Whole Genome Analysis of Proteus mirabilis in a Poultry Breeder Farm Reveals the Dissemination of blaNDM and blaCTX-M Mediated by Diverse Mobile Genetic Elements
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Identification of P. mirabilis
2.2. Antimicrobial Susceptibility Testing
2.3. Whole Genome Sequencing and Bioinformatics Analysis
2.4. Genetic Environment Analysis of blaCTX-M and blaNDM
3. Results
3.1. Isolation and Antibiotic Resistance of P. mirabilis
3.2. ARGs of P. mirabilis Isolates
3.3. Phylogeny Analysis of P. mirabilis Isolates
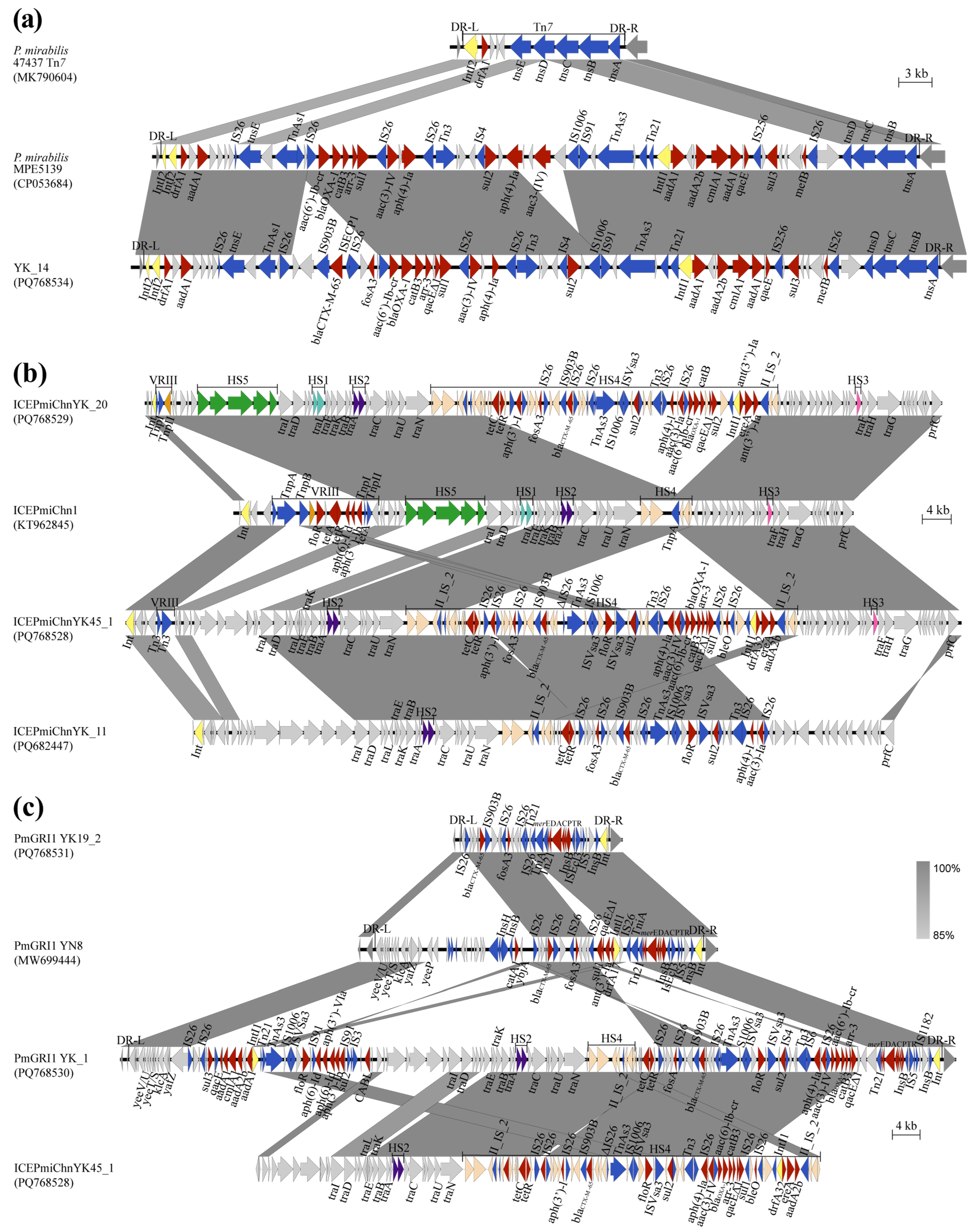
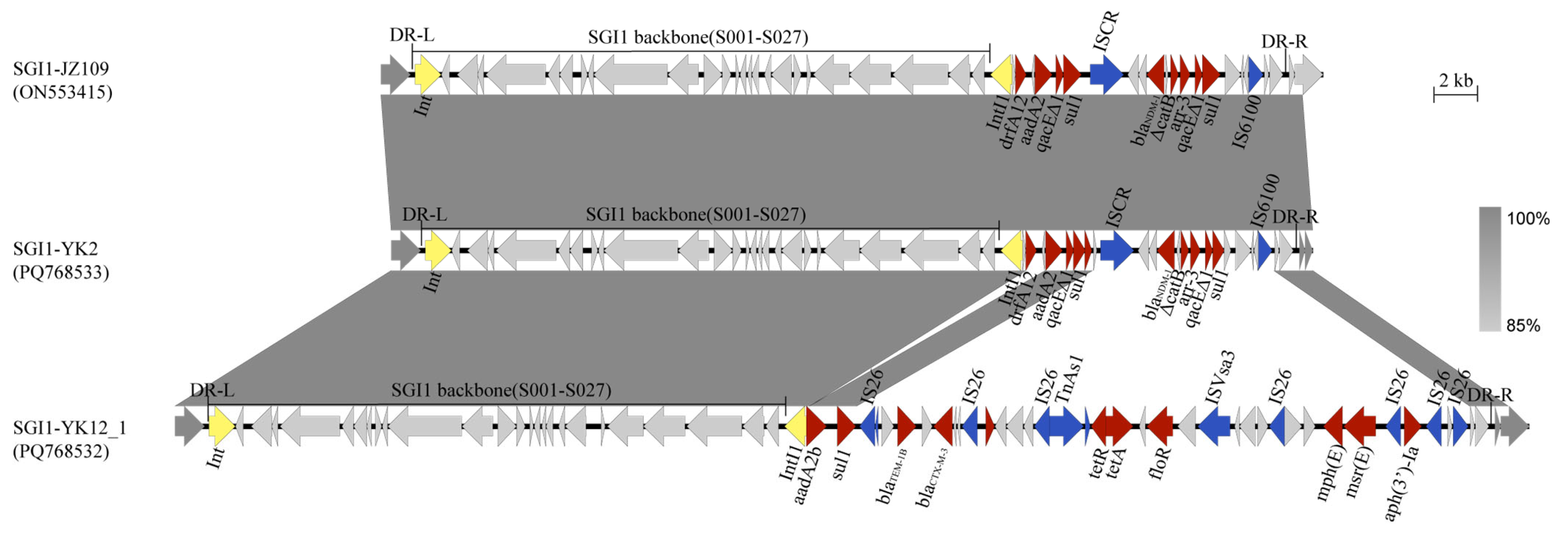
3.4. Genetic Environments of blaCTX-M and blaNDM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Dai, H.; Lu, B.; Li, Z.; Huang, Z.; Cai, H.; Yu, K.; Wang, D. Multilocus sequence analysis for the taxonomic updating and identification of the genus Proteus and reclassification of Proteus genospecies 5 O’Hara et al. 2000, Proteus cibarius Hyun et al. 2016 as later heterotypic synonyms of Proteus terrae Behrendt et. BMC Microbiol. 2020, 20, 152. [Google Scholar] [CrossRef] [PubMed]
- Girlich, D.; Dortet, L.; Poirel, L.; Nordmann, P. Integration of the blaNDM-1 carbapenemase gene into Proteus genomic island 1 (PGI1-PmPEL) in a Proteus mirabilis clinical isolate. J. Antimicrob. Chemother. 2015, 70, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Schaffer, J.N.; Pearson, M.M. Proteus mirabilis and Urinary Tract Infections. Microbiol. Spectr. 2015, 3, 383–433. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, S.; Stickler, D.J.; Mobley, H.L.; Shirtliff, M. Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin. Microbiol. Rev. 2008, 21, 26–59. [Google Scholar] [CrossRef]
- Armbruster, C.E.; Mobley, H.L.; Pearson, M.M. Pathogenesis of Proteus mirabilis infection. EcoSal Plus 2018, 8, 10–1128. [Google Scholar] [CrossRef]
