resistance in weedy rice to imidazolinone herbicides in Malaysia. PLoS ONE 2020, 15, e0227397. [Google Scholar]
- Ahmed, Q.N.; Zainudin, H.; Othman, A.S. Morphological relationships between weedy rice morphotypes and commercial rice varieties in Perak, Malaysia. Int. J. Bio-Resour. Stress Manag. 2011, 2, 86–92. [Google Scholar]
- Hamid, Z.A.A.; Mansor, M.; Man, A. Life cycle and morphological characteristics of weedy rice (Oryza sativa complex, locally called padi angin), a noxious weed of ricefields in Malaysia. J. Biosains 2007, 18, 55–79. [Google Scholar]
- Puteh, A.B.; Rosli, R.; Mohamad, R.B. Dormancy and cardinal temperatures during seed germination of five weedy rice (Oryza spp.) strains. Pertanika J. Trop. Agric. Sci. 2010, 33, 243–250. [Google Scholar]
- Mansor, M.; Rezaul Karim, S.M.; Abidin, Z. Effects of farmers’ cultural practices on the weedy rice infestation and rice production. Sci. Res. Essays 2012, 7, 609–615. [Google Scholar] [CrossRef]
- Mohamed, Z.; Terani, R.; Shamsudin, M.N.; Latif, I.A. Paddy farmers’ sustainability practices in granary areas in Malaysia. Resour. 2016, 5, 17. [Google Scholar] [CrossRef]
- Terano, R.; Mohamed, Z.; Din, N.S.Z. Determinants of farmers’ adoption of clearfield production system in Malaysia. Agric. Agric. Sci. Procedia 2016, 9, 103–107. [Google Scholar] [CrossRef]
- Mahmod, I.F.; Jeyasimman, S.; Mispan, M.S.; Supandi, F.; Khatib, A.; Saiman, M.Z. Comparative metabolomics analysis of weedy rice (Oryza spp.) across peninsular Malaysia. Agriculture 2023, 13, 1230. [Google Scholar] [CrossRef]
- Baki, B.B. Weed Ecology and Management in Rice Ecosystems; Universiti Malaya Press: Kuala Lumpur, Malaysia, 2006. [Google Scholar]
- Dai, X.; Shen, L. Advances and trends in omics technology development. Front. Med. 2022, 9, 911861. [Google Scholar] [CrossRef]
- Maroli, A.S.; Gaines, T.A.; Foley, M.E.; Duke, S.O.; Doğramacı, M.; Anderson, J.V.; Horvath, D.P.; Chao, W.S.; Tharayil, N. Omics in weed science: A perspective from genomics, transcriptomics, and metabolomics approaches. Weed Sci. 2018, 66, 681–695. [Google Scholar] [CrossRef]
- Van Emon, J.M. The omics revolution in agricultural research. J. Agric. Food Chem. 2016, 64, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Patterson, E.; Saski, C.; Küpper, A.; Beffa, R.; Gaines, T.A. Omics potential in herbicide-resistant weed management. Plants 2019, 8, 607. [Google Scholar] [CrossRef] [PubMed]
- Goad, D.M.; Jia, Y.; Gibbons, A.; Liu, Y.; Gealy, D.; Caicedo, A.L.; Olsen, K.M. Identification of novel QTL conferring sheath blight resistance in two weedy rice mapping populations. Rice 2020, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.L.; Wu, D.H.; Wu, C.C.; Huang, Y.F. Explore the genetics of weedy traits using rice 3K database. Bot. Stud. 2021, 62, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Fu, K.; Mou, C.; Yu, J.; Zhu, X.; Huang, Y.; Zhao, C.; Hao, Q.; Zhang, F.; Song, W.; et al. Fine mapping of qSdr9, a novel locus for seed dormancy (SD) in weedy rice, and development of NILs with a strong SD allele. Mol. Breed. 2020, 40, 1–11. [Google Scholar] [CrossRef]
- Yean, R.A.; Dilipkumar, M.; Rahman, S.; Song, B.K. A two-in-one strategy: Target and nontarget site mechanisms both play important role in IMI-resistant weedy rice. Int. J. Mol. Sci. 2021, 22, 982. [Google Scholar] [CrossRef] [PubMed]
- Buffon, G.; Blasi, E.A.D.R.; Lamb, T.I.; Adamski, J.M.; Schwambach, J.; Ricachenevsky, F.K.; Bertolazi, A.; Silveira, V.; Lopes, M.C.B.; Sperotto, R.A. Oryza sativa cv. Nipponbare and Oryza barthii as unexpected tolerance and susceptibility sources against Schizotetranychus oryzae (Acari: Tetranychidae) mite infestation. Front. Plant Sci. 2021, 12, 613568. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.; Agostinetto, D.; Langaro, A.C.; Garcia, J.R.; Lamego, F.P. Physiological and molecular responses in rice, weedy rice and barnyardgrass exposed to supra-optimal temperatures. Planta Daninha 2019, 37, e019182522. [Google Scholar] [CrossRef]
