Harnessing Genetic Tools for Sustainable Bioenergy: A Review of Sugarcane Biotechnology in Biofuel Production
Abstract
:1. Introduction
2. Sugarcane Overview
2.1. Genomic Diversity and Hybridization
2.2. Genomic Structure and Repetitive Elements
2.3. Sugarcane, a Potential Biofuel Crop
2.4. Sugarcane Biomass Potential and Bioenergy
2.5. Genetic Studies on Sugarcane Biomass Yield
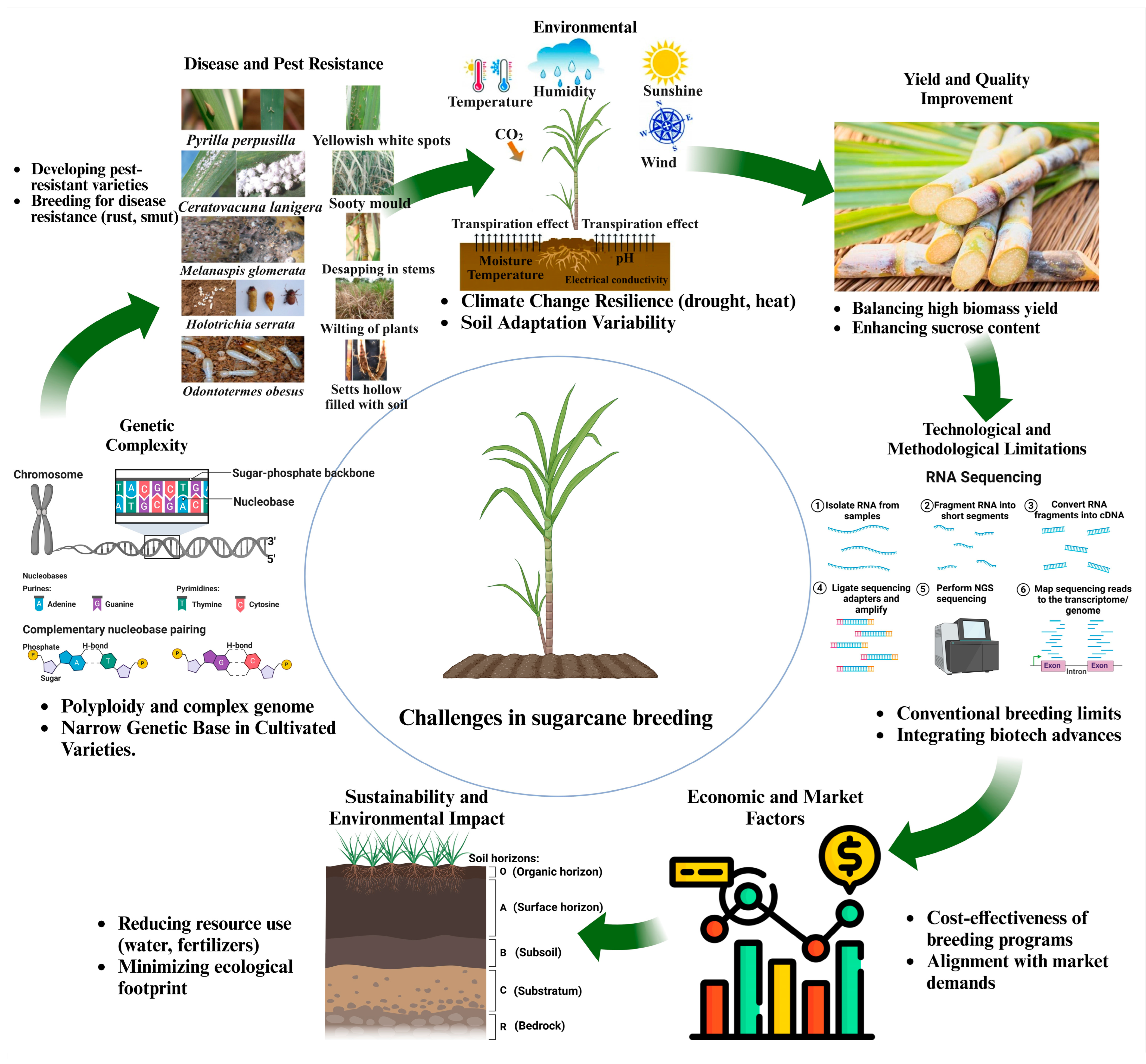
3. Challenges of Sugarcane Biomass Improvement
3.1. Narrow Genetic Bases of Current Cultivars
3.2. Flowering Synchronization and Fertility Challenges in Sugarcane Breeding
3.3. Long Breeding/Selection Cycle
3.4. Commercial Sugarcane Cultivars Genomic Complexity and Genome Size
4. Improving Sugarcane as a Bioenergy Crop through Genetic Engineering Approaches
4.1. RNA Interference (RNAi) Technology
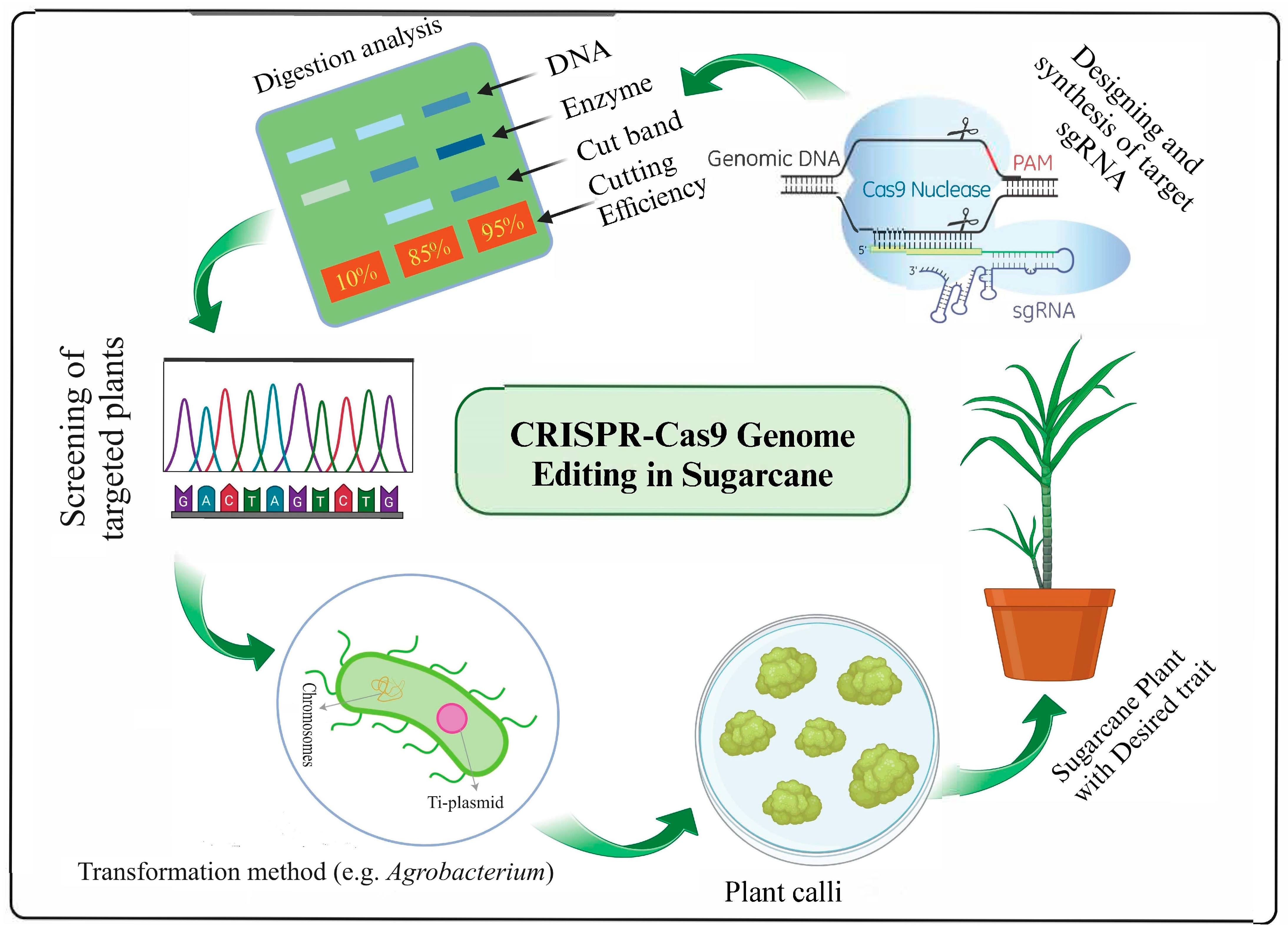
4.2. Gene Editing (GE) Technology
5. Prospects and Challenges of Genome Editing in Sugarcane
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tayyab, M.; Noman, A.; Islam, W.; Waheed, S.; Arafat, Y.; Ali, F.; Zaynab, M.; Lin, S.; Zhang, H.; Lin, W. Bioethanol production from lignocellulosic biomass by environment-friendly pretreatment methods: A review. Appl. Ecol. Environ. Res. 2018, 16, 225–249. [Google Scholar] [CrossRef]
- KN, Y.; TM, M.U.; Sachdeva, S.; Thakur, S. Lignocellulosic Biorefinery Technologies: A Perception into Recent Advances in Biomass Fractionation, Biorefineries, Economic Hurdles and Market Outlook. Fermentation 2023, 9, 238. [Google Scholar] [CrossRef]
- Grippi, D.; Clemente, R.; Bernal, M.P. Chemical and bioenergetic characterization of biofuels from plant biomass: Perspectives for southern europe. Appl. Sci. 2020, 10, 3571. [Google Scholar] [CrossRef]
- Huang, J.; Khan, M.T.; Perecin, D.; Coelho, S.T.; Zhang, M. Sugarcane for bioethanol production: Potential of bagasse in Chinese perspective. Renew. Sustain. Energy Rev. 2020, 133, 110296. [Google Scholar] [CrossRef]
- Thakur, A.; Kumar, A.; Kaya, S.; Vo, D.-V.N.; Sharma, A. Suppressing inhibitory compounds by nanomaterials for highly efficient biofuel production: A review. Fuel 2022, 312, 122934. [Google Scholar] [CrossRef]
- Bhat, R.A.; Singh, D.V.; Tonelli, F.M.P.; Hakeem, K.R. Metabolic Routes to Biofuels Extraction. In Plant and Algae Biomass: Feasible Sources for Biofuel Production; Springer: Berlin/Heidelberg, Germany, 2022; pp. 87–105. [Google Scholar]
- Bakari, H.; Djomdi; Ruben, Z.F.; Roger, D.D.; Cedric, D.; Guillaume, P.; Pascal, D.; Philippe, M.; Gwendoline, C. Sorghum (Sorghum bicolor L. Moench) and Its Main Parts (By-Products) as Promising Sustainable Sources of Value-Added Ingredients. Waste Biomass Valoriz. 2023, 14, 1023–1044. [Google Scholar] [CrossRef]
- Miranda, N.T.; Motta, I.L.; Maciel Filho, R.; Maciel, M.R.W. Sugarcane bagasse pyrolysis: A review of operating conditions and products properties. Renew. Sustain. Energy Rev. 2021, 149, 111394. [Google Scholar] [CrossRef]
- Canabarro, N.I.; Silva-Ortiz, P.; Nogueira, L.; Cantarella, H.; Maciel-Filho, R.; Souza, G.M. Sustainability assessment of ethanol and biodiesel production in Argentina, Brazil, Colombia, and Guatemala. Renew. Sustain. Energy Rev. 2023, 171, 113019. [Google Scholar] [CrossRef]
- Tesfaw, A. The current trends of bioethanol production from cheese whey using yeasts: Biological and economical perspectives. Front. Energy Res. 2023, 11, 1183035. [Google Scholar] [CrossRef]
- Soni, S.K.; Sharma, A.; Soni, R. Microbial Enzyme Systems in the Production of Second Generation Bioethanol. Sustainability 2023, 15, 3590. [Google Scholar] [CrossRef]
- Broda, M.; Yelle, D.J.; Serwańska, K. Bioethanol production from lignocellulosic biomass—Challenges and solutions. Molecules 2022, 27, 8717. [Google Scholar] [CrossRef]
- Bhattarai, K.; Conway, D.; Bhattarai, K.; Conway, D. Agriculture and Environment. In Contemporary Environmental Problems in Nepal: Geographic Perspectives; Springer: Berlin/Heidelberg, Germany, 2021; pp. 335–445. [Google Scholar]
- Leontopoulos, S.; Arabatzis, G. The contribution of energy crops to biomass production. In Low Carbon Energy Technologies in Sustainable Energy Systems; Elsevier: Amsterdam, The Netherlands, 2021; pp. 47–113. [Google Scholar]
- Vandenberghe, L.; Valladares-Diestra, K.; Bittencourt, G.; Torres, L.Z.; Vieira, S.; Karp, S.; Sydney, E.; de Carvalho, J.; Soccol, V.T.; Soccol, C. Beyond sugar and ethanol: The future of sugarcane biorefineries in Brazil. Renew. Sustain. Energy Rev. 2022, 167, 112721. [Google Scholar] [CrossRef]
