Metagenomic Analysis of Seasonal Variations in Viral Dynamics and Diversity in Seawater of Jeju Island, Republic of Korea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation of Viral Community DNA
2.3. Data Analysis
2.3.1. Sequence Quality Control
2.3.2. De Novo Sequence Data Analysis
3. Results
3.1. Sampling Site and Environmental Characteristics
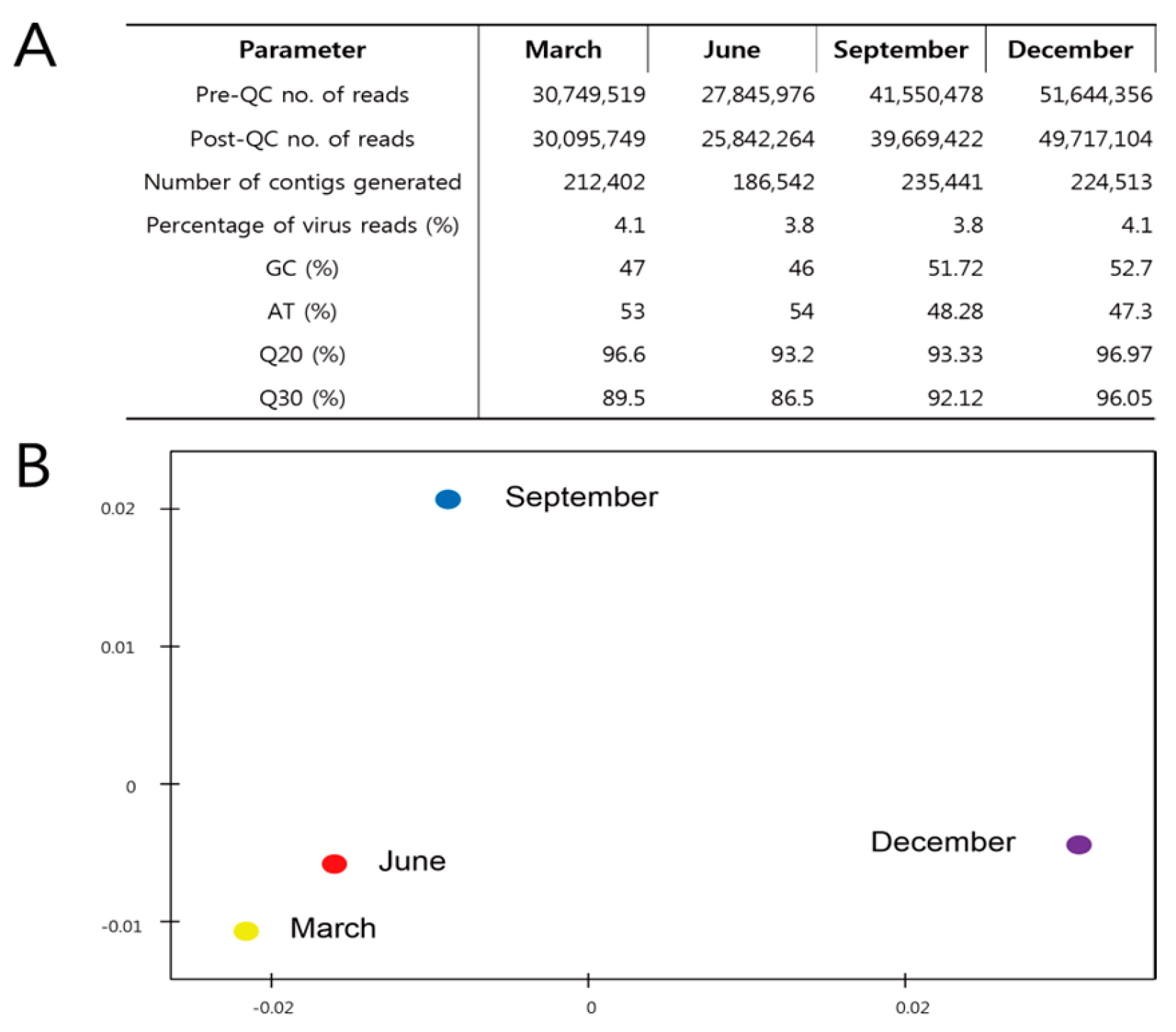
3.2. Metagenome Comparisons
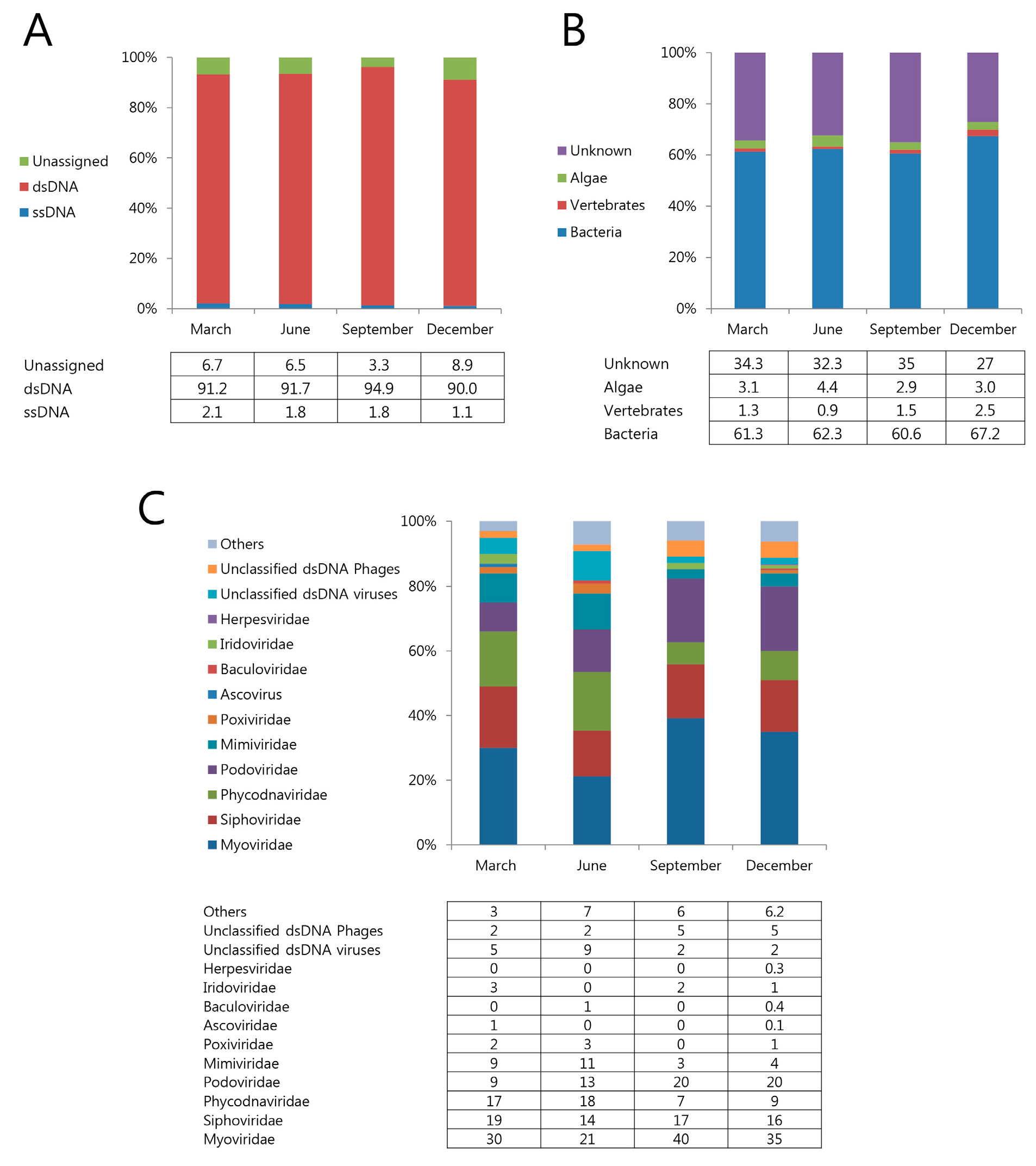
3.3. Marine Viral Community Diversity and Composition
3.4. Seasonal Change of Marine Viruses
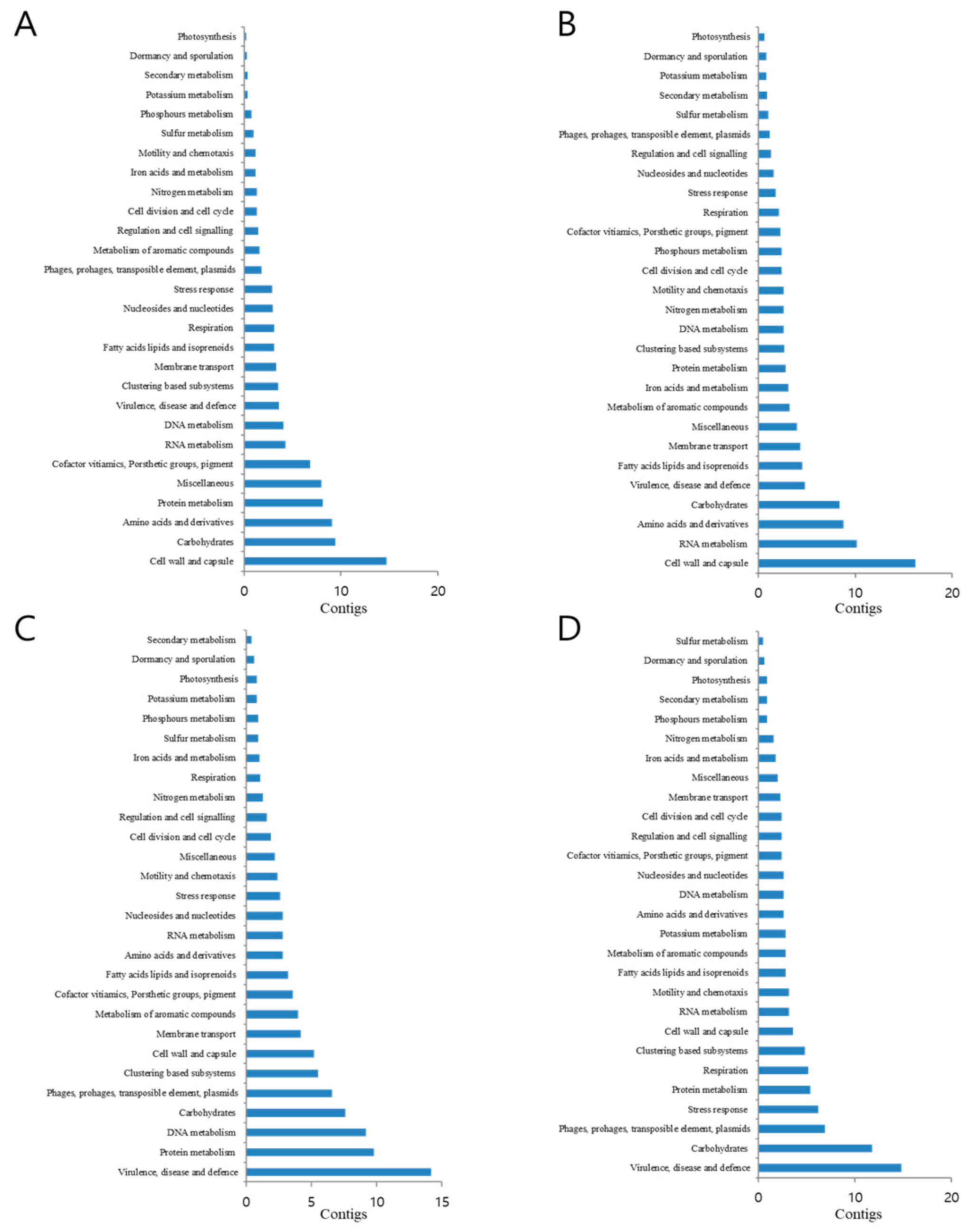
3.5. Functional Classification of Identified Metagenomic Contigs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Welsh, J.E.; Steenhuis, P.; de Moraes, K.R.; van der Meer, J.; Thieltges, D.W.; Brussaard, C.P. Marine virus predation by non-host organisms. Sci. Rep. 2020, 10, 5221. [Google Scholar] [CrossRef]
- Wommack, K.E.; Colwell, R.R. Virioplankton: Viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 2000, 64, 69–114. [Google Scholar] [CrossRef] [PubMed]
- Parada, V.; Sintes, E.; van Aken, H.M.; Weinbauer, M.G.; Herndl, G.J. Viral abundance, decay, and diversity in the meso-and bathypelagic waters of the North Atlantic. Appl. Environ. Microbiol. 2007, 73, 4429–4438. [Google Scholar] [CrossRef]