- Pan, Y.; Liang, H.; Gao, L.; Dai, G.; Chen, W.; Yang, X.; Wu, H.; Qing, D.; Gao, J.; Huang, J.; et al. Transcriptomic profiling of germinating seeds under cold stress and characterization of the cold-tolerant gene LTG5 in rice. BMC Plant Biol. 2020, 20, 1–17. [Google Scholar] [CrossRef]
- Subudhi, P.K.; Garcia, R.S.; Coronejo, S.; Tapia, R. Comparative transcriptomics of rice genotypes with contrasting responses to nitrogen stress reveals genes influencing nitrogen uptake through the regulation of root architecture. Int. J. Mol. Sci. 2020, 21, 5759. [Google Scholar] [CrossRef]
- Xiong, Q.; Chen, X.; Shen, T.; Zhong, L.; Zhu, C.; Peng, X.; He, X.; Fu, J.; Ouyang, L.; Bian, J.; et al. Metabolomics and proteomics analyses of grain yield reduction in rice under abrupt drought-flood alternation. bioRxiv 2018, 271940. [Google Scholar] [CrossRef]
- Wang, X.Q.; Kwon, S.W.; Park, Y.J. Evaluation of genetic diversity and linkage disequilibrium in Korea-bred rice varieties using SSR markers. Electron. J. Biotechnol. 2013, 16, 5. [Google Scholar]
- He, Z.; Jiang, X.; Ratnasekera, D.; Grassi, F.; Perera, U.; Lu, B.R. Seedmediated gene flow promotes genetic diversity of weedy rice within populations: Implications for weed management. PLoS ONE 2014, 9, e112778. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Li, T.Y.; Li, D.D.; Wang, Z.Y.; Li, S.; Li, D.P.; Han, X.; Liu, J.M.; Xuan, Y.H. Overexpression of loose plant architecture 1 increases planting density and resistance to sheath blight disease via activation of PIN-FORMED 1a in rice. Plant Biotechnol. J. 2019, 17, 855. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhu, J.; Fu, F.; Ye, C.Y.; Wang, W.; Mao, L.; Lin, Z.; Chen, L.; Zhang, H.; Guo, L.; et al. Genome re-sequencing suggested a weedy rice origin from domesticated indica–japonica hybridization: A case study from southern China. Planta 2014, 240, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhou, Y.; Mao, L.; Ye, C.; Wang, W.; Zhang, J.; Yu, Y.; Fu, F.; Wang, Y.; Qian, F.; et al. Genomic variation associated with local adaptation of weedy rice during de-domestication. Nat. Commun. 2017, 8, 15323. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Jia, L.; Wu, D.; Weng, X.; Chen, L.; Sun, J.; Chen, M.; Mao, L.; Jiang, B.; Ye, C.; et al. Diverse genetic mechanisms underlie worldwide convergent rice feralization. Genome Biol. 2020, 21, 70. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Qian, Q.; Ma, D.R.; Xu, Z.J.; Liu, D.; Du, H.B.; Chen, W.F. Introgression and selection shaping the genome and adaptive loci of weedy rice in northern China. New Phytol. 2013, 197, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Thurber, C.S.; Jia, M.H.; Jia, Y.; Caicedo, A.L. Similar traits, different genes? Examining convergent evolution in related weedy rice population. Mol. Ecol. 2013, 22, 685–698. [Google Scholar] [CrossRef]
- Vigueira, C.C.; Qi, X.; Song, B.K.; Li, L.F.; Caicedo, A.L.; Jia, Y.; Olsen, K.M. Call of the wild rice: Oryza rufipogon shapes weedy rice evolution in Southeast Asia. Evol. Appl. 2018, 12, 93–104. [Google Scholar] [CrossRef]
- Islam, M.S.; Coronejo, S.; Subudhi, P.K. Whole-genome sequencing reveals uniqueness of black-hulled and straw-hulled weedy rice genomes. Theor. Appl. Genet. 2020, 133, 2461–2475. [Google Scholar] [CrossRef]
- Wang, W.; Zhao, M.; Zhang, G.; Liu, Z.; Hua, Y.; Jia, X.; Song, J.; Ma, D.; Sun, J. Weedy rice as a novel gene resource: A genome-wide association study of anthocyanin biosynthesis and an evaluation of nutritional quality. Front. Plant Sci. 2020, 11, 878. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Wang, Z.; Guo, J.Y.; Xia, Q.Y.; Zhao, H.; Zhang, Y.L.; Guo, A.P.; Lu, B.R. Increases in genetic diversity of weedy rice associated with ambient temperatures and limited gene flow. Biology 2021, 10, 71. [Google Scholar] [CrossRef]