- Talukdar, D.; Verma, D.K.; Malik, K.; Mohapatra, B.; Yulianto, R. Sugarcane as a potential biofuel crop. In Sugarcane Biotechnology: Challenges and Prospects; Springer: Berlin/Heidelberg, Germany, 2017; pp. 123–137. [Google Scholar]
- Rosero-Chasoy, G.; Rodríguez-Jasso, R.M.; Aguilar, C.N.; Buitrón, G.; Chairez, I.; Ruiz, H.A. Microbial co-culturing strategies for the production high value compounds, a reliable framework towards sustainable biorefinery implementation—An overview. Bioresour. Technol. 2021, 321, 124458. [Google Scholar] [CrossRef]
- Mahmud, M.A.; Anannya, F.R. Sugarcane bagasse-A source of cellulosic fiber for diverse applications. Heliyon 2021, 7, e07771. [Google Scholar] [CrossRef] [PubMed]
- Li, A.-M.; Liao, F.; Wang, M.; Chen, Z.-L.; Qin, C.-X.; Huang, R.-Q.; Verma, K.K.; Li, Y.-R.; Que, Y.-X.; Pan, Y.-Q. Transcriptomic and Proteomic Landscape of Sugarcane Response to Biotic and Abiotic Stressors. Int. J. Mol. Sci. 2023, 24, 8913. [Google Scholar] [CrossRef] [PubMed]
- Velvizhi, G.; Balakumar, K.; Shetti, N.P.; Ahmad, E.; Pant, K.K.; Aminabhavi, T.M. Integrated biorefinery processes for conversion of lignocellulosic biomass to value added materials: Paving a path towards circular economy. Bioresour. Technol. 2022, 343, 126151. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Xu, F.; Wang, Z.; Wu, Q.; Cheng, W.; Que, Y.; Xu, L. Mapping of QTLs and Screening Candidate Genes Associated with the Ability of Sugarcane Tillering and Ratooning. Int. J. Mol. Sci. 2023, 24, 2793. [Google Scholar] [CrossRef]
- Manimekalai, R.; Suresh, G.; Govinda Kurup, H.; Athiappan, S.; Kandalam, M. Role of NGS and SNP genotyping methods in sugarcane improvement programs. Crit. Rev. Biotechnol. 2020, 40, 865–880. [Google Scholar] [CrossRef]
- Mohan, C.; Easterling, M.; Yau, Y.-Y. Gene editing technologies for sugarcane improvement: Opportunities and limitations. Sugar Tech 2022, 24, 369–385. [Google Scholar] [CrossRef]
- Wang, T.; Fang, J.; Zhang, J. Advances in sugarcane genomics and genetics. Sugar Tech 2022, 24, 354–368. [Google Scholar] [CrossRef]
- Neris, D.; Mattiello, L.; Zuñiga, G.; Purgatto, E.; Menossi, M. Reduction of ethylene biosynthesis in sugarcane induces growth and investment in the non-enzymatic antioxidant apparatus. Plant Cell Rep. 2022, 41, 979–993. [Google Scholar] [CrossRef] [PubMed]
- Amalraj, V.A.; Balasundaram, N. On the taxonomy of the members of ‘Saccharum complex’. Genet. Resour. Crop Evol. 2006, 53, 35–41. [Google Scholar] [CrossRef]
- Daniels, J.; Roach, B.T. Taxonomy and evolution. In Developments in Crop Science; Elsevier: Amsterdam, The Netherlands, 1987; Volume 11, pp. 7–84. [Google Scholar]
- Manners, J.; McIntyre, L.; Casu, R.; Cordeiro, G.; Jackson, M.; Aitken, K.; Jackson, P.; Bonnett, G.; Lee, S.; Henry, R. Can genomics revolutionize genetics and breeding in sugarcane. In Proceedings of the 4th International Crop Science Congress, Brisbane, Australia, 26 September–1 October 2004; pp. 1–4. [Google Scholar]
- Irvine, J.E. Saccharum species as horticultural classes. Theor. Appl. Genet. 1999, 98, 186–194. [Google Scholar] [CrossRef]
- Zhang, Q.; Qi, Y.; Pan, H.; Tang, H.; Wang, G.; Hua, X.; Wang, Y.; Lin, L.; Li, Z.; Li, Y. Genomic insights into the recent chromosome reduction of autopolyploid sugarcane Saccharum spontaneum. Nat. Genet. 2022, 54, 885–896. [Google Scholar] [CrossRef] [PubMed]
- RR, P.; CN, B. Studies in Saccharum spontaneum distribution and geographical association of chromosome numbers. Cytologia 1960, 25, 152–172. [Google Scholar]
- D’Hont, A.; Ison, D.; Alix, K.; Roux, C.; Glaszmann, J.C. Determination of basic chromosome numbers in the genus Saccharum by physical mapping of ribosomal RNA genes. Genome 1998, 41, 221–225. [Google Scholar] [CrossRef]
- Piperidis, G.; Piperidis, N.; D’Hont, A. Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol. Genet. Genom. 2010, 284, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Bremer, G. Problems in breeding and cytology of sugar cane. Euphytica 1961, 10, 59–78. [Google Scholar] [CrossRef]
- D’Hont, A.; Grivet, L.; Feldmann, P.; Glaszmann, J.; Rao, S.; Berding, N. Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp.) by molecular cytogenetics. Mol. Gen. Genet. MGG 1996, 250, 405–413. [Google Scholar] [CrossRef]
- Piperidis, G.; D’hont, A. Chromosome composition analysis of various Saccharum interspecific hybrids by genomic in situ hybridisation (GISH). In Proceedings of the International Society of Sugar Cane Technologists, XXIV Congress, Brisbane, Australia, 17–21 September 2001; Volume 2, pp. 565–566. [Google Scholar]
- Cuadrado, A.; Acevedo, R.; Moreno Díaz de la Espina, S.; Jouve, N.; De La Torre, C. Genome remodelling in three modern S. officinarum × S. spontaneum sugarcane cultivars. J. Exp. Bot. 2004, 55, 847–854. [Google Scholar] [CrossRef]