- Wu, K.; Wang, J.; Yan, Z.; Zhu, Y.; Wang, S.; Fu, B.; Sun, C.; Li, R.; Fox, E.M.; Fanning, S.; et al. Genomic adaptation of Clostridium perfringens to human intestine. iMetaOmics 2024, 1, e38. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, C.; Zhang, R.; Chen, Y.; Shen, Y.; Hu, F.; Liu, D.; Lu, J.; Guo, Y.; Xia, X.; et al. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: An epidemiological comparative study. Lancet Infect. Dis. 2020, 20, 1161–1171. [Google Scholar] [CrossRef]
- Wu, K.; Zhang, Y.; Xu, W.; Lin, X.; Li, C.; Wang, J.; Li, R.; Tang, Y.; Lei, C.; Wang, H. Transmission of carbapenem-resistant enterobacterales producing NDM-5 during the broiler breeding process in China. Vet. Microbiol. 2024, 298, 110282. [Google Scholar] [CrossRef]
- Naghavi, M.; Vollset, S.E.; Ikuta, K.S.; Swetschinski, L.R.; Gray, A.P.; Wool, E.E.; Robles Aguilar, G.; Mestrovic, T.; Smith, G.; Han, C.; et al. Global burden of bacterial antimicrobial resistance 1990–2021: A systematic analysis with forecasts to 2050. Lancet 2024, 404, 1199–1226. [Google Scholar] [CrossRef]
- Wise, M.G.; Karlowsky, J.A.; Mohamed, N.; Hermsen, E.D.; Kamat, S.; Townsend, A.; Brink, A.; Soriano, A.; Paterson, D.L.; Moore, L.S.P.; et al. Global trends in carbapenem- and difficult-to-treat-resistance among World Health Organization priority bacterial pathogens: ATLAS surveillance program 2018–2022. J. Glob. Antimicrob. Resist. 2024, 37, 168–175. [Google Scholar] [CrossRef]
- Tseng, C.-H.; Liu, C.-W.; Liu, P.-Y. Extended-Spectrum β-Lactamases (ESBL) Producing Bacteria in Animals. Antibiotics 2023, 12, 661. [Google Scholar] [CrossRef] [PubMed]
- Codjoe, F.; Donkor, E. Carbapenem Resistance: A Review. Med. Sci. 2017, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Beltrão, E.M.B.; Oliveira, É.M.d.; Scavuzzi, A.M.L.; Firmo, E.F.; Lopes, A.C.d.S. Virulence factors of Proteus mirabilis clinical isolates carrying blaKPC-2 and blaNDM-1 and first report blaOXA-10 in Brazil. J. Infect. Chemother. 2022, 28, 363–372. [Google Scholar] [CrossRef] [PubMed]