- Han, B.; Ma, X.; Cui, D.; Wang, Y.; Geng, L.; Cao, G.; Zhang, H.; Koh, H.J.; Han, L. Analysis of evolutionary relationships provides new clues to the origins of weedy rice. Ecol. Evol. 2020, 10, 891–900. [Google Scholar] [CrossRef]
- Lang, H.; He, Y.; Sun, J.; Li, F.; Ma, D. Integrative hormone and transcriptome analyses underline the role of abscisic acid in seed shattering of weedy rice. Plant Growth Regul. 2020, 94, 261–273. [Google Scholar] [CrossRef]
- Bevilacqua, C.B.; Basu, S.; Pereira, A.; Tseng, T.M.; Zimmer, P.D.; Burgos, N.R. Analysis of stress-responsive gene expression in cultivated and weedy rice differing in cold stress tolerance. PLoS ONE 2015, 10, e0132100. [Google Scholar] [CrossRef] [PubMed]
- Guan, S.; Xu, Q.; Ma, D.; Zhang, W.; Xu, Z.; Zhao, M.; Guo, Z. Transcriptomics profiling in response to cold stress in cultivated rice and weedy rice. Gene 2019, 685, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.H.; Kim, D.Y.; Moon, Y.S.; Pack, I.S.; Jeong, S.C.; Kim, H.B.; Kim, C.G. Performance of hybrids between abiotic stress-tolerant transgenic rice and its weedy relatives under water-stressed conditions. Sci. Rep. 2020, 10, 9319. [Google Scholar] [CrossRef]
- Han, B.; Ma, X.; Cui, D.; Geng, L.; Cao, G.; Zhang, H.; Han, L. Parallel reaction monitoring revealed tolerance to drought proteins in weedy rice (Oryza sativa f. spontanea). Sci. Rep. 2020, 10, 12935. [Google Scholar] [CrossRef]
- Nam, M.H.; Bang, E.; Kwon, T.Y.; Kim, Y.; Kim, E.H.; Cho, K.; Park, W.J.; Kim, B.G.; Yoon, I.S. Metabolite profiling of diverse rice germplasm and identification of conserved metabolic markers of rice roots in response to long-term mild salinity stress. Int. J. Mol. Sci. 2015, 16, 21959–21974. [Google Scholar] [CrossRef]
- Yan, H.; Ma, L.; Wang, Z.; Lin, Z.; Su, J.; Lu, B.R. Multiple tissue-specific expression of rice seed-shattering gene SH4 regulated by its promoter pSH4. Rice 2015, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Tian, L.; Li, J.; Wang, C.; Lee, D.; Peng, R.; Chen, L. Morphological characterization of weedy rice populations from different regions of Asia. Mol. Plant Breed. 2017, 8, 52–64. [Google Scholar] [CrossRef]
- Fogliatto, S.; Vidotto, F.; Ferrero, A. Germination of weedy rice in response to field conditions during winter. Weed Technol. 2011, 25, 252–261. [Google Scholar] [CrossRef]
- Fukuda, A.; Shimizu, H.; Shiratsuchi, H.; Yamaguchi, H.; Ohdaira, Y.; Mochida, H. Complementary genes that cause black ripening hulls in F1 plants of crosses between indica and japonica rice cultivars. Plant Prod. Sci. 2012, 15, 270–273. [Google Scholar] [CrossRef]
- Gu, X.Y.; Foley, M.E.; Horvath, D.P.; Anderson, J.V.; Feng, J.; Zhang, L.; Mowry, C.R.; Ye, H.; Suttle, J.C.; Kadowaki, K.I.; et al. Association between seed dormancy and pericarp color is controlled by a pleiotropic gene that regulates abscisic acid and flavonoid synthesis in weedy red rice. Genetics 2011, 189, 1515–1524. [Google Scholar] [CrossRef] [PubMed]
- Prathepa, P. Pericarp colour and haplotype diversity in weedy rice (O. sativa f. spontanea) from Thailand. Pak. J. Biol. Sci. 2009, 12, 1075–1079. [Google Scholar]
- Vigueira, C.C.; Li, W.; Olsen, K.M. The role of Bh4 in parallel evolution of hull colour in domesticated and weedy rice. J. Evol. Biol. 2013, 26, 1738–1749. [Google Scholar] [CrossRef]
- Roma-Burgos, N.; San Sudo, M.P.; Olsen, K.M.; Werle, I.; Song, B.K. Weedy rice (Oryza spp.): What’s in a name? Weed Sci. 2021, 69, 505–513. [Google Scholar] [CrossRef]
- Thurber, C.S.; Hepler, P.K.; Caicedo, A.L. Timing is everything: Early degradation of abscission layer is associated with increased seed shattering in US weedy rice. BMC Plant Biol. 2011, 11, 14. [Google Scholar] [CrossRef]
- Akasaka, M.; Konishi, S.; Izawa, T.; Ushiki, J. Histological and genetic characteristics associated with the seed-shattering habit of weedy rice (Oryza sativa L.) from Okayama, Japan. Breed. Sci. 2011, 61, 168–173. [Google Scholar] [CrossRef]