- Grivet, L.; Arruda, P. Sugarcane genomics: Depicting the complex genome of an important tropical crop. Curr. Opin. Plant Biol. 2002, 5, 122–127. [Google Scholar] [CrossRef] [PubMed]
- Ming, R.; Moore, P.H.; Wu, K.K.; D’Hont, A.; Glaszmann, J.C.; Tew, T.L.; Mirkov, T.E.; Da Silva, J.; Jifon, J.; Rai, M. Sugarcane improvement through breeding and biotechnology. Plant Breed. Rev. 2010, 27, 15–118. [Google Scholar]
- Raboin, L.-M.; Pauquet, J.; Butterfield, M.; D’Hont, A.; Glaszmann, J.-C. Analysis of genome-wide linkage disequilibrium in the highly polyploid sugarcane. Theor. Appl. Genet. 2008, 116, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.; Garcia, A.A.F.; Oliveira, K.; Matsuoka, S.; Arizono, H.; de Souza, C., Jr.; De Souza, A. Analysis of genetic similarity detected by AFLP and coefficient of parentage among genotypes of sugar cane (Saccharum spp.). Theor. Appl. Genet. 2002, 104, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Souza, G.M.; Berges, H.; Bocs, S.; Casu, R.; D’Hont, A.; Ferreira, J.E.; Henry, R.; Ming, R.; Potier, B.; Van Sluys, M.-A. The sugarcane genome challenge: Strategies for sequencing a highly complex genome. Trop. Plant Biol. 2011, 4, 145–156. [Google Scholar] [CrossRef]
- D’Hont, A.; Glaszmann, J.C. Sugarcane genome analysis with molecular markers: A first decade of research. In Proceedings of the International Society of Sugar Cane Technologists, XXIV Congress, Brisbane, Australia, 17–21 September 2001; Volume 2, pp. 556–559. [Google Scholar]
- Shabbir, R.; Javed, T.; Afzal, I.; Sabagh, A.E.; Ali, A.; Vicente, O.; Chen, P. Modern biotechnologies: Innovative and sustainable approaches for the improvement of sugarcane tolerance to environmental stresses. Agronomy 2021, 11, 1042. [Google Scholar] [CrossRef]
- Jannoo, N.; Grivet, L.; Chantret, N.; Garsmeur, O.; Glaszmann, J.C.; Arruda, P.; D’Hont, A. Orthologous comparison in a gene-rich region among grasses reveals stability in the sugarcane polyploid genome. Plant J. 2007, 50, 574–585. [Google Scholar] [CrossRef]
- McCormick, R.F.; Truong, S.K.; Sreedasyam, A.; Jenkins, J.; Shu, S.; Sims, D.; Kennedy, M.; Amirebrahimi, M.; Weers, B.D.; McKinley, B. The Sorghum bicolor reference genome: Improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 2018, 93, 338–354. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, X.; Tang, H.; Zhang, Q.; Hua, X.; Ma, X.; Zhu, F.; Jones, T.; Zhu, X.; Bowers, J. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat. Genet. 2018, 50, 1565–1573. [Google Scholar] [CrossRef]
- Healey, A.; Garsmeur, O.; Lovell, J.; Shengquiang, S.; Sreedasyam, A.; Jenkins, J.; Plott, C.; Piperidis, N.; Pompidor, N.; Llaca, V. The complex polyploid genome architecture of sugarcane. Nature 2024, 628, 804–810. [Google Scholar] [CrossRef]
- Zhang, J.; Nagai, C.; Yu, Q.; Pan, Y.-B.; Ayala-Silva, T.; Schnell, R.J.; Comstock, J.C.; Arumuganathan, A.K.; Ming, R. Genome size variation in three Saccharum species. Euphytica 2012, 185, 511–519. [Google Scholar] [CrossRef]
- Karamchandani, B.M.; Chakraborty, S.; Dalvi, S.G.; Satpute, S.K. Chitosan and its derivatives: Promising biomaterial in averting fungal diseases of sugarcane and other crops. J. Basic Microbiol. 2022, 62, 533–554. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Yan, M.; Wang, J.; Zhao, L.; Zhou, F.; Hu, Z.; Jin, S.; Diao, Y. The complete chloroplast genome sequences of five Miscanthus species, and comparative analyses with other grass plastomes. Ind. Crops Prod. 2021, 162, 113248. [Google Scholar] [CrossRef]
- Meena, M.R.; Kumar, R.; Ramaiyan, K.; Chhabra, M.L.; Raja, A.K.; Krishnasamy, M.; Kulshreshtha, N.; Pandey, S.K.; Ram, B. Biomass potential of novel interspecific and intergeneric hybrids of Saccharum grown in sub-tropical climates. Sci. Rep. 2020, 10, 21560. [Google Scholar] [CrossRef] [PubMed]
- Ain, N.-U.; Haider, F.U.; Fatima, M.; Habiba; Zhou, Y.; Ming, R. Genetic determinants of biomass in C4 crops: Molecular and agronomic approaches to increase biomass for biofuels. Front. Plant Sci. 2022, 13, 839588. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Machín, L.; Piloto-Rodríguez, R.; Rubio-González, A.; Iturria-Quintero, P.; Ronsse, F. Pretreatment of Sugarcane Residues for Combustion in Biomass Power Stations: A Review. Sugar Tech 2022, 24, 732–745. [Google Scholar] [CrossRef]
- Hans, M.; Lugani, Y.; Chandel, A.K.; Rai, R.; Kumar, S. Production of first-and second-generation ethanol for use in alcohol-based hand sanitizers and disinfectants in India. Biomass Convers. Biorefinery 2021, 13, 7423–7440. [Google Scholar] [CrossRef] [PubMed]