- Kanzari, L.; Ferjani, S.; Saidani, M.; Hamzaoui, Z.; Jendoubi, A.; Harbaoui, S.; Ferjani, A.; Rehaiem, A.; Boutiba Ben Boubaker, I.; Slim, A. First report of extensively-drug-resistant Proteus mirabilis isolate carrying plasmid-mediated blaNDM-1 in a Tunisian intensive care unit. Int. J. Antimicrob. Agents 2018, 52, 906–909. [Google Scholar] [CrossRef]
- Girlich, D.; Dortet, L.; Poirel, L.; Nordmann, P. Integration of the blaNDM-1 carbapenemase gene into Salmonella genomic island 1 (SGI1-W) in a Proteus mirabilis clinical isolate. J. Antimicrob. Chemother. 2014, 70, 363–372. [Google Scholar]
- Kang, Q.; Wang, X.; Zhao, J.; Liu, Z.; Ji, F.; Chang, H.; Yang, J.; Hu, S.; Jia, T.; Wang, X. Multidrug-resistant Proteus mirabilis isolates carrying blaOXA-1 and blaNDM-1 from wildlife in China: Increasing public health risk. Integr. Zool. 2021, 16, 798–809. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, H.; Chen, G.; Dong, N. Isolation of four carbapenem-resistant gram-negative species from a single fly. Anim. Dis. 2024, 4, 4. [Google Scholar] [CrossRef]
- Florensa, A.F.; Kaas, R.S.; Clausen, P.T.L.C.; Aytan-Aktug, D.; Aarestrup, F.M. ResFinder–an open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb. Genom. 2022, 8, 000748. [Google Scholar] [CrossRef]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Li, Z.; Peng, C.; Zhang, G.; Shen, Y.; Zhang, Y.; Liu, C.; Liu, M.; Wang, F. Prevalence and characteristics of multidrug-resistant Proteus mirabilis from broiler farms in Shandong Province, China. Poult. Sci. 2022, 101, 101710. [Google Scholar] [CrossRef]
- Ramatla, T.; Ramaili, T.; Lekota, K.; Mileng, K.; Ndou, R.; Mphuthi, M.; Khasapane, N.; Syakalima, M.; Thekisoe, O. Antibiotic resistance and virulence profiles of Proteus mirabilis isolated from broiler chickens at abattoir in South Africa. Vet. Med. Sci. 2024, 10, e1371. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Shen, J.; Xu, Y.; Ding, P.; Gao, X.; Pan, Y.; Wu, H.; Hu, G.; He, D. Epidemic characteristics of the SXT/R391 integrated conjugative elements in multidrug-resistant Proteus mirabilis isolated from chicken farm. Poult. Sci. 2023, 102, 102640. [Google Scholar] [CrossRef] [PubMed]