- Hua, L.; Wang, D.R.; Tan, L.; Fu, Y.; Liu, F.; Xiao, L.; Zhu, Z.; Fu, Q.; Sun, X.; Gu, P.; et al. LABA1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell 2015, 27, 1875–1888. [Google Scholar] [CrossRef]
- Luo, J.; Liu, H.; Zhou, T.; Gu, B.; Huang, X.; Shangguan, Y.; Zhu, J.; Li, Y.; Zhao, Y.; Wang, Y.; et al. An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 2013, 25, 3360–3376. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, K.; Takeuchi, Y.; Ebana, K.; Miyao, A.; Hirochika, H.; Hara, N.; Kobayashi, M.; Ban, Y.; Hattori, T.; Yano, M. Molecular cloning of Sdr4, a regulator involved in seed dormancy and domestication of rice. Proc. Natl. Acad. Sci. USA 2010, 107, 5792–5797. [Google Scholar] [CrossRef] [PubMed]
- Reagon, M.; Thurber, C.S.; Olsen, K.M.; Jia, Y.; Caicedo, A.L. The long and the short of it: SD1 polymorphism and the evolution of growth trait divergence in US weedy rice. Mol. Ecol. 2011, 20, 3743–3756. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, N.; Asami, H.; Hirabayashi, H.; Uchino, A.; Imaizumi, T.; Ishimaru, K. A Rice Ancestral Genetic Resource Conferring Ideal Plant Shapes for Vegetative Growth and Weed Suppression. Front. Plant Sci. 2021, 12, 748531. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Numaguchi, K.; Miura, K.; Yoshida, K.; Thanh, P.T.; Htun, T.M.; Yamasaki, M.; Komeda, N.; Matsumoto, T.; Terauchi, R.; et al. OsLG1 regulates a closed panicle trait in domesticated rice. Nat. Genet. 2013, 45, 462–465. [Google Scholar] [CrossRef] [PubMed]
- Subudhi, P.K.; De Leon, T.B.; Tapia, R.; Chai, C.; Karan, R.; Ontoy, J.; Singh, P.K. Genetic interaction involving photoperiod-responsive Hd1 promotes early flowering under long-day conditions in rice. Sci. Rep. 2018, 8, 2081. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, J.; Wu, X.; Dong, L. Na+/K+ balance and transport regulatory mechanisms in weedy and cultivated rice (Oryza sativa L.) under salt stress. BMC Plant Biol. 2018, 18, 375. [Google Scholar] [CrossRef] [PubMed]
- Piveta, L.B.; Roma-Burgos, N.; Noldin, J.A.; Viana, V.E.; Oliveira, C.D.; Lamego, F.P.; Avila, L.A.D. Molecular and physiological responses of rice and weedy rice to heat and drought stress. Agriculture 2020, 11, 9. [Google Scholar] [CrossRef]
- Kaspary, T.E.; Cutti, L.; Rafaeli, R.S.; Delatorre, C.A.; Merotto, A., Jr. Genes related to flooding tolerance during germination and early growth of weedy rice. Weed Res. 2020, 60, 435–449. [Google Scholar] [CrossRef]
- Jin, M.; Chen, L.; Deng, X.W.; Tang, X. Development of herbicide resistance genes and their application in rice. Crop J. 2022, 10, 26–35. [Google Scholar] [CrossRef]
- Jia, Y.; Gealy, D. Weedy red rice has novel sources of resistance to biotic stress. Crop J. 2018, 6, 443–450. [Google Scholar] [CrossRef]
- Zhu, Y.; Ellstrand, N.C.; Lu, B.R. Sequence polymorphisms in wild, weedy, and cultivated rice suggest seed-shattering locus sh4 played a minor role in Asian rice domestication. Ecol. Evol. 2012, 2, 2106–2113. [Google Scholar] [CrossRef]
- Amarasinghe, Y.P.J.; Kuwata, R.; Nishimura, A.; Phan, P.D.T.; Ishikawa, R.; Ishii, T. Evaluation of domestication loci associated with awnlessness in cultivated rice, Oryza sativa. Rice 2020, 13, 26. [Google Scholar] [CrossRef]
- Wu, C.C.; Wei, F.J.; Chiou, W.Y.; Tsai, Y.C.; Wu, H.P.; Gotarkar, D.; Wei, Z.H.; Lai, B.H.; Hsing, Y.I.C. Studies of rice Hd1 haplotypes worldwide reveal adaptation of flowering time to different environments. PLoS ONE 2020, 15, e0239028. [Google Scholar] [CrossRef]
- He, Q.; Kim, K.W.; Park, Y.J. Population genomics identifies the origin and signatures of selection of Korean weedy rice. Plant Biotech. J. 2017, 15, 357–366. [Google Scholar] [CrossRef]
- Lee, S.; Jia, Y.; Jia, M.; Gealy, D.R.; Olsen, K.M.; Caicedo, A.L. Molecular evolution of the rice blast resistance gene Pi-ta in invasive weedy rice in the USA. PLoS ONE 2011, 6, e26260. [Google Scholar] [CrossRef]