- Kandel, R.; Yang, X.; Song, J.; Wang, J. Potentials, challenges, and genetic and genomic resources for sugarcane biomass improvement. Front. Plant Sci. 2018, 9, 151. [Google Scholar] [CrossRef]
- Waclawovsky, A.J.; Sato, P.M.; Lembke, C.G.; Moore, P.H.; Souza, G.M. Sugarcane for bioenergy production: An assessment of yield and regulation of sucrose content. Plant Biotechnol. J. 2010, 8, 263–276. [Google Scholar] [CrossRef]
- Whittaker, A.; Botha, F.C. Carbon partitioning during sucrose accumulation in sugarcane internodal tissue. Plant Physiol. 1997, 115, 1651–1659. [Google Scholar] [CrossRef]
- Pereira, S.C.; Maehara, L.; Machado, C.M.M.; Farinas, C.S. 2G ethanol from the whole sugarcane lignocellulosic biomass. Biotechnol. Biofuels 2015, 8, 44. [Google Scholar] [CrossRef] [PubMed]
- Seabra, J.E.; Tao, L.; Chum, H.L.; Macedo, I.C. A techno-economic evaluation of the effects of centralized cellulosic ethanol and co-products refinery options with sugarcane mill clustering. Biomass Bioenergy 2010, 34, 1065–1078. [Google Scholar] [CrossRef]
- Alonso Pippo, W.; Luengo, C.; Alonsoamador Morales Alberteris, L.; Garzone, P.; Cornacchia, G. Energy recovery from sugarcane-trash in the light of 2nd generation biofuels. Part 1: Current situation and environmental aspects. Waste Biomass Valoriz. 2011, 2, 1–16. [Google Scholar] [CrossRef]
- Macrelli, S.; Mogensen, J.; Zacchi, G. Techno-economic evaluation of 2nd generation bioethanol production from sugar cane bagasse and leaves integrated with the sugar-based ethanol process. Biotechnol. Biofuels 2012, 5, 22. [Google Scholar] [CrossRef]
- van der Weijde, T.; Alvim Kamei, C.L.; Torres, A.F.; Vermerris, W.; Dolstra, O.; Visser, R.G.; Trindade, L.M. The potential of C4 grasses for cellulosic biofuel production. Front. Plant Sci. 2013, 4, 107. [Google Scholar] [CrossRef] [PubMed]
- Somerville, C.; Youngs, H.; Taylor, C.; Davis, S.C.; Long, S.P. Feedstocks for lignocellulosic biofuels. Science 2010, 329, 790–792. [Google Scholar] [CrossRef] [PubMed]
- Kalak, T. Potential Use of Industrial Biomass Waste as a Sustainable Energy Source in the Future. Energies 2023, 16, 1783. [Google Scholar] [CrossRef]
- Ragauskas, A.J.; Williams, C.K.; Davison, B.H.; Britovsek, G.; Cairney, J.; Eckert, C.A.; Frederick, W.J., Jr.; Hallett, J.P.; Leak, D.J.; Liotta, C.L. The path forward for biofuels and biomaterials. Science 2006, 311, 484–489. [Google Scholar] [CrossRef]
- Carvalho-Netto, O.V.; Bressiani, J.A.; Soriano, H.L.; Fiori, C.S.; Santos, J.M.; Barbosa, G.V.; Xavier, M.A.; Landell, M.G.; Pereira, G.A. The potential of the energy cane as the main biomass crop for the cellulosic industry. Chem. Biol. Technol. Agric. 2014, 1, 20. [Google Scholar] [CrossRef]
- Miocque, J. Evaluation of growth and yield of green matter of sugarcane from Araraquara, SP region.= Avaliação de crescimento e de produtividade de matéria verde da cana-de-açúcar na região de Araraquara-SP. STAB-Sugar Ethanol Byprod. = Açúcar Álcool E Subprodutos 1999, 17, 45–47. [Google Scholar]
- Alexander, A.G. The Energy Cane Alternative; AE Fok Kam: Elsevier Science Publishers: Amsterdam, The Nederland, 1985; Volume 8. [Google Scholar]
- Bischoff, K.P.; Gravois, K.A.; Reagan, T.E.; Hoy, J.W.; Kimbeng, C.A.; LaBorde, C.M.; Hawkins, G.L. Registration of ‘L 79–1002’sugarcane. J. Plant Regist. 2008, 2, 211–217. [Google Scholar] [CrossRef]
- Knoll, J.E.; Anderson, W.F.; Richard Jr, E.P.; Doran-Peterson, J.; Baldwin, B.; Hale, A.L.; Viator, R.P. Harvest date effects on biomass quality and ethanol yield of new energycane (Saccharum hyb.) genotypes in the Southeast USA. Biomass Bioenergy 2013, 56, 147–156. [Google Scholar] [CrossRef]
- Matsuoka, S.; Stolf, R. Sugarcane tillering and ratooning: Key factors for a profitable cropping. Sugarcane Prod. Cultiv. Uses 2012, 5, 137–157. [Google Scholar]
- Tew, T.L.; Cobill, R.M. Genetic improvement of sugarcane (Saccharum spp.) as an energy crop. In Genetic Improvement of Bioenergy Crops; Springer: Berlin/Heidelberg, Germany, 2008; pp. 273–294. [Google Scholar]
- Matsuoka, S.; Kennedy, A.J.; Santos, E.G.D.d.; Tomazela, A.L.; Rubio, L.C.S. Energy cane: Its concept, development, characteristics, and prospects. Adv. Bot. 2014, 2014, 597275. [Google Scholar] [CrossRef]
- Sandhu, H.S.; Gilbert, R. Production of Biofuel Crops in Florida: Sugarcane/Energy Cane; University of Florida: Gainesville, FL, USA, 2014. [Google Scholar]
- Van Antwerpen, R.; Berry, S.; Van Antwerpen, T.; Smithers, J.; Joshi, S.; Van Der Laan, M. Sugarcane as an energy crop: Its role in biomass economy. Biofuel Crop Sustain. 2013, 1, 53–108. [Google Scholar]