- Mushegian, A. Are there 1031 virus particles on earth, or more, or fewer? J. Bacteriol. 2020, 202, e00052-20. [Google Scholar] [CrossRef]
- Biggs, T.E.; Huisman, J.; Brussaard, C.P. Viral lysis modifies seasonal phytoplankton dynamics and carbon flow in the Southern Ocean. ISME J. 2021, 15, 3615–3622. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.-C.; Fuhrman, J.A. Effects of phytoplankton, viral communities, and warming on free-living and particle-associated marine prokaryotic community structure. Nat. Commun. 2022, 13, 7905. [Google Scholar] [CrossRef]
- Middelboe, M.; Jorgensen, N.; Kroer, N. Effects of viruses on nutrient turnover and growth efficiency of noninfected marine bacterioplankton. Appl. Environ. Microbiol. 1996, 62, 1991–1997. [Google Scholar] [CrossRef] [PubMed]
- Middelboe, M.; Lyck, P.G. Regeneration of dissolved organic matter by viral lysis in marine microbial communities. Aquat. Microb. Ecol. 2002, 27, 187–194. [Google Scholar] [CrossRef]
- Handelsman, J. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 2004, 68, 669–685. [Google Scholar] [CrossRef]
- Jin, H.; Tian, L.; Bing, W.; Zhao, J.; Ren, L. Bioinspired marine antifouling coatings: Status, prospects, and future. Prog. Mater. Sci. 2022, 124, 100889. [Google Scholar] [CrossRef]