- Wang, W.; Xia, H.; Yang, X.; Xu, T.; Si, H.J.; Cai, X.X.; Wang, F.; Su, J.; Snow, A.A.; Lu, B.R. A novel 5-enolpyruvoylshikimate-3-phosphate (EPSP) synthase transgene for glyphosate resistance stimulates growth and fecundity in weedy rice (Oryza sativa) without herbicide. New Phytol. 2014, 202, 679–688. [Google Scholar] [CrossRef]
- Katam, R.; Lin, C.; Grant, K.; Katam, C.S.; Chen, S. Advances in plant metabolomics and its applications in stress and single-cell biology. Int. J. Mol. Sci. 2022, 23, 6985. [Google Scholar] [CrossRef]
- Obata, T.; Fernie, A.R. The use of metabolomics to dissect plant response to abiotic stresses. Cell Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Zhang, X.; Luo, J.; Liu, H.; Wen, W.; Luo, H.; Yan, J.; Xiao, Y. Metabolomics analysis reveals differences in evolution between maize and rice. Plant J. 2020, 103, 1710–1722. [Google Scholar] [CrossRef] [PubMed]
- Razzaq, A.; Sadia, B.; Raza, A.; Khalid Hameed, M.; Saleem, F. Metabolomics: A way forward for crop improvement. Metabolites 2019, 9, 303. [Google Scholar] [CrossRef] [PubMed]
- Uawisetwathana, U.; Karoonuthaisiri, N. Metabolomics for rice quality and traceability: Feasibility and future aspects. Curr. Opin. Food Sci. 2019, 28, 58–66. [Google Scholar] [CrossRef]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-based plant metabolomics: Where do we stand, where do we go? Trends Biotechnol. 2011, 29, 267–275. [Google Scholar] [CrossRef]
- Kusano, M.; Yang, Z.; Okazaki, Y. Using metabolomic approaches to explore chemical diversity in rice. Mol. Plant 2015, 8, 58–67. [Google Scholar] [CrossRef]
- Hu, C.; Shi, J.; Quan, S.; Cui, B.; Kleessen, S.; Nikoloski, Z.; Tohge, T.; Alexander, D.; Guo, L.; Lin, H.; et al. Metabolic variation between japonica and indica rice cultivars as revealed by non-targeted metabolomics. Sci. Rep. 2014, 4, 5067. [Google Scholar] [CrossRef]
- Ishikawa, T.; Takahara, K.; Hirabayashi, T.; Matsumura, H.; Fujisawa, S.; Terauchi, R.; Uchimiya, H.; Kawai-Yamada, M. Metabolome analysis of response to oxidative stress in rice suspension cells overexpressing cell death suppressor Bax inhibitor-1. Plant Cell Physiol. 2010, 51, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Sana, T.R.; Fischer, S.; Wohlgemuth, G.; Katrekar, A.; Jung, K.H.; Ronald, P.C.; Fiehn, O. Metabolomic and transcriptomic analysis of the rice response to the bacterial blight pathogen Xanthomonas oryzae pv. oryzae. Metabolomics 2010, 6, 451–465. [Google Scholar] [CrossRef]
- Mumm, R.; Hageman, J.A.; Calingacion, M.N.; De Vos, R.C.H.; Jonker, H.H.; Erban, A.; Kopka, J.; Hansen, T.H.; Laursen, K.H.; Schjoerring, J.K.; et al. Multi-platform metabolomics analyses of a broad collection of fragrant and non-fragrant rice varieties reveals the high complexity of grain quality characteristics. Metabolomics 2016, 12, 38. [Google Scholar] [CrossRef]
- Pramai, P.; Hamid, N.A.A.; Mediani, A.; Maulidiani, M.; Abas, F.; Jiamyangyuen, S. Metabolite profiling, antioxidant, and α-glucosidase inhibitory activities of germinated rice: Nuclear-magnetic-resonance-based metabolomics study. J. Food Drug Anal. 2018, 26, 47–57. [Google Scholar] [CrossRef]
- Zarei, I.; Luna, E.; Leach, J.E.; McClung, A.; Vilchez, S.; Koita, O.; Ryan, E.P. Comparative rice bran metabolomics across diverse cultivars and functional rice gene-bran metabolite relationships. Metabolites 2018, 8, 63. [Google Scholar] [CrossRef]
- Nadir, S.; Xiong, H.B.; Zhu, Q.; Zhang, X.L.; Xu, H.Y.; Li, J.; Dongchen, W.; Henry, D.; Guo, X.Q.; Khan, S.; et al. Weedy rice in sustainable rice production. A review. Agron. Sustain. Dev. 2017, 37, 1–14. [Google Scholar] [CrossRef]
- Lawas, L.M.F.; Erban, A.; Kopka, J.; Jagadish, S.V.K.; Zuther, E.; Hincha, D.K. Metabolic responses of rice source and sink organs during recovery from combined drought and heat stress in the field. Gigascience 2019, 8, giz102. [Google Scholar] [CrossRef] [PubMed]