- Zhou, H.; Liu, G.; Lu, J.; He, J. Genetic analysis of sugarcane biomass yield and its component traits using ADAA model. J. Trop. Agric. 2009, 47, 70–73. [Google Scholar]
- Sanghera, G.; Tyagi, V.; Kumar, R.; Thind, K.; Sharma, B. Quality parameters and their association with cane yield in sugarcane under subtropical conditions. In Proceedings of the National Symposium Crop Improvement Inclusive Sustainable Development Held at Punjab Agricultural University, Ludhiana, India, 7–9 November 2014; pp. 796–798. [Google Scholar]
- Hoarau, J.-Y.; Grivet, L.; Offmann, B.; Raboin, L.-M.; Diorflar, J.-P.; Payet, J.; Hellmann, M.; D’Hont, A.; Glaszmann, J.-C. Genetic dissection of a modern sugarcane cultivar (Saccharum spp.). II. Detection of QTLs for yield components. Theor. Appl. Genet. 2002, 105, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Aitken, K.; Jackson, P.; Piperidis, G.; McIntyre, L. QTL identified for yield components in a cross between a sugarcane (Saccharum spp.) cultivar Q165A and a S. officinarum clone IJ76-514. In Proceedings of the New Directions for a Diverse Planet, 4th International Crop Science Congress, Brisbane, Australia, 26 September–1 October 2004. [Google Scholar]
- Bilal, M.; Saeed, M.; Nasir, I.A.; Tabassum, B.; Zameer, M.; Khan, A.; Tariq, M.; Javed, M.A.; Husnain, T. Association mapping of cane weight and tillers per plant in sugarcane. Biotechnol. Biotechnol. Equip. 2015, 29, 617–623. [Google Scholar] [CrossRef]
- Hoang, N.V.; Furtado, A.; O’Keeffe, A.J.; Botha, F.C.; Henry, R.J. Association of gene expression with biomass content and composition in sugarcane. PLoS ONE 2017, 12, e0183417. [Google Scholar] [CrossRef]
- Hart, G.; Schertz, K.; Peng, Y.; Syed, N. Genetic mapping of Sorghum bicolor (L.) Moench QTLs that control variation in tillering and other morphological characters. Theor. Appl. Genet. 2001, 103, 1232–1242. [Google Scholar] [CrossRef]
- Jordan, D.; Casu, R.; Besse, P.; Carroll, B.; Berding, N.; McIntyre, C. Markers associated with stalk number and suckering in sugarcane colocate with tillering and rhizomatousness QTLs in sorghum. Genome 2004, 47, 988–993. [Google Scholar] [CrossRef]
- Rogers, J.S. The inheritance of inflorescence characters in maize-teosinte hybrids. Genetics 1950, 35, 541. [Google Scholar] [CrossRef]
- Whipple, C.J.; Kebrom, T.H.; Weber, A.L.; Yang, F.; Hall, D.; Meeley, R.; Schmidt, R.; Doebley, J.; Brutnell, T.P.; Jackson, D.P. grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc. Natl. Acad. Sci. USA 2011, 108, E506–E512. [Google Scholar] [CrossRef]
- Kebrom, T.H.; Burson, B.L.; Finlayson, S.A. Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 2006, 140, 1109–1117. [Google Scholar] [CrossRef]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef]
- Takeda, T.; Suwa, Y.; Suzuki, M.; Kitano, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M.; Ueguchi, C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003, 33, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Merrick, P.; Zhang, Z.; Ji, C.; Yang, B.; Fei, S.z. Targeted mutagenesis in tetraploid switchgrass (Panicum virgatum L.) using CRISPR/Cas9. Plant Biotechnol. J. 2018, 16, 381–393. [Google Scholar] [CrossRef]
- Pribil, M.; Hermann, S.; Dun, G.; Karno, X.; Ngo, C.; O’neill, S.; Wang, L.; Bonnett, G.; Chandler, P.; Beveridge, C. Altering sugarcane shoot architecture through genetic engineering: Prospects for increasing cane and sugar yield. In Proceedings of the Australian Society of Sugar Cane Technologists; Australian Society of Sugar Cane Technologists: Brisbane, Australia, 2007; Volume 29, pp. 251–257. [Google Scholar]
- Li, Q.; Song, J.; Peng, S.; Wang, J.P.; Qu, G.Z.; Sederoff, R.R.; Chiang, V.L. Plant biotechnology for lignocellulosic biofuel production. Plant Biotechnol. J. 2014, 12, 1174–1192. [Google Scholar] [CrossRef]
- Jung, J.H.; Vermerris, W.; Gallo, M.; Fedenko, J.R.; Erickson, J.E.; Altpeter, F. RNA interference suppression of lignin biosynthesis increases fermentable sugar yields for biofuel production from field-grown sugarcane. Plant Biotechnol. J. 2013, 11, 709–716. [Google Scholar] [CrossRef]
- Manickavasagam, M.; Ganapathi, A.; Anbazhagan, V.; Sudhakar, B.; Selvaraj, N.; Vasudevan, A.; Kasthurirengan, S. Agrobacterium-mediated genetic transformation and development of herbicide-resistant sugarcane (Saccharum species hybrids) using axillary buds. Plant Cell Rep. 2004, 23, 134–143. [Google Scholar] [CrossRef]
- Lakshmanan, P.; Geijskes, R.J.; Aitken, K.S.; Grof, C.L.; Bonnett, G.D.; Smith, G.R. Sugarcane biotechnology: The challenges and opportunities. In Vitro Cell. Dev. Biol.-Plant 2005, 41, 345–363. [Google Scholar] [CrossRef]
