A Skeleton-Based Method of Root System 3D Reconstruction and Phenotypic Parameter Measurement from Multi-View Image Sequence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
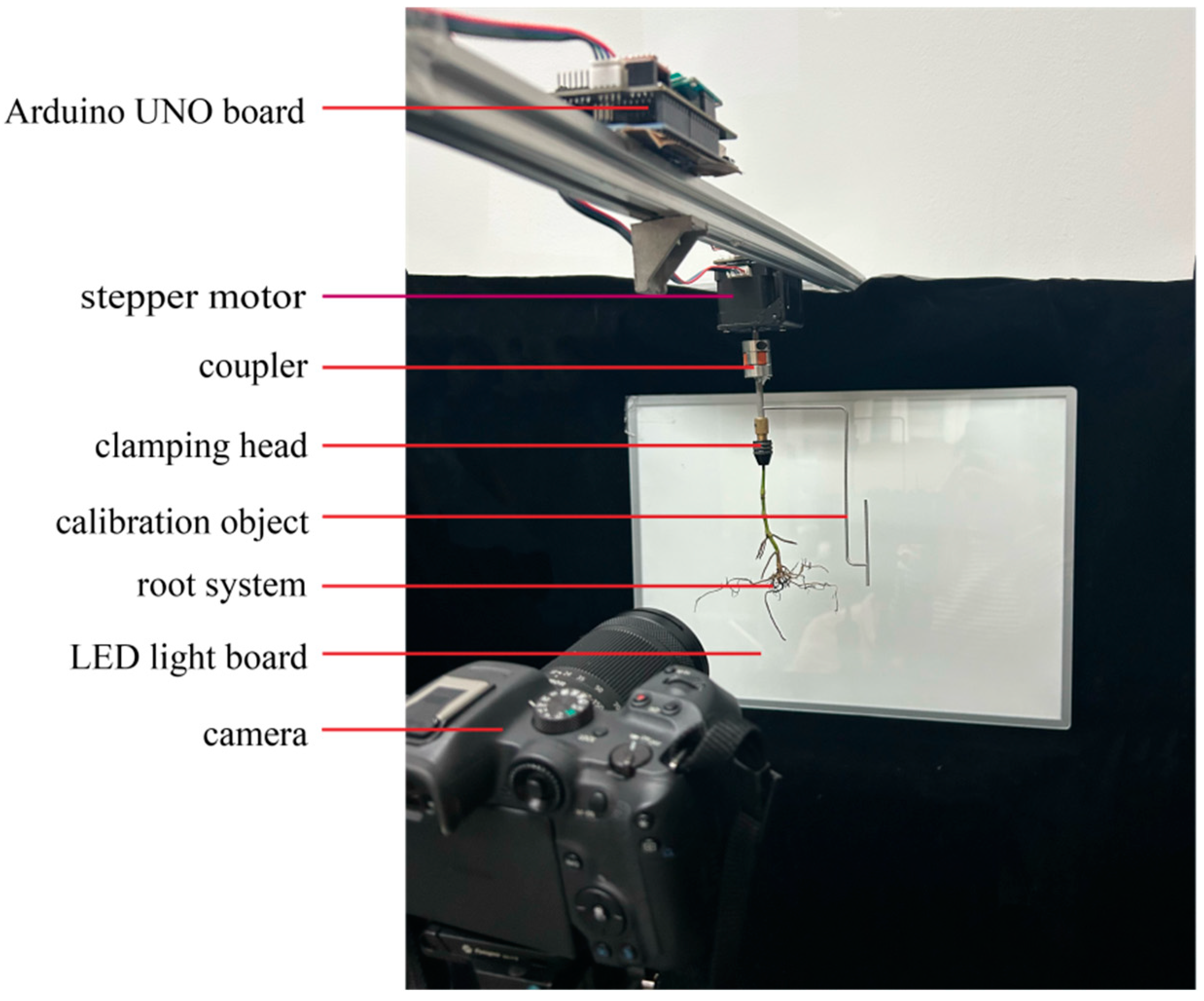
2.2. Experiment System
2.3. Image Preprocessing
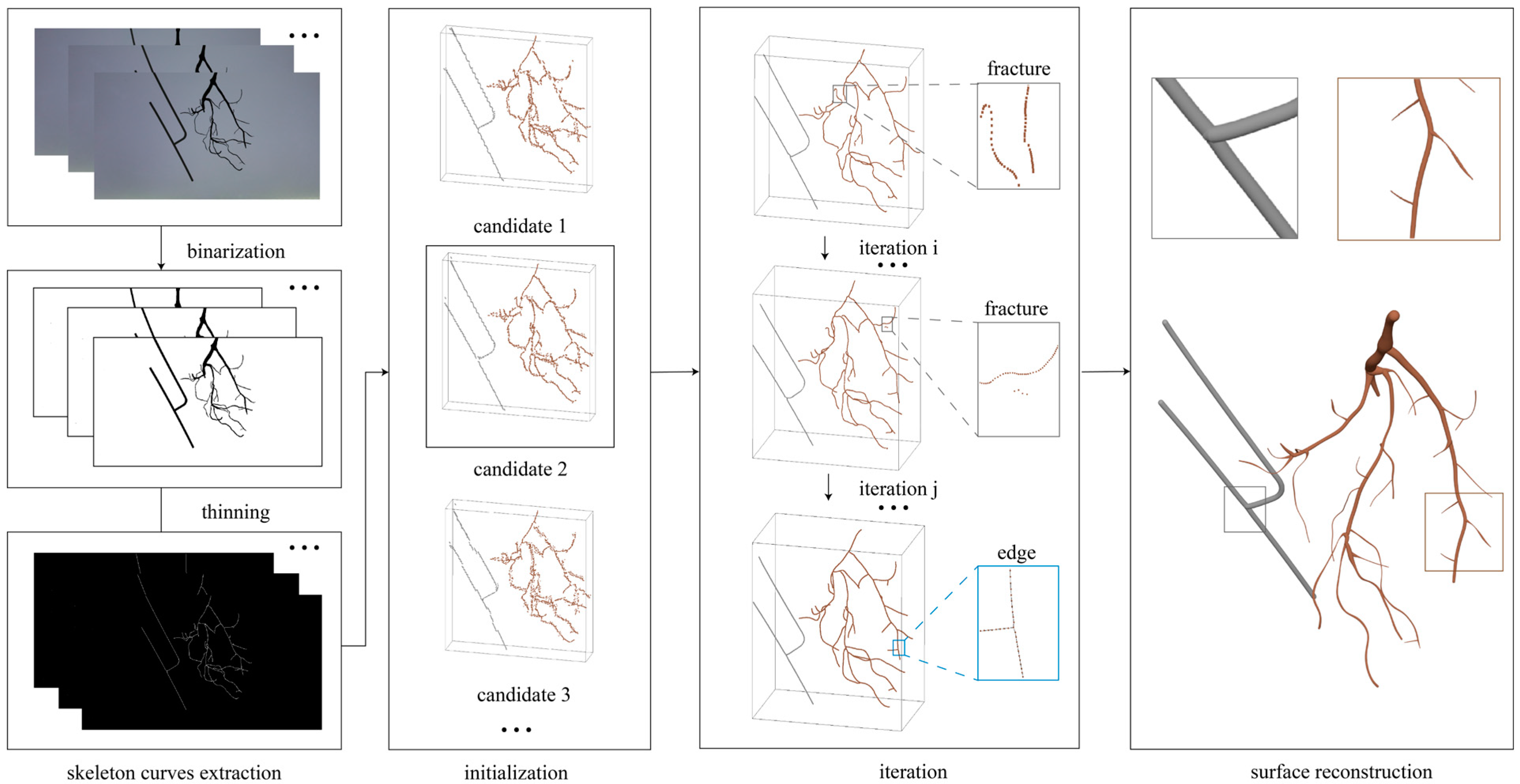
2.4. Skeleton-Based 3D Reconstruction
- (1)
- The binary masks of the pixels showing the target object in the foreground were processed by thinning method to extract on pixel wide skeleton curves which were henceforth denoted by . And the input views were denoted by .
- (2)
- By using the optical flow method and bundle adjustment from the first few pictures, several 3D points candidates were initialized, and the best-performing 3D points candidates, henceforth denoted by , were selected as a basis for subsequent iterations.
- (3)
- The camera poses and the curve network were computed by minimizing an objective function (Equation (2)) that measured the sum of the squared 2D distances between the projection of the curve network, , and the corresponding 2D skeletal curve, , across all input pictures. And, a commonly used formulation for curve fitting was utilized to efficiently minimize the distance error term (Equation (3)). The function was minimized iteratively in an alternating fashion that first optimized the camera poses while fixing the curve points, and then optimized the curve points while fixing the camera poses. During the iteration, had to be matched with the points , in the view of , and the matching was performed by combining a distance-based criterion with a constraint on curve consistency. During the initialization and iteration, , which recorded the edges by pairing points, was constructed and updated based on 3D points through the variant of Kruskal’s algorithm, which determined whether the points were connected based on the distance between them and the length of the loop formed by the edges. At the same time, the 3D points were uniformly resampled. Also at the same time, there may be self-occlusion due to the root structure. To determine whether the 3D points were subject to self-occlusion in certain view , the neighboring pixels of the matching point for each point would be examined with a 3 3 local window, and a 3D point set that matches the pixels would be generated. Then, the spatial compactness factor, , was computed from the average distance between the points in P and their centroid. If , was labeled as self-occluded, setting to 0, or setting to 1.where was 0 if the point was self-occluded, and, otherwise, was 0. was distance error, and was the normal direction of the point . was the tangent direction of the point . was regularization term, and was the focal length of the camera.
- (4)
- To reconstruct the root surface, the root system was considered to be composed of generalized cylinders, so the radius of each point was the key which was calculated using the corresponding image observations from all multi-view binary images. Specifically, the radius of was the average of the radii over all the input images.where was the distances from the projection point of , in each view , to both sides of the defined strip. was the depth of with respect to the view .