- Lei, C.-W.; Zhang, A.-Y.; Liu, B.-H.; Wang, H.-N.; Guan, Z.-B.; Xu, C.-W.; Xia, Q.-Q.; Cheng, H.; Zhang, D.-D. Molecular Characteristics of Salmonella Genomic Island 1 in Proteus mirabilis Isolates from Poultry Farms in China. Antimicrob. Agents Chemother. 2014, 58, 7570–7572. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, H.; Wu, K.; Zhang, T.; Mou, Y.; Liu, L.; Wang, X.; Xu, W.; Chen, W.; Chen, X.; Wang, H.; et al. Whole Genome Analysis of Proteus mirabilis in a Poultry Breeder Farm Reveals the Dissemination of blaNDM and blaCTX-M Mediated by Diverse Mobile Genetic Elements. Agriculture 2025, 15, 555. https://doi.org/10.3390/agriculture15050555
Hu H, Wu K, Zhang T, Mou Y, Liu L, Wang X, Xu W, Chen W, Chen X, Wang H, et al. Whole Genome Analysis of Proteus mirabilis in a Poultry Breeder Farm Reveals the Dissemination of blaNDM and blaCTX-M Mediated by Diverse Mobile Genetic Elements. Agriculture. 2025; 15(5):555. https://doi.org/10.3390/agriculture15050555
Chicago/Turabian StyleHu, Haibin, Ke Wu, Tiejun Zhang, Yuhuan Mou, Luya Liu, Xiaoqin Wang, Wei Xu, Wenping Chen, Xiaojiao Chen, Hongning Wang, and et al. 2025. "Whole Genome Analysis of Proteus mirabilis in a Poultry Breeder Farm Reveals the Dissemination of blaNDM and blaCTX-M Mediated by Diverse Mobile Genetic Elements" Agriculture 15, no. 5: 555. https://doi.org/10.3390/agriculture15050555
APA StyleHu, H., Wu, K., Zhang, T., Mou, Y., Liu, L., Wang, X., Xu, W., Chen, W., Chen, X., Wang, H., & Lei, C. (2025). Whole Genome Analysis of Proteus mirabilis in a Poultry Breeder Farm Reveals the Dissemination of blaNDM and blaCTX-M Mediated by Diverse Mobile Genetic Elements. Agriculture, 15(5), 555. https://doi.org/10.3390/agriculture15050555