- Arceneaux, G. Cultivated sugarcanes of the world and their botanical derivation. Proc. Int. Soc. Sugar Cane Technol. 1967, 12, 844–854. [Google Scholar]
- Roach, B. Origin and improvement of the genetic base of sugarcane. Proc. Aust. Soc. Sugar Cane Technol. 1989, 11, 34–47. [Google Scholar]
- Dumont, T.; Barau, L.; Thong-Chane, A.; Dijoux, J.; Mellin, M.; Daugrois, J.; Hoarau, J.-Y. Sugarcane breeding in Reunion: Challenges, achievements and future prospects. Sugar Tech 2022, 24, 181–192. [Google Scholar] [CrossRef]
- Singh, P.; Singh, S.P.; Tiwari, A.K.; Sharma, B.L. Genetic diversity of sugarcane hybrid cultivars by RAPD markers. 3 Biotech 2017, 7, 222. [Google Scholar] [CrossRef] [PubMed]
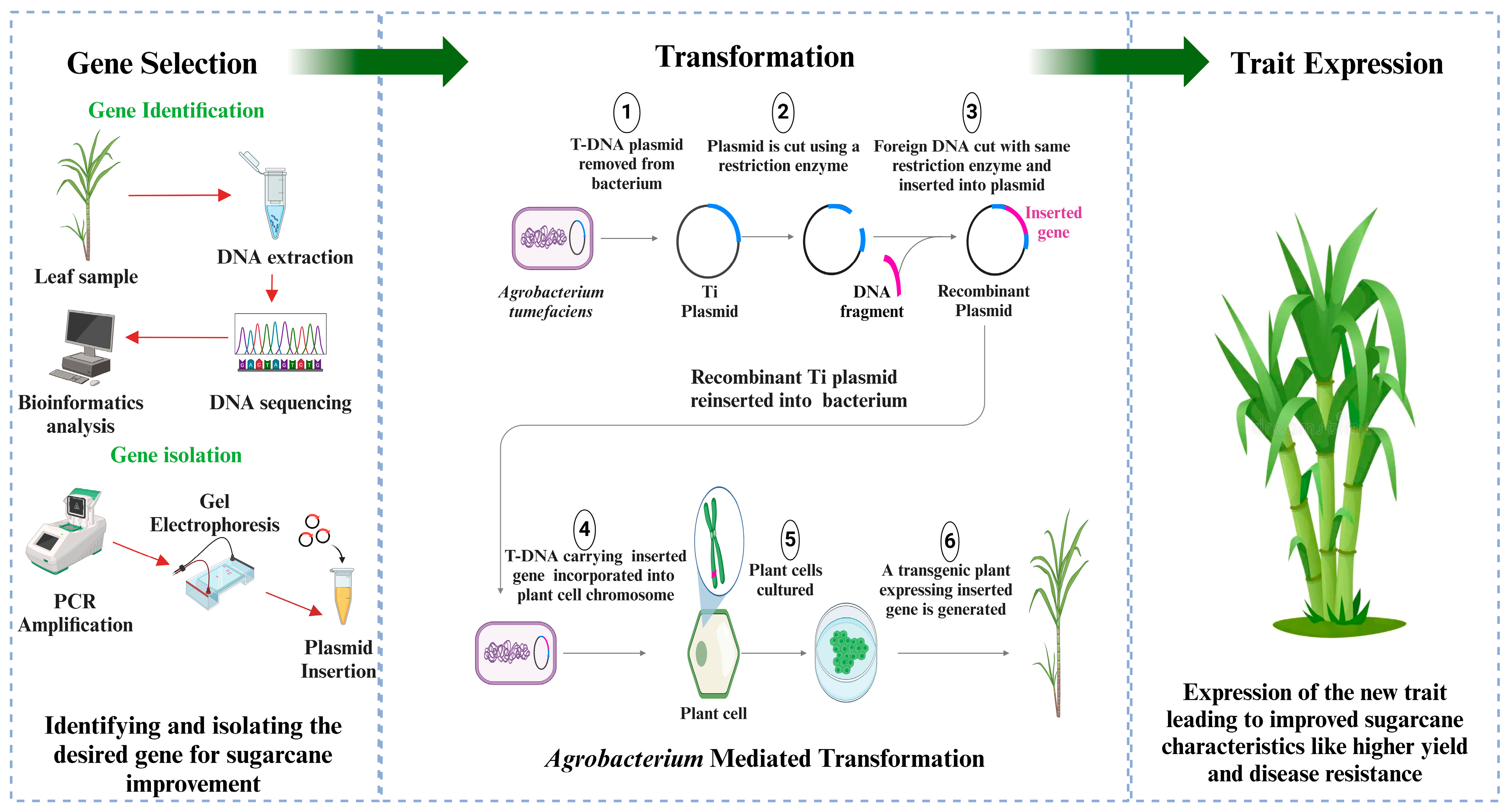
- Budeguer, F.; Enrique, R.; Perera, M.F.; Racedo, J.; Castagnaro, A.P.; Noguera, A.S.; Welin, B. Genetic transformation of sugarcane, current status and future prospects. Front. Plant Sci. 2021, 12, 768609. [Google Scholar] [CrossRef] [PubMed]
- Babu, K.H.; Devarumath, R.; Thorat, A.; Nalavade, V.; Saindane, M.; Appunu, C.; Suprasanna, P. Sugarcane transgenics: Developments and opportunities. Genet. Modif. Crops Curr. Status Prospect. Chall. 2021, 1, 241–265. [Google Scholar]
- Nuss, K. Flowering of sugarcane in a photoperiod house from 1971 to 1981. Proc. S. Afr. Sugar Technol. Assoc. 1982, 56, 140–142. [Google Scholar]
- Moore, P.H.; Nuss, K. Flowering and flower synchronization. Dev. Crop Sci. 1987, 11, 273–311. [Google Scholar]
- Pittelkow, C.M.; Linquist, B.A.; Lundy, M.E.; Liang, X.; Van Groenigen, K.J.; Lee, J.; Van Gestel, N.; Six, J.; Venterea, R.T.; Van Kessel, C. When does no-till yield more? A global meta-analysis. Field Crops Res. 2015, 183, 156–168. [Google Scholar] [CrossRef]
- Herridge, D.F.; Peoples, M.B.; Boddey, R.M. Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 2008, 311, 1–18. [Google Scholar] [CrossRef]
- Skinner, J. Controlled pollination of sugarcane. Bur. Sugar Exp. Stn. Tech Commun. 1959, 1, 7–20. [Google Scholar]
- Tew, T.L.; Pan, Y.B. Microsatellite (Simple Sequence Repeat) Marker–based Paternity Analysis of a Seven-Parent Sugarcane Polycross. Crop Sci. 2010, 50, 1401–1408. [Google Scholar] [CrossRef]
- Racedo, J.; Gutiérrez, L.; Perera, M.F.; Ostengo, S.; Pardo, E.M.; Cuenya, M.I.; Welin, B.; Castagnaro, A.P. Genome-wide association mapping of quantitative traits in a breeding population of sugarcane. BMC Plant Biol. 2016, 16, 142. [Google Scholar] [CrossRef] [PubMed]
- Gazaffi, R.; Oliveira, K.M.; Souza, A.; Garcia, A.A.F. Sugarcane: Breeding methods and genetic mapping. In Sugarcane Bioethanol—R&D for Productivity and Sustainability; Blucher Open Access: São Paulo, Brazil, 2014; pp. 333–344. [Google Scholar]
- Sreenivasan, T.; Ahloowalia, B.; Heinz, D. Sugarcane improvement through breeding. Cytogenetics 1987, 5, 211–253. [Google Scholar]
- Lu, Y.; D’Hont, A.; Walker, D.; Rao, P.; Feldmann, P.; Glaszmann, J.-C. Relationships among ancestral species of sugarcane revealed with RFLP using single copy maize nuclear probes. Euphytica 1994, 78, 7–18. [Google Scholar] [CrossRef]
- Jannoo, N.; Grivet, L.; Seguin, M.; Paulet, F.; Domaingue, R.; Rao, P.; Dookun, A.; D’Hont, A.; Glaszmann, J.-C. Molecular investigation of the genetic base of sugarcane cultivars. Theor. Appl. Genet. 1999, 99, 171–184. [Google Scholar] [CrossRef]
- Matsuoka, S.; Ferro, J.; Arruda, P. The Brazilian experience of sugarcane ethanol industry. In Vitro Cell. Dev. Biol.-Plant 2009, 45, 372–381. [Google Scholar] [CrossRef]
- Gouy, M.; Rousselle, Y.; Bastianelli, D.P.; Lecomte, L.; Bonnal, D.; Roques, J.-C.; Efile, S.; Rocher, J.; Daugrois, L.; Toubi, S.N.; et al. Experimental assessment of the accuracy of genomic selection in sugarcane. Theor. Appl. Genet. 2013, 126, 2575–2586. [Google Scholar] [CrossRef]
- Harrison, M.D.; Geijskes, J.; Coleman, H.D.; Shand, K.; Kinkema, M.; Palupe, A.; Hassall, R.; Sainz, M.; Lloyd, R.; Miles, S. Accumulation of recombinant cellobiohydrolase and endoglucanase in the leaves of mature transgenic sugar cane. Plant Biotechnol. J. 2011, 9, 884–896. [Google Scholar] [CrossRef]
- Jaffe, G. Withering on the Vine; Center for Science in the Public Interest: Washington, DC, USA, 2005. [Google Scholar]
- DoE, U. Breaking the Biological Barriers to Cellulosic Ethanol: A Joint Research Agenda. In Report from the December 2005 Workshop, DOE/SC-0095; US Department of Energy Office of Science: Washington, DC, USA, 2016. Available online: www.genomicscience.energy.gov/biofuels/ (accessed on 28 February 2024).