2.5. Scale Alignment for Phenotypic Parameters Measurement
2.6. Skeleton-Based Point Completion
- (1)
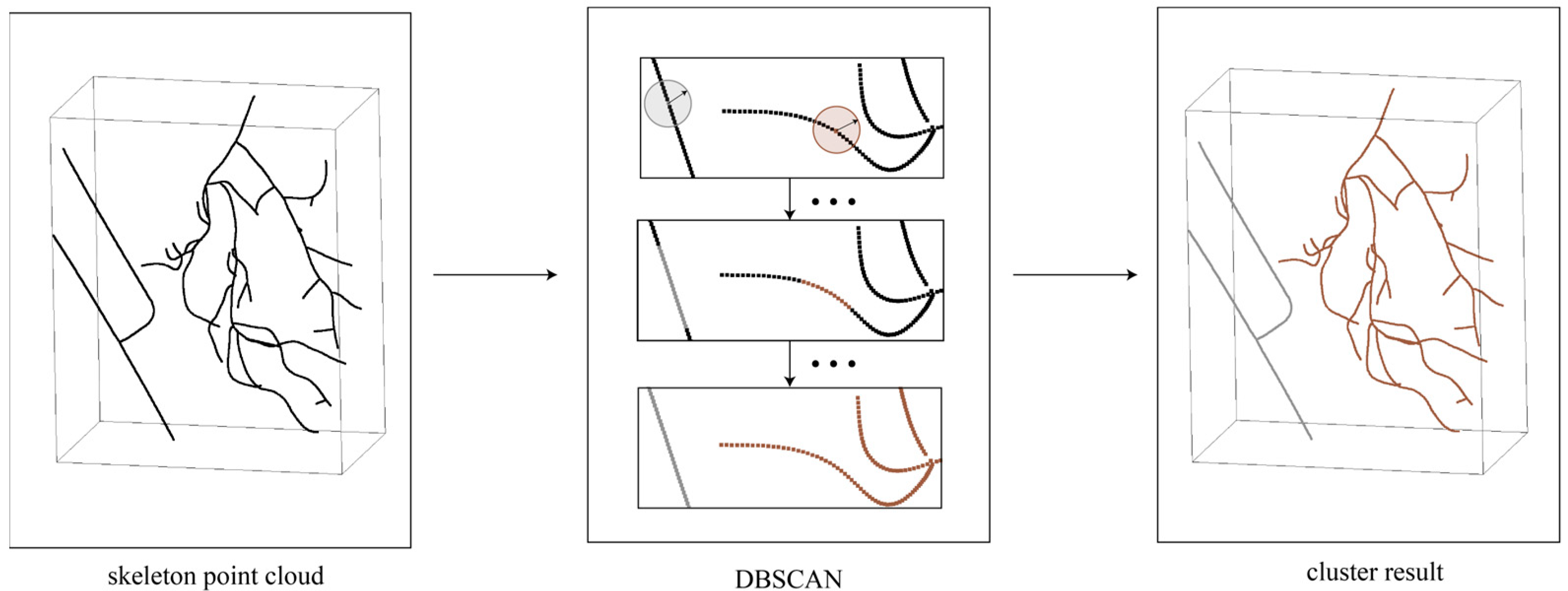
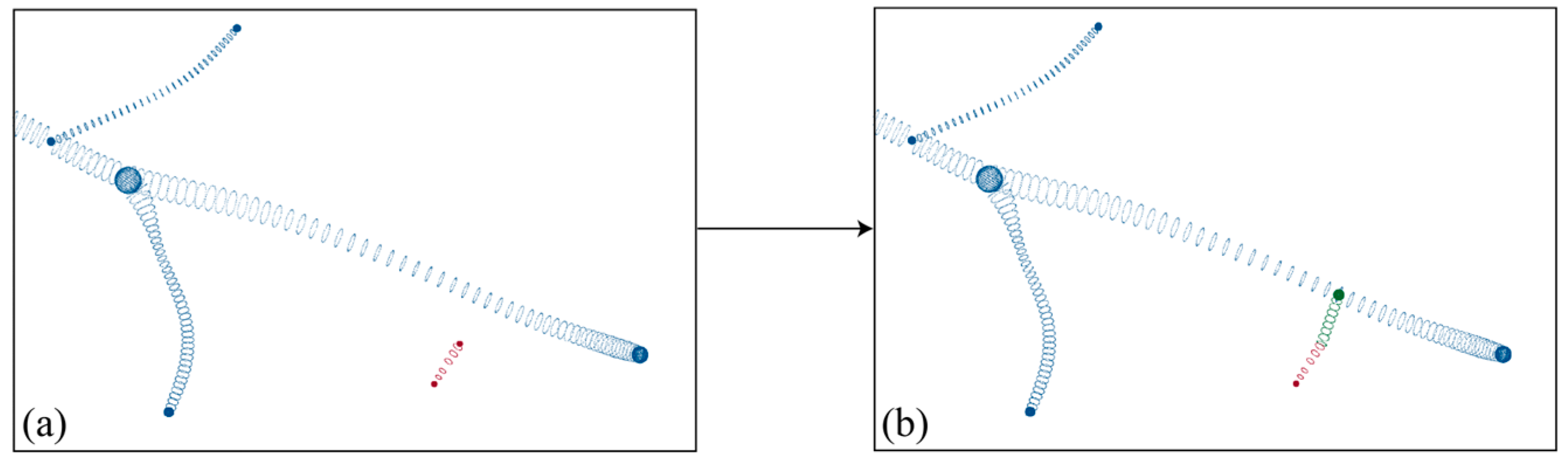
- The connectivity of the reconstructed skeleton point cloud was determined based on the DFS algorithm, dividing the skeleton point cloud with a missing region into independent skeleton point clouds, as in Figure 8a,b. Based on the number of points, independent skeletons were classified into a primary skeleton and sub-skeletons, while also saving all endpoint coordinates, henceforth denoted by .