- Yasmin, S.; Hafeez, F.Y.; Mirza, M.S.; Rasul, M.; Arshad, H.M.; Zubair, M.; Iqbal, M. Biocontrol of bacterial leaf blight of rice and profiling of secondary metabolites produced by rhizospheric Pseudomonas aeruginosa BRp3. Front. Microb. 2017, 8, 1895. [Google Scholar] [CrossRef] [PubMed]
- Pipatpongpinyo, W.; Korkmaz, U.; Wu, H.; Kena, A.; Ye, H.; Feng, J.; Gu, X.Y. Assembling seed dormancy genes into a system identified their effects on seedbank longevity in weedy rice. Heredity 2020, 124, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Howel, K.A.; Narsai, R.; Carroll, A.; Ivanova, A.; Lohse, M.; Usadel, B.; Millar, A.H.; Whelan, J. Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcriptions factors and the role of RNA instability in the germination process. Plant Physiol. 2009, 149, 961–980. [Google Scholar] [CrossRef]
- Frank, T.; Reichardt, B.; Shu, Q.; Engel, K.H. Metabolite profiling of coloured rice (Oryza sativa L.) grains. J. Cereal Sci. 2012, 55, 112–119. [Google Scholar] [CrossRef]
- Lee, D.Y.; Shrestha, S.; Seo, W.D.; Lee, M.H.; Jeong, T.S.; Cho, J.H.; Song, Y.C.; Kang, H.W.; Rho, Y.D.; Baek, N.I. Structural and quantitative analysis of antioxidant and low-density lipoprotein-antioxidant flavonoids from the grains of sugary rice. J. Med. Food 2012, 15, 399–405. [Google Scholar] [CrossRef]
- Pereira-Caro, G.; Cros, G.; Yokota, T.; Crozier, A. Phytochemical profiles of black, red, brown, and white rice from the Camargue region of France. J. Agric. Food Chem. 2013, 61, 7976–7986. [Google Scholar] [CrossRef]
- Kim, G.R.; Jung, E.S.; Lee, S.; Lim, S.H.; Ha, S.H.; Lee, C.H. Combined mass spectrometry-based metabolite profiling of different pigmented rice (Oryza sativa L.) seeds and correlation with antioxidant activities. Molecules 2014, 19, 15673–15686. [Google Scholar] [CrossRef]
- Wijaya, D.N.; Susanto, F.A.; Purwestri, Y.A.; Ismoyowati, D.; Nuringtyas, T.R. NMR metabolite comparison of local pigmented rice in Yogyakarta. Indones. J. Biotechnol. 2017, 22, 68–75. [Google Scholar] [CrossRef]
- Lin, Z.; Zhang, X.; Wang, Z.; Jiang, Y.; Liu, Z.; Alexander, D.; Li, G.; Wang, S.; Ding, Y. Metabolomic analysis of pathways related to rice grain chalkiness by a notched-belly mutant with high occurrence of white-belly grains. BMC Plant Biol. 2017, 17, 39. [Google Scholar] [CrossRef]
- Ranjitha, H.P.; Gowda, R.; Nethra, N.; Amruta, N.; Kandikattu, H.K. Biochemical and metabolomics on rice cultivars. Rice Sci. 2019, 26, 189–194. [Google Scholar] [CrossRef]
- Song, E.H.; Kim, H.J.; Jeong, J.; Chung, H.J.; Kim, H.Y.; Bang, E.; Hong, Y.S. A 1H HR-MAS NMR-based metabolomic study for metabolic characterization of rice grain from various Oryza sativa L. cultivars. J. Agric. Food Chem. 2016, 64, 3009–3016. [Google Scholar] [CrossRef]
- Heuberger, A.L.; Lewis, M.R.; Chen, M.H.; Brick, M.A.; Leach, J.E.; Ryan, E.P. Metabolomic and functional genomic analyses reveal varietal differences in bioactive compounds of cooked rice. PLoS ONE 2010, 5, e12915. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Chen, W.; Gao, Y.; Liu, X.; Zhang, H.; Xu, C.; Yu, S.; Zhang, Q.; Luo, J. Genetic analysis of the metabolome exemplified using a rice population. Proc. Natl. Acad. Sci. USA 2013, 110, 20320–20325. [Google Scholar] [CrossRef] [PubMed]
- Jom, K.N.; Lorjaroenphon, Y.; Udompijitkul, P. Differentiation of four varieties of germinating Thai colored indica rice (Oryza sativa L.) by metabolite profiling. Food Sci. Tech. Res. 2016, 22, 65–73. [Google Scholar] [CrossRef]
- Kim, T.J.; Kim, S.Y.; Park, Y.J.; Lim, S.H.; Ha, S.H.; Park, S.U.; Lee, B.; Kim, J.K. Metabolite profiling reveals distinct modulation of complex metabolic networks in non-pigmented, black, and red rice (Oryza sativa L.) Cultivars. Metabolites 2021, 11, 367. [Google Scholar] [CrossRef]
- Yang, Y.; Saand, M.A.; Huang, L.; Abdelaal, W.B.; Zhang, J.; Wu, Y.; Li, J.; Sirohi, M.H.; Wang, F. Applications of multi-omics technologies for crop improvement. Front. Plant Sci. 2021, 12, 563953. [Google Scholar] [CrossRef]