- Radhesh Krishnan, S.; Mohan, C. Methods of sugarcane transformation. In Sugarcane Biotechnology: Challenges and Prospects; Springer: Berlin/Heidelberg, Germany, 2017; pp. 51–60. [Google Scholar]
- Jung, J.H.; Fouad, W.M.; Vermerris, W.; Gallo, M.; Altpeter, F. RNAi suppression of lignin biosynthesis in sugarcane reduces recalcitrance for biofuel production from lignocellulosic biomass. Plant Biotechnol. J. 2012, 10, 1067–1076. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Altpeter, F. TALEN mediated targeted mutagenesis of the caffeic acid O-methyltransferase in highly polyploid sugarcane improves cell wall composition for production of bioethanol. Plant Mol. Biol. 2016, 92, 131–142. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Bonawitz, N.D.; Chapple, C. The genetics of lignin biosynthesis: Connecting genotype to phenotype. Annu. Rev. Genet. 2010, 44, 337–363. [Google Scholar] [CrossRef] [PubMed]
- Van Acker, R.; Leplé, J.-C.; Aerts, D.; Storme, V.; Goeminne, G.; Ivens, B.; Légée, F.; Lapierre, C.; Piens, K.; Van Montagu, M.C. Improved saccharification and ethanol yield from field-grown transgenic poplar deficient in cinnamoyl-CoA reductase. Proc. Natl. Acad. Sci. USA 2014, 111, 845–850. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Dixon, R.A. Lignin modification improves fermentable sugar yields for biofuel production. Nat. Biotechnol. 2007, 25, 759–761. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; He, X.; Poovaiah, C.R.; Wuddineh, W.A.; Ma, J.; Mann, D.G.; Wang, H.; Jackson, L.; Tang, Y.; Neal Stewart Jr, C. Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol. 2012, 193, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Fornalé, S.; Capellades, M.; Encina, A.; Wang, K.; Irar, S.; Lapierre, C.; Ruel, K.; Joseleau, J.-P.; Berenguer, J.; Puigdomènech, P. Altered lignin biosynthesis improves cellulosic bioethanol production in transgenic maize plants down-regulated for cinnamyl alcohol dehydrogenase. Mol. Plant 2012, 5, 817–830. [Google Scholar] [CrossRef]
- Sattler, S.E.; Funnell-Harris, D.L. Modifying lignin to improve bioenergy feedstocks: Strengthening the barrier against pathogens? Front. Plant Sci. 2013, 4, 70. [Google Scholar] [CrossRef]
- Poovaiah, C.R.; Nageswara-Rao, M.; Soneji, J.R.; Baxter, H.L.; Stewart, C.N., Jr. Altered lignin biosynthesis using biotechnology to improve lignocellulosic biofuel feedstocks. Plant Biotechnol. J. 2014, 12, 1163–1173. [Google Scholar] [CrossRef]
- Baulcombe, D. RNA silencing in plants. Nature 2004, 431, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Kannan, B.; Dermawan, H.; Moxley, G.W.; Altpeter, F. Precision breeding for RNAi suppression of a major 4-coumarate: Coenzyme A ligase gene improves cell wall saccharification from field grown sugarcane. Plant Mol. Biol. 2016, 92, 505–517. [Google Scholar] [CrossRef] [PubMed]
- Bewg, W.P.; Poovaiah, C.; Lan, W.; Ralph, J.; Coleman, H.D. RNAi downregulation of three key lignin genes in sugarcane improves glucose release without reduction in sugar production. Biotechnol. Biofuels 2016, 9, 270. [Google Scholar] [CrossRef] [PubMed]
- Boettcher, M.; McManus, M.T. Choosing the right tool for the job: RNAi, TALEN, or CRISPR. Mol. Cell 2015, 58, 575–585. [Google Scholar] [CrossRef]
- Smith, I.; Greenside, P.G.; Natoli, T.; Lahr, D.L.; Wadden, D.; Tirosh, I.; Narayan, R.; Root, D.E.; Golub, T.R.; Subramanian, A. Evaluation of RNAi and CRISPR technologies by large-scale gene expression profiling in the Connectivity Map. PLoS Biol. 2017, 15, e2003213. [Google Scholar] [CrossRef] [PubMed]
- Puchta, H.; Dujon, B.; Hohn, B. Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res. 1993, 21, 5034–5040. [Google Scholar] [CrossRef]
- Li, T.; Liu, B.; Spalding, M.H.; Weeks, D.P.; Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 2012, 30, 390–392. [Google Scholar] [CrossRef] [PubMed]
- Kannan, B.; Jung, J.H.; Moxley, G.W.; Lee, S.M.; Altpeter, F. TALEN-mediated targeted mutagenesis of more than 100 COMT copies/alleles in highly polyploid sugarcane improves saccharification efficiency without compromising biomass yield. Plant Biotechnol. J. 2018, 16, 856–866. [Google Scholar] [CrossRef]
- Khan, Q.; Qin, Y.; Guo, D.-J.; Yang, L.-T.; Song, X.-P.; Xing, Y.-X.; Li, Y.-R. A Review of the diverse genes and molecules involved in sucrose metabolism and innovative approaches to improve sucrose content in sugarcane. Agronomy 2023, 13, 2957. [Google Scholar] [CrossRef]
- Hussin, S.H.; Liu, X.; Li, C.; Diaby, M.; Jatoi, G.H.; Ahmed, R.; Imran, M.; Iqbal, M.A. An Updated Overview on Insights into Sugarcane Genome Editing via CRISPR/Cas9 for Sustainable Production. Sustainability 2022, 14, 12285. [Google Scholar] [CrossRef]
- Zhou, X.; Rao, S.; Wrightstone, E.; Sun, T.; Lui, A.C.W.; Welsch, R.; Li, L. Phytoene synthase: The key rate-limiting enzyme of carotenoid biosynthesis in plants. Front. Plant Sci. 2022, 13, 884720. [Google Scholar] [CrossRef] [PubMed]
- Surya Krishna, S.; Viswanathan, R.; Valarmathi, R.; Lakshmi, K.; Appunu, C. CRISPR/Cas-Mediated Genome Editing Approach for Improving Virus Resistance in Sugarcane. Sugar Tech 2023, 25, 735–750. [Google Scholar] [CrossRef]
- Genetu, A.; Getahum, T. Review on Adaptation and Breeding Mechanisms of Salinity Stress for Sugarcane. J. Gene Engg Bio Res. 2024, 6, 01–13. [Google Scholar]
- Jaganathan, D.; Ramasamy, K.; Sellamuthu, G.; Jayabalan, S.; Venkataraman, G. CRISPR for crop improvement: An update review. Front. Plant Sci. 2018, 9, 985. [Google Scholar] [CrossRef]
- Puchta, H. Applying CRISPR/Cas for genome engineering in plants: The best is yet to come. Curr. Opin. Plant Biol. 2017, 36, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Waterworth, W.M.; Drury, G.E.; Bray, C.M.; West, C.E. Repairing breaks in the plant genome: The importance of keeping it together. New Phytol. 2011, 192, 805–822. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.-K.; Puchta, H. CRISPR/Cas-mediated gene targeting in plants: Finally a turn for the better for homologous recombination. Plant Cell Rep. 2019, 38, 443–453. [Google Scholar] [CrossRef]
- Zhang, Y.; Malzahn, A.A.; Sretenovic, S.; Qi, Y. The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 2019, 5, 778–794. [Google Scholar] [CrossRef]
- Wiedenheft, B.; Sternberg, S.H.; Doudna, J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012, 482, 331–338. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc. B 2019, 374, 20180087. [Google Scholar] [CrossRef]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Nekrasov, V. Plant genome editing made easy: Targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 2013, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, C.; Lillemo, M.; Hvoslef-Eide, T.A. Global regulation of genetically modified crops amid the gene edited crop boom–a review. Front. Plant Sci. 2021, 12, 630396. [Google Scholar] [CrossRef] [PubMed]
- Wolt, J.D.; Wang, K.; Yang, B. The regulatory status of genome-edited crops. Plant Biotechnol. J. 2016, 14, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Lin, C.-Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef]
- Eid, A.; Mohan, C.; Sanchez, S.; Wang, D.; Altpeter, F. Multiallelic, targeted mutagenesis of magnesium chelatase with CRISPR/Cas9 provides a rapidly scorable phenotype in highly polyploid sugarcane. Front. Genome Ed. 2021, 3, 654996. [Google Scholar] [CrossRef] [PubMed]
- Oz, M.T.; Altpeter, A.; Karan, R.; Merotto, A.; Altpeter, F. CRISPR/Cas9-mediated multi-allelic gene targeting in sugarcane confers herbicide tolerance. Front. Genome Ed. 2021, 3, 673566. [Google Scholar] [CrossRef] [PubMed]
- Zaman, Q.U.; Li, C.; Cheng, H.; Hu, Q. Genome editing opens a new era of genetic improvement in polyploid crops. Crop J. 2019, 7, 141–150. [Google Scholar] [CrossRef]
- Premachandran, M.; Prathima, P.; Lekshmi, M. Sugarcane and polyploidy: A review. J. Sugarcane Res 2011, 1, 1–15. [Google Scholar]