- (2)
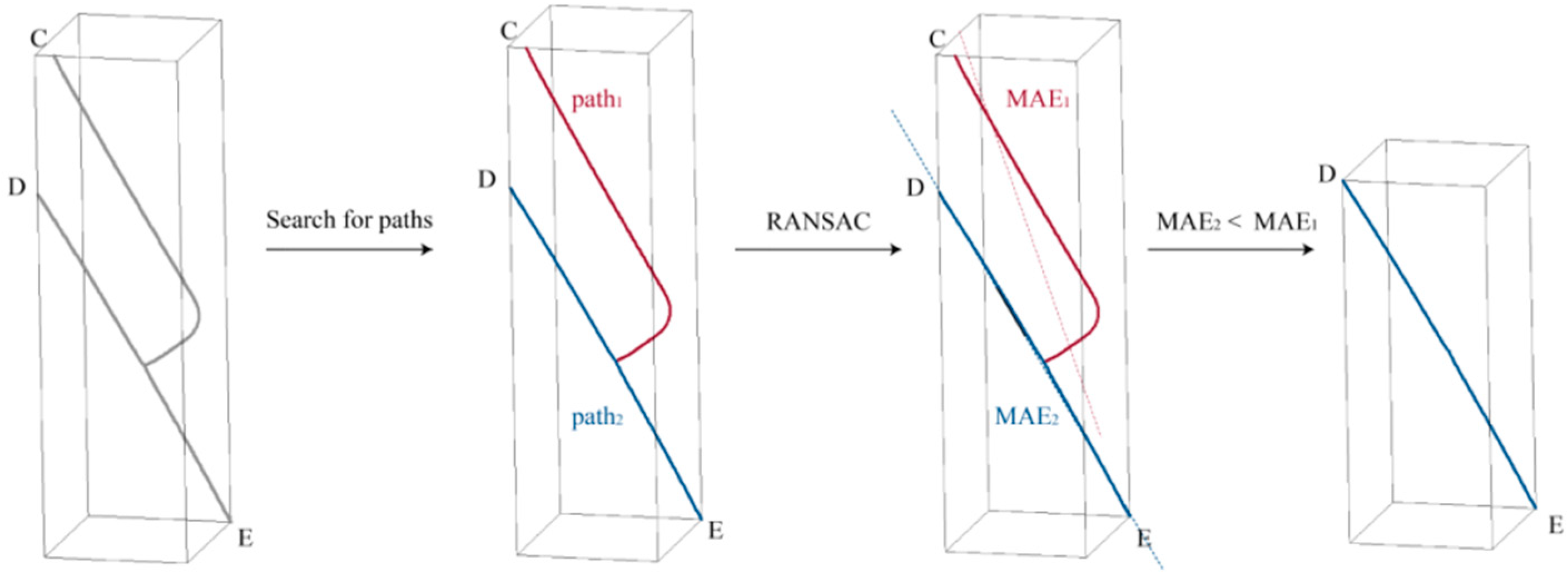
- For each sub-skeleton, through all endpoints are iterated through, and the tangent vector is found for . As in Figure 8b, for each , all points in the primary skeleton are traversed to find a point that satisfied Equation (7). If such a point is found, it is considered a candidate connection point, and the length of is recorded. After completing the traversal, the shortest is retained as the connection line, and is checked with regard to it being in the endpoint set. If so, it is removed. To ensure an even distribution of the skeleton point cloud, points were uniformly sampled along the connection line at intervals of the average point distance, as in Figure 8c. At the same time, to ensure the smoothness of the skeleton, if is an endpoint, the points sampled along , and points of this sub-skeleton, are used for curve fitting. Then, points are uniformly sampled along the curve again, and the sampled points are added to the skeleton points. After traversing all sub-skeletons, the skeleton point completion was completed as Figure 8d.
- (3)
- To generate the surface, the radii of the points sampled on the connection line were also required. To ensure the smoothness of the generated surface, if is an endpoint, the radius along the connection line is then set to vary linearly with the radius of and ; if is not an endpoint, the radius along the connection line is considered to be the same as at point . Based on the completed skeleton points and the radius, surface generation can be accomplished, as seen in Figure 9.