- Barding, G.A.; Beni, S.; Fukao, T.; Bailey-Serres, J.; Larive, C.K. Comparison of GC-MS and NMR for metabolite profiling of rice subjected to submergence stress. Proteome Res. 2012, 12, 898–909. [Google Scholar] [CrossRef]
- Barding, G.A.; Fukao, T.; Beni, S.; Bailey-Serres, J.; Larive, C.K. Differential metabolomic regulation governed by the rice SUB1A gene during submergence stress and identification of alanylglycine by 1H NMR spectroscopy. J. Proteome Res. 2011, 11, 320–330. [Google Scholar] [CrossRef]
- Jung, E.S.; Lee, S.; Lim, S.H.; Ha, S.H.; Liu, K.H.; Lee, C.H. Metabolite profiling of the short-term responses of rice leaves (Oryza sativa cv. Ilmi) cultivated under different LED lights and its correlations with antioxidant activities. Plant Sci. 2013, 210, 61–69. [Google Scholar] [CrossRef]
- Degenkolbe, T.; Do, P.T.; Kopka, J.; Zuther, E.; Hincha, D.K.; Kohl, K.I. Identification of drought tolerance markers in a diverse population of rice cultivars by expression and metabolite profiling. PLoS ONE 2013, 8, e63637. [Google Scholar] [CrossRef]
- Uawisetwathana, U.; Graham, S.F.; Kamolsukyunyong, W.; Sukhaket, W.; Klanchui, A.; Toojinda, T.; Vanavichit, A.; Karoonuthaisiri, N.; Elliot, C.T. Quantitative 1H NMR metabolome profiling of Thai jasmin rice (Oryza sativa) reveals primary metabolic response during brown planthopper infestation. Metabolomics 2015, 11, 1640–1655. [Google Scholar] [CrossRef]
- Maruyama, K.; Urano, K.; Yoshiwara, K.; Morishita, Y.; Sakurai, N.; Suzuki, H.; Kojima, M.; Sakakibara, H.; Shibata, D.; Saito, K.; et al. Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiol. 2014, 164, 1759–1771. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.; Yue, L.; Xia, X.; Liu, X.; Zhang, W. Comparative metaabolomics analysis of different resistant rice varieties in response to the brown planthopper Nilaparvata lugens Hemiptera: Delphacidae. Metabolomics 2019, 15, 62. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Wang, W.; Zhang, F.; Deng, J.; Li, Z.; Fu, B. Comparative metabolite profiling of two rice genotypes with contrasting salt stress tolerance at the seedling stage. PLoS ONE 2014, 9, e108020. [Google Scholar] [CrossRef]
- Casartelli, A.; Riewe, D.; Hubberten, H.M.; Altman, T.; Hoefgen, R.; Heuer, S. Exploring traditional aus-type rice for metabolites conferring drought tolerance. Rice 2018, 11, 9. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.H.; Wang, C.; Li, S.X.; Su, Z.Z.; Zhao, H.N.; Mao, L.J.; Feng, X.X.; Liu, P.P.; Chen, X.; Synder, J.H.; et al. Friend or foe: Differential responses of rice to invasion by mutualistic or pathogenic fungi revealed by RNAseq and metabolite profiling. Sci. Rep. 2015, 5, 13624. [Google Scholar] [CrossRef]
- Pinu, F.R.; Beale, D.J.; Paten, A.M.; Kouremenos, K.; Swarup, S.; Schirra, H.J.; Wishart, D. Systems biology and multi-omics integration: Viewpoints from the metabolomics research community. Metabolites 2019, 9, 76. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, F.; Deng, Y.; Sun, L.; Mao, M.; Chen, R.; Qiang, Q.; Zhou, J.; Long, T.; Zhao, X.; et al. Integrated metabolomics and transcriptomics analyses reveal the metabolic differences and molecular basis of nutritional quality in landraces and cultivated rice. Metabolites 2022, 12, 384. [Google Scholar] [CrossRef]