- Meng, X.; Yu, H.; Zhang, Y.; Zhuang, F.; Song, X.; Gao, S.; Gao, C.; Li, J. Construction of a genome-wide mutant library in rice using CRISPR/Cas9. Mol. Plant 2017, 10, 1238–1241. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, J.; Zhou, J.; Wang, L.; Yang, S.; Hurst, L.D.; Li, W.-H.; Tian, D. Identifying a large number of high-yield genes in rice by pedigree analysis, whole-genome sequencing, and CRISPR-Cas9 gene knockout. Proc. Natl. Acad. Sci. USA 2018, 115, E7559–E7567. [Google Scholar] [CrossRef] [PubMed]
- Riaño-Pachón, D.M.; Mattiello, L. Draft genome sequencing of the sugarcane hybrid SP80-3280. F1000Research 2017, 6, 861. [Google Scholar] [CrossRef] [PubMed]
- Garsmeur, O.; Droc, G.; Aitken, K.S.; Potier, B.; Van Sluys, M.-A.; Hervouet, C.; Van der Vossen, E.; Henry, R.; Schmutz, J.; D’Hont, A. A monoploid reference sequence for the highly complex genome of sugarcane. In Proceedings of the ISSCT Joint Breeding & Germplasm and Molecular Biology Workshops, Okinawa, Japan, 22 October 2018. [Google Scholar]
- Augustine, S.M.; Narayan, J.A.; Syamaladevi, D.P.; Appunu, C.; Chakravarthi, M.; Ravichandran, V.; Subramonian, N. Erianthus arundinaceus HSP70 (EaHSP70) overexpression increases drought and salinity tolerance in sugarcane (Saccharum spp. hybrid). Plant Sci. 2015, 232, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Augustine, S.M.; Ashwin Narayan, J.; Syamaladevi, D.P.; Appunu, C.; Chakravarthi, M.; Ravichandran, V.; Tuteja, N.; Subramonian, N. Introduction of pea DNA helicase 45 into sugarcane (Saccharum spp. hybrid) enhances cell membrane thermostability and upregulation of stress-responsive genes leads to abiotic stress tolerance. Mol. Biotechnol. 2015, 57, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Augustine, S.M.; Ashwin Narayan, J.; Syamaladevi, D.P.; Appunu, C.; Chakravarthi, M.; Ravichandran, V.; Tuteja, N.; Subramonian, N. Overexpression of EaDREB2 and pyramiding of EaDREB2 with the pea DNA helicase gene (PDH45) enhance drought and salinity tolerance in sugarcane (Saccharum spp. hybrid). Plant Cell Rep. 2015, 34, 247–263. [Google Scholar] [CrossRef] [PubMed]
- Joyce, P.; Kuwahata, M.; Turner, N.; Lakshmanan, P. Selection system and co-cultivation medium are important determinants of Agrobacterium-mediated transformation of sugarcane. Plant Cell Rep. 2010, 29, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.-J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J. Morphogenic regulators Baby boom and Wuschel improve monocot transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef] [PubMed]
- Lowe, K.; La Rota, M.; Hoerster, G.; Hastings, C.; Wang, N.; Chamberlin, M.; Wu, E.; Jones, T.; Gordon-Kamm, W. Rapid genotype “independent” Zea mays L.(maize) transformation via direct somatic embryogenesis. In Vitro Cell. Dev. Biol.-Plant 2018, 54, 240–252. [Google Scholar] [CrossRef]
- Gazaffi, R.; Oliveira, K.; Souza, A.; Garcia, A. The importance of the germplasm in developing agro-energetic profile sugarcane cultivars. In Sugar Cane Bioethanol: R&D for Productivity and Sustainability; Blucher: São Paulo, Brazil, 2010; pp. 333–343. [Google Scholar]
- Ma, X.; Zhu, Q.; Chen, Y.; Liu, Y.-G. CRISPR/Cas9 platforms for genome editing in plants: Developments and applications. Mol. Plant 2016, 9, 961–974. [Google Scholar] [CrossRef]
- Garsmeur, O.; Droc, G.; Antonise, R.; Grimwood, J.; Potier, B.; Aitken, K.; Jenkins, J.; Martin, G.; Charron, C.; Hervouet, C. A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat. Commun. 2018, 9, 2638. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xing, H.-L.; Wang, Z.-P.; Zhang, H.-Y.; Yang, F.; Wang, X.-C.; Chen, Q.-J. Potential high-frequency off-target mutagenesis induced by CRISPR/Cas9 in Arabidopsis and its prevention. Plant Mol. Biol. 2018, 96, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Souza, G.M.; Van Sluys, M.-A.; Lembke, C.G.; Lee, H.; Margarido, G.R.A.; Hotta, C.T.; Gaiarsa, J.W.; Diniz, A.L.; Oliveira, M.d.M.; Ferreira, S.d.S. Assembly of the 373k gene space of the polyploid sugarcane genome reveals reservoirs of functional diversity in the world’s leading biomass crop. GigaScience 2019, 8, giz129. [Google Scholar] [CrossRef] [PubMed]
- Altpeter, F.; Springer, N.M.; Bartley, L.E.; Blechl, A.E.; Brutnell, T.P.; Citovsky, V.; Conrad, L.J.; Gelvin, S.B.; Jackson, D.P.; Kausch, A.P. Advancing crop transformation in the era of genome editing. Plant Cell 2016, 28, 1510–1520. [Google Scholar] [CrossRef]
- Healey, A.; Furtado, A.; Cooper, T.; Henry, R.J. Protocol: A simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 2014, 10, 21. [Google Scholar] [CrossRef]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef]
- Kim, H.; Kim, S.-T.; Ryu, J.; Kang, B.-C.; Kim, J.-S.; Kim, S.-G. CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 2017, 8, 14406. [Google Scholar] [CrossRef]
- Liang, Z.; Chen, K.; Li, T.; Zhang, Y.; Wang, Y.; Zhao, Q.; Liu, J.; Zhang, H.; Liu, C.; Ran, Y. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 2017, 8, 14261. [Google Scholar] [CrossRef]
- Andersson, M.; Turesson, H.; Olsson, N.; Fält, A.S.; Ohlsson, P.; Gonzalez, M.N.; Samuelsson, M.; Hofvander, P. Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol. Plant. 2018, 164, 378–384. [Google Scholar] [CrossRef]
- Schausberger, P. Herbivore-associated bacteria as potential mediators and modifiers of induced plant defense against spider mites and thrips. Front. Plant Sci. 2018, 9, 1107. [Google Scholar] [CrossRef]
- Park, J.; Choi, S.; Park, S.; Yoon, J.; Park, A.Y.; Choe, S. DNA-free genome editing via ribonucleoprotein (RNP) delivery of CRISPR/Cas in lettuce. In Plant Genome Editing with CRISPR Systems; Springer: Berlin/Heidelberg, Germany, 2019; pp. 337–354. [Google Scholar]
- Mohan, C. Genome editing in sugarcane: Challenges ahead. Front. Plant Sci. 2016, 7, 1542. [Google Scholar] [CrossRef] [PubMed]
| Gene of Interest | Modification Method | Trait Affected | Observed Outcome | References |
|---|---|---|---|---|
| Sucrose Synthase (SuSy) gene | CRISPR-Cas9 | Sugar Content | 18% increase in sucrose content in stalks; enhanced sweetness | [137] |
| Drought Tolerance Gene (DTG) | CRISPR-Cas9 | Drought Tolerance | Increased resilience to water scarcity by 30% during dry seasons | [138] |
| Lignin Biosynthesis Genes (LBG) | TALEN | Lignin Content | Reduced lignin content by 25%, facilitating easier biofuel production | [136] |
| Phytoene Synthase (PSY) gene | Gene overexpression | Biomass Yield | 22% higher biomass yield through enhanced photosynthesis efficiency | [139] |
| Disease Resistance Gene (DRG1) | CRISPR-Cas9 | Disease Resistance | Improved resistance against rust and smut diseases by over 40% | [140] |
| Salinity Tolerance Gene (STG) | CRISPR-Cas9 | Salinity Tolerance | Enhanced growth under high salinity conditions by 35% | [141] |
| Caffeic acid O-methyltransferase (COMT) | RNAi | Lignin Content | Reduced lignin content, improved digestibility | [93] |
| 4-Coumarate-CoA Ligase (4CL) | RNAi | Lignin Content | Reduced lignin content, improved biofuel production | [130,131] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmad, K.; Ming, R. Harnessing Genetic Tools for Sustainable Bioenergy: A Review of Sugarcane Biotechnology in Biofuel Production. Agriculture 2024, 14, 1312. https://doi.org/10.3390/agriculture14081312
Ahmad K, Ming R. Harnessing Genetic Tools for Sustainable Bioenergy: A Review of Sugarcane Biotechnology in Biofuel Production. Agriculture. 2024; 14(8):1312. https://doi.org/10.3390/agriculture14081312
Chicago/Turabian StyleAhmad, Kashif, and Ray Ming. 2024. "Harnessing Genetic Tools for Sustainable Bioenergy: A Review of Sugarcane Biotechnology in Biofuel Production" Agriculture 14, no. 8: 1312. https://doi.org/10.3390/agriculture14081312