- Sanseverino, I.; Pretto, P.; António, D.C.; Lahm, A.; Facca, C.; Loos, R.; Skejo, H.; Beghi, A.; Pandolfi, F.; Genoni, P. Metagenomics analysis to investigate the microbial communities and their functional profile during cyanobacterial blooms in Lake Varese. Microb. Ecol. 2022, 83, 850–868. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M. Marine viruses: Truth or dare. Mar. Sci. 2012, 4, 425–428. [Google Scholar] [CrossRef]
- Kristensen, D.M.; Mushegian, A.R.; Dolja, V.V.; Koonin, E.V. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2010, 18, 11–19. [Google Scholar] [CrossRef]
- John, S.G.; Mendez, C.B.; Deng, L.; Poulos, B.; Kauffman, A.K.M.; Kern, S.; Brum, J.; Polz, M.F.; Boyle, E.A.; Sullivan, M.B. A simple and efficient method for concentration of ocean viruses by chemical flocculation. Environ. Microbiol. Rep. 2011, 3, 195–202. [Google Scholar] [CrossRef]
- Zheng, Y.; Gao, Z.; Wu, S.; Ruan, A. Community Structure, Drivers, and Potential Functions of Different Lifestyle Viruses in Chaohu Lake. Viruses 2024, 16, 590. [Google Scholar] [CrossRef]
- Martínez Martínez, J.; Martinez-Hernandez, F.; Martinez-Garcia, M. Single-virus genomics and beyond. Nat. Rev. Microbiol. 2020, 18, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Kieft, K.; Adams, A.; Salamzade, R.; Kalan, L.; Anantharaman, K. vRhyme enables binning of viral genomes from metagenomes. Nucleic Acids Res. 2022, 50, e83. [Google Scholar] [CrossRef]