2.7. Evaluation of Reconstruction Quality
3. Results
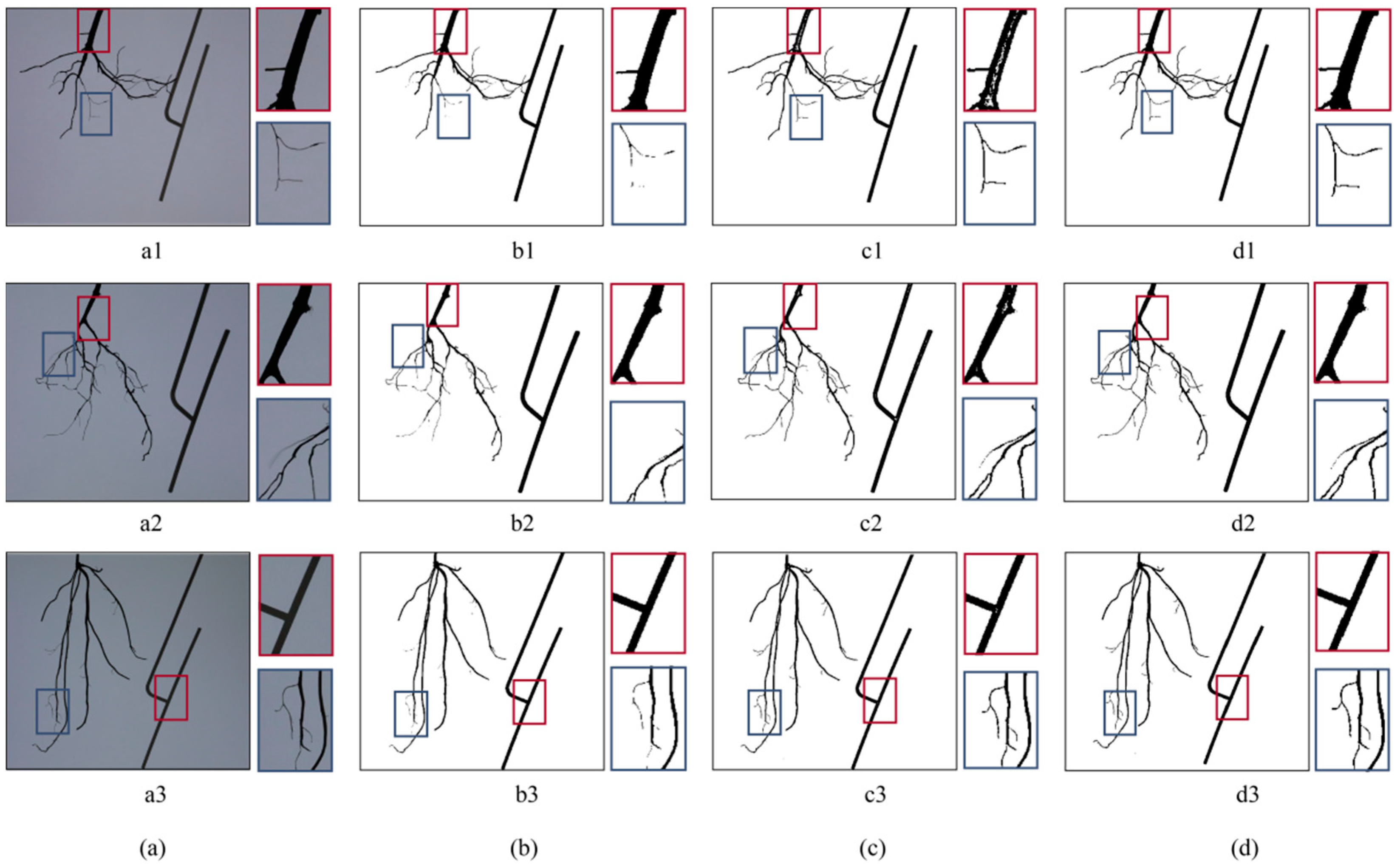
3.1. Segmentation of Target Object
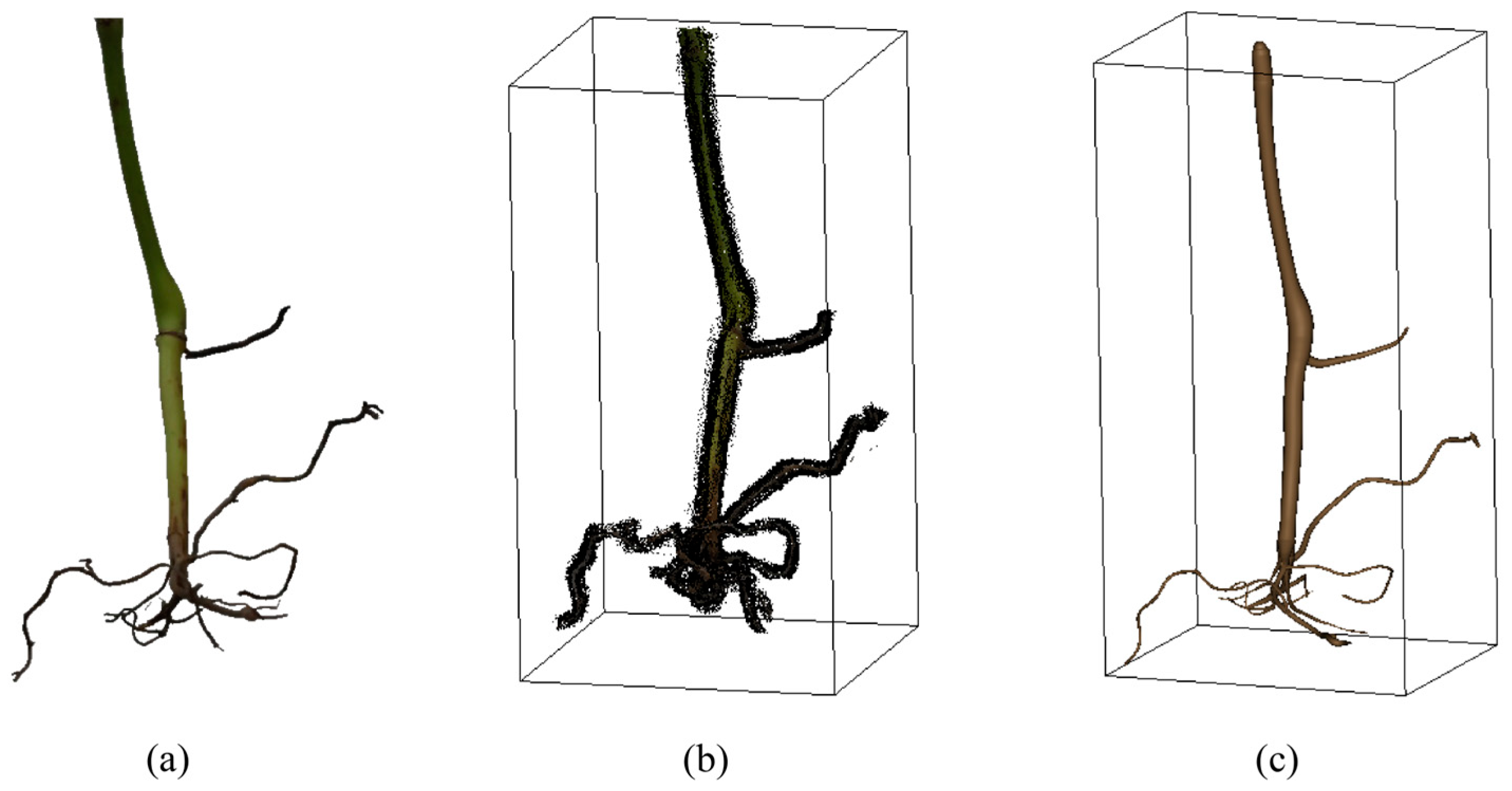
3.2. Three-Dimensional Reconstruction Results
3.3. Reconstruction Quality
3.3.1. Reconstruction Performance
3.3.2. Phenotypic Parameter Measurement Performance
3.4. Reconstruction Time Cost
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Uga, Y.; Sugimoto, K.; Ogawa, S.; Rane, J.; Ishitani, M.; Hara, N.; Kitomi, Y.; Inukai, Y.; Ono, K.; Kanno, N.; et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013, 45, 1097. [Google Scholar] [CrossRef] [PubMed]
- Mahanta, D.; Rai, R.K.; Mishra, S.D.; Raja, A.; Purakayastha, T.J.; Varghese, E. Influence of phosphorus and biofertilizers on soybean and wheat root growth and properties. Field Crops Res. 2014, 166, 1–9. [Google Scholar] [CrossRef]
- Bonato, T.; Beggio, G.; Pivato, A.; Piazza, R. Maize plant (Zea mays) uptake of organophosphorus and novel brominated flame retardants from hydroponic cultures. Chemosphere 2022, 287, 132456. [Google Scholar] [CrossRef] [PubMed]
- Alemu, A.; Feyissa, T.; Maccaferri, M.; Sciara, G.; Tuberosa, R.; Ammar, K.; Badebo, A.; Acevedo, M.; Letta, T.; Abeyo, B. Genome-wide association analysis unveils novel QTLs for seminal root system architecture traits in Ethiopian durum wheat. BMC Genom. 2021, 22, 20. [Google Scholar] [CrossRef]
- Mathieu, L.; Lobet, G.; Tocquin, P.; Perilleux, C. “Rhizoponics”: A novel hydroponic rhizotron for root system analyses on mature Arabidopsis thaliana plants. Plant Methods 2015, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Junker, A.; Seiler, C.; Altmann, T. Phenotyping roots in darkness: Disturbance-free root imaging with near infrared illumination. Funct. Plant Biol. 2018, 45, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Goclawski, J.; Sekulska-Nalewajko, J.; Gajewska, E.; Wielanek, M. An automatic segmentation method for scanned images of wheat root systems with dark discolourations. Int. J. Appl. Math. Comput. Sci. 2009, 19, 679–689. [Google Scholar] [CrossRef]
- Yugan, C.; Xuecheng, Z. Plant root image processing and analysis based on 2D scanner. In Proceedings of the 2010 IEEE Fifth International Conference on Bio-Inspired Computing: Theories and Applications (BIC-TA), Changsha, China, 23–26 September 2010; pp. 1216–1220. [Google Scholar]
- Arnold, T.; Bodner, G. Study of visible imaging and near-infrared imaging spectroscopy for plant root phenotyping. In Proceedings of the Conference on Sensing for Agriculture and Food Quality and Safety X, Orlando, FL, USA, 17–18 April 2018. [Google Scholar]
- Narisetti, N.; Henke, M.; Seiler, C.; Junker, A.; Ostermann, J.; Altmann, T.; Gladilin, E. Fully-automated root image analysis (faRIA). Sci. Rep. 2021, 11, 16047. [Google Scholar] [CrossRef]
- Smith, A.G.; Petersen, J.; Selvan, R.; Rasmussen, C.R. Segmentation of roots in soil with U-Net. Plant Methods 2020, 16, 13. [Google Scholar] [CrossRef]