- Chen, Y.S.; Ho, T.H.D.; Liu, L.; Lee, D.H.; Lee, C.H.; Chen, Y.R.; Lin, S.Y.; Lu, C.A.; Yu, S.M. Sugar starvation-regulated MYBS2 and 14–3-3 protein interactions enhance plant growth, stress tolerance, and grain weight in rice. Proc. Natl. Acad. Sci. USA 2019, 116, 21925–21935. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Wang, J.; Wang, W.; Wang, Y.; Xu, J.; Li, Z.; Zhou, X.; Fu, B. Integrated analysis of the transcriptome and metabolome revealed the molecular mechanisms underlying the enhanced salt tolerance of rice due to the application of exogenous melatonin. Front. Plant Sci. 2021, 11, 618680. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Su, C.; Cui, Z.; Huang, L.; Gu, S.; Jiang, S.; Feng, J.; Xu, H.; Zhang, W.; Jiang, L.; et al. Transcriptomics and metabolomics reveal tolerance new mechanism of rice roots to Al stress. Front. Genet. 2023, 13, 1063984. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, S.; Datta, K.; Molla, K.A.; Gayen, D.; Das, K.; Sarkar, S.N.; Datta, S.K. Proteo-metabolomic investigation of transgenic rice unravels metabolic alterations and accumulation of novel proteins potentially involved in defence against Rhizoctonia solani. Sci. Rep. 2019, 9, 10461. [Google Scholar] [CrossRef] [PubMed]
- Vo, K.T.X.; Rahman, M.M.; Rahman, M.M.; Trinh, K.T.T.; Kim, S.T.; Jeon, J. proteomics and metabolomics studies on the biotic stress responses of rice: An update. Rice 2021, 14, 30. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Liu, Y.; Gui, R.; Wang, Z.; Li, Z.; Han, Y.; Guo, X.; Sun, J. Comparative multi-omics analysis of hypoxic germination tolerance in weedy rice embryos and coleoptiles. Genomics 2021, 113, 3337–3348. [Google Scholar] [CrossRef] [PubMed]
- Rai, N.; Zhang, Y.; Ram, B.G.; Schumacher, L.; Yellavajjala, R.K.; Bajwa, S.; Sun, X. Application of deep learning in precision weed management: A review. Comp. Elec. Agric. 2023, 206, 107698. [Google Scholar] [CrossRef]
- Vasileiou, M.; Kyrgiakos, L.N.; Kleisiari, C.; Kleftodimos, G.; Vlontzos, G.; Belhouchette, H.; Pardalos, P.M. Transforming weed management in sustainable agriculture with artificial intelligence: A systematic literature review towards weed identification and deep learning. Crop Prot. 2024, 176, 106522. [Google Scholar] [CrossRef]
- Dadashzadeh, M.; Abbaspour-Gilandeh, Y.; Mesri-Gundoshmian, T.; Sabzi, S.; Arribas, J.I. A strereoscopic video computer vision system for weed discrimination in rice field under both natural and controlled light conditions by machine learning. Measurement 2024, 237, 115072. [Google Scholar] [CrossRef]
| Years | Agronomical Practices | Rice Morphology | Problems | Rice Varieties Development | Weedy Rice Issues |
|---|---|---|---|---|---|
| Pre-1970 |
|
|
| Mahsuri, Murni, Masria, Jaya, Sri Malaysia 11, Pulut Malaysia 1, Setanjung, Sekembang, Sekencang | No reports of weedy rice |
| 1971–1975 |
|
|
| Kadaria, Pulut Siding, Manik, Muda, Seberang, Makmur, MR81, MR84 | |
| 1976–1985 |
|
|
| MR103, MR106, PH9, MR123, MR127, MR159, MR167, MR185, MR211, MRQ50 | |
| 1986–1997 |
|
| MR219, MR220, MRQ74, MR232, MR220CL1, MR220CL2, MRM16, MR253, MR263 | First report of weedy rice | |
| 1998–2010 |
| Weedy rice mimicking cultivated rice. | |||
| 2011–2020 |
|
|
| MR269, MR284, MARDI Sempadan 303, MARDI Sebernas 207, MR1A, MRQ76 | Weedy rice resistance to herbicide and tolerance to stress conditions |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahmod, I.F.; Syed Bakar, S.N.; Mispan, M.S.; Supandi, F.; Mohamed, Z.; Saiman, M.Z. Weedy Rice Infestation in Malaysia: What Do We Know and Where Do We Go? Agriculture 2024, 14, 1307. https://doi.org/10.3390/agriculture14081307
Mahmod IF, Syed Bakar SN, Mispan MS, Supandi F, Mohamed Z, Saiman MZ. Weedy Rice Infestation in Malaysia: What Do We Know and Where Do We Go? Agriculture. 2024; 14(8):1307. https://doi.org/10.3390/agriculture14081307
Chicago/Turabian StyleMahmod, Intan Filzah, Sharifah Nurnabilah Syed Bakar, Muhamad Shakirin Mispan, Farahaniza Supandi, Zulqarnain Mohamed, and Mohd Zuwairi Saiman. 2024. "Weedy Rice Infestation in Malaysia: What Do We Know and Where Do We Go?" Agriculture 14, no. 8: 1307. https://doi.org/10.3390/agriculture14081307
APA StyleMahmod, I. F., Syed Bakar, S. N., Mispan, M. S., Supandi, F., Mohamed, Z., & Saiman, M. Z. (2024). Weedy Rice Infestation in Malaysia: What Do We Know and Where Do We Go? Agriculture, 14(8), 1307. https://doi.org/10.3390/agriculture14081307