- Gong, L.; Du, X.; Zhu, K.; Lin, C.; Lin, K.; Wang, T.; Lou, Q.; Yuan, Z.; Huang, G.; Liu, C. Pixel level segmentation of early-stage in-bag rice root for its architecture analysis. Comput. Electron. Agric. 2021, 186, 106197. [Google Scholar] [CrossRef]
- Thesma, V.; Mohammadpour Velni, J. Plant Root Phenotyping Using Deep Conditional GANs and Binary Semantic Segmentation. Sensors 2023, 23, 309. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; Bian, Y.; Niu, Z.; Taha, M.F.; He, Y.; Qiu, Z. Fast neural distance field-based three-dimensional reconstruction method for geometrical parameter extraction of walnut shell from multiview images. Comput. Electron. Agric. 2024, 224, 109189. [Google Scholar] [CrossRef]
- Zhou, L.; Jin, S.; Wang, J.; Zhang, H.; Shi, M.; Zhou, H. 3D positioning of Camellia oleifera fruit-grabbing points for robotic harvesting. Biosyst. Eng. 2024, 246, 110–121. [Google Scholar] [CrossRef]
- Gregory, P.J.; Hutchison, D.J.; Read, D.B.; Jenneson, P.M.; Gilboy, W.B.; Morton, E.J. Non-invasive imaging of roots with high resolution X-ray micro-tomography. Plant Soil 2003, 255, 351–359. [Google Scholar] [CrossRef]
- Hou, L.; Gao, W.; der Bom van, F.; Weng, Z.; Doolette, C.L.; Maksimenko, A.; Hausermann, D.; Zheng, Y.; Tang, C.; Lombi, E.; et al. Use of X-ray tomography for examining root architecture in soils. Geoderma 2022, 405, 115405. [Google Scholar] [CrossRef]
- Jiang, Z.; Leung, A.K.; Liu, J. Segmentation uncertainty of vegetated porous media propagates during X-ray CT image-based analysis. Plant Soil 2024. [Google Scholar] [CrossRef]
- Metzner, R.; Eggert, A.; van Dusschoten, D.; Pflugfelder, D.; Gerth, S.; Schurr, U.; Uhlmann, N.; Jahnke, S. Direct comparison of MRI and X-ray CT technologies for 3D imaging of root systems in soil: Potential and challenges for root trait quantification. Plant Methods 2015, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- van Dusschoten, D.; Metzner, R.; Kochs, J.; Postma, J.A.; Pflugfelder, D.; Buehler, J.; Schurr, U.; Jahnke, S. Quantitative 3D Analysis of Plant Roots Growing in Soil Using Magnetic Resonance Imaging. Plant Physiol. 2016, 170, 1176–1188. [Google Scholar] [CrossRef]
- Heeren, B.; Paulus, S.; Goldbach, H.; Kuhlmann, H.; Mahlein, A.-K.; Rumpf, M.; Wirth, B. Statistical shape analysis of tap roots: A methodological case study on laser scanned sugar beets. BMC Bioinform. 2020, 21, 335. [Google Scholar] [CrossRef]
- Todo, C.; Ikeno, H.; Yamase, K.; Tanikawa, T.; Ohashi, M.; Dannoura, M.; Kimura, T.; Hirano, Y. Reconstruction of Conifer Root Systems Mapped with Point Cloud Data Obtained by 3D Laser Scanning Compared with Manual Measurement. Forests 2021, 12, 1117. [Google Scholar] [CrossRef]
- Kargar, A.R.; MacKenzie, R.A.; Apwong, M.; Hughes, E.; van Aardt, J. Stem and root assessment in mangrove forests using a low-cost, rapid-scan terrestrial laser scanner. Wetl. Ecol. Manag. 2020, 28, 883–900. [Google Scholar] [CrossRef]
- Pflugfelder, D.; Kochs, J.; Koller, R.; Jahnke, S.; Mohl, C.; Pariyar, S.; Fassbender, H.; Nagel, K.A.; Watt, M.; van Dusschoten, D.; et al. The root system architecture of wheat establishing in soil is associated with varying elongation rates of seminal roots: Quantification using 4D magnetic resonance imaging. J. Exp. Bot. 2022, 73, 2050–2060. [Google Scholar] [CrossRef] [PubMed]
- Schneider, H.M.; Postma, J.A.; Kochs, J.; Pflugfelder, D.; Lynch, J.P.; van Dusschoten, D. Spatio-Temporal Variation in Water Uptake in Seminal and Nodal Root Systems of Barley Plants Grown in Soil. Front. Plant Sci. 2020, 11, 1247. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Chen, S.; Wu, B.; Liu, Y.; Tang, W.; Liu, F.; He, Y.; Zhang, C. Detection of oilseed rape clubroot based on low-field nuclear magnetic resonance imaging. Comput. Electron. Agric. 2024, 218, 108687. [Google Scholar] [CrossRef]
- Wu, Q.; Wu, J.; Hu, P.; Zhang, W.; Ma, Y.; Yu, K.; Guo, Y.; Cao, J.; Li, H.; Li, B.; et al. Quantification of the three-dimensional root system architecture using an automated rotating imaging system. Plant Methods 2023, 19, 11. [Google Scholar] [CrossRef]
- Sunvittayakul, P.; Kittipadakul, P.; Wonnapinij, P.; Chanchay, P.; Wannitikul, P.; Sathitnaitham, S.; Phanthanong, P.; Changwitchukarn, K.; Suttangkakul, A.; Ceballos, H.; et al. Cassava root crown phenotyping using three-dimension (3D) multi-view stereo reconstruction. Sci. Rep. 2022, 12, 10030. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Wang, Y.; Parikh, D.; Khan, A.; Lu, G. Simultaneous Direct Depth Estimation and Synthesis Stereo for Single Image Plant Root Reconstruction. IEEE Trans. Image Process. 2021, 30, 4883–4893. [Google Scholar] [CrossRef]
- Masuda, T. 3D Shape Reconstruction of Plant Roots in a Cylindrical Tank From Multiview Images. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 2149–2157. [Google Scholar]
- Liu, L.; Chen, N.; Ceylan, D.; Theobalt, C.; Wang, W.; Mitra, N.J.; Assoc Comp, M. CURVEFUSION: Reconstructing Thin Structures from RGBD Sequences. In Proceedings of the 11th ACM SIGGRAPH Conference and Exhibition on Computer Graphics and Interactive Techniques in Asia (SA), Tokyo, Japan, 4–7 December 2018. [Google Scholar]
- Martin, T.; Montes, J.; Bazin, J.-C.; Popa, T. Topology-aware reconstruction of thin tubular structures. In Proceedings of the SIGGRAPH Asia 2014 Technical Briefs, Shenzhen, China, 3–6 December 2014; p. 12. [Google Scholar]
- Li, S.; Yao, Y.; Fang, T.; Quan, L. Reconstructing Thin Structures of Manifold Surfaces by Integrating Spatial Curves. In Proceedings of the 31st IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 2887–2896. [Google Scholar]
- Tabb, A. Shape from Silhouette Probability Maps: Reconstruction of thin objects in the presence of silhouette extraction and calibration error. In Proceedings of the 26th IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Portland, OR, USA, 23–28 June 2013; pp. 161–168. [Google Scholar]
- Tabb, A.; Medeiros, H. A robotic vision system to measure tree traits. In Proceedings of the 2017 IEEE/RSJ International Conference on Intelligent Robots and Systems (IROS), 24–28 September 2017; pp. 6005–6012. [Google Scholar]
- Wang, P.; Liu, L.; Chen, N.; Chu, H.-K.; Theobalt, C.; Wang, W. Vid2Curve: Simultaneous Camera Motion Estimation and Thin Structure Reconstruction from an RGB Video. ACM Trans. Graph. 2020, 39, 132-1. [Google Scholar] [CrossRef]
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Olson, E. AprilTag: A robust and flexible visual fiducial system. In Proceedings of the IEEE International Conference on Robotics and Automation (ICRA), Shanghai, China, 9–13 May 2011. [Google Scholar]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. In Proceedings of the Knowledge Discovery and Data Mining, Portland, OR, USA, 2–4 August 1996. [Google Scholar]
- Fischler, M.A.; Bolles, R.C. Random sample consensus: A paradigm for model fitting with applications to image analysis and automated cartography. Commun. ACM 1981, 24, 381–395. [Google Scholar] [CrossRef]
- Okamoto, Y.; Ikeno, H.; Hirano, Y.; Tanikawa, T.; Yamase, K.; Todo, C.; Dannoura, M.; Ohashi, M. 3D reconstruction using Structure-from-Motion: A new technique for morphological measurement of tree root systems. Plant Soil 2022, 477, 829–841. [Google Scholar] [CrossRef]







| Species | SPE | FPE | ||
|---|---|---|---|---|
| AVG (Pixels) | SD (Pixels) | AVG (Pixels) | SD (Pixels) | |
| Ob | 0.588 | 0.079 | 0.480 | 0.029 |
| Sg | 0.585 | 0.109 | 0.472 | 0.041 |
| Sb | 0.532 | 0.052 | 0.451 | 0.020 |
| Total | 0.570 | 0.090 | 0.468 | 0.034 |
| Species | Root Number | Root Length | |||
|---|---|---|---|---|---|
| Precision | Recall | MAE (cm) | MAPE | RMSE (cm) | |
| Ob | 0.95 | 0.95 | 0.71 | 1.51% | 0.92 |
| Sg | 0.97 | 0.96 | 1.47 | 3.61% | 1.61 |
| Sb | 0.97 | 0.96 | 0.90 | 1.66% | 1.37 |
| Total | 0.97 | 0.96 | 1.06 | 2.38% | 1.35 |
| AVG | MAX | SD | |
|---|---|---|---|
| Image capture | 14.07 s | 23.00 s | 3.30 s |
| Image preprocessing | 13.50 s | 16.63 s | 1.36 s |
| 3D reconstruction | 1.93 min | 3.56 min | 0.46 min |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, C.; Huang, T.; Niu, Z.; Sun, X.; He, Y.; Qiu, Z. A Skeleton-Based Method of Root System 3D Reconstruction and Phenotypic Parameter Measurement from Multi-View Image Sequence. Agriculture 2025, 15, 343. https://doi.org/10.3390/agriculture15030343
Xu C, Huang T, Niu Z, Sun X, He Y, Qiu Z. A Skeleton-Based Method of Root System 3D Reconstruction and Phenotypic Parameter Measurement from Multi-View Image Sequence. Agriculture. 2025; 15(3):343. https://doi.org/10.3390/agriculture15030343
Chicago/Turabian StyleXu, Chengjia, Ting Huang, Ziang Niu, Xinyue Sun, Yong He, and Zhengjun Qiu. 2025. "A Skeleton-Based Method of Root System 3D Reconstruction and Phenotypic Parameter Measurement from Multi-View Image Sequence" Agriculture 15, no. 3: 343. https://doi.org/10.3390/agriculture15030343
APA StyleXu, C., Huang, T., Niu, Z., Sun, X., He, Y., & Qiu, Z. (2025). A Skeleton-Based Method of Root System 3D Reconstruction and Phenotypic Parameter Measurement from Multi-View Image Sequence. Agriculture, 15(3), 343. https://doi.org/10.3390/agriculture15030343