- Schulz, F.; Abergel, C.; Woyke, T. Giant virus biology and diversity in the era of genome-resolved metagenomics. Nat. Rev. Microbiol. 2022, 20, 721–736. [Google Scholar] [CrossRef]
- Kim, M.-S.; Whon, T.W.; Bae, J.-W. Comparative viral metagenomics of environmental samples from Korea. Genom. Inform. 2013, 11, 121. [Google Scholar] [CrossRef]
- Deghorain, M.; Van Melderen, L. The Staphylococci phages family: An overview. Viruses 2012, 4, 3316–3335. [Google Scholar] [CrossRef] [PubMed]
- van Vliet, D.M.; von Meijenfeldt, F.B.; Dutilh, B.E.; Villanueva, L.; Sinninghe Damsté, J.S.; Stams, A.J.; Sánchez-Andrea, I. The bacterial sulfur cycle in expanding dysoxic and euxinic marine waters. Environ. Microbiol. 2021, 23, 2834–2857. [Google Scholar] [CrossRef]
- Tripp, H.J.; Kitner, J.B.; Schwalbach, M.S.; Dacey, J.W.; Wilhelm, L.J.; Giovannoni, S.J. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 2008, 452, 741–744. [Google Scholar] [CrossRef] [PubMed]
- Davison, A.J.; Trus, B.L.; Cheng, N.; Steven, A.C.; Watson, M.S.; Cunningham, C.; Le Deuff, R.-M.; Renault, T. A novel class of herpesvirus with bivalve hosts. J. Gen. Virol. 2005, 86, 41–53. [Google Scholar] [CrossRef]
- Davison, A.J. Herpesvirus systematics. Vet. Microbiol. 2010, 143, 52–69. [Google Scholar] [CrossRef]
- Hyatt, A.; Gould, A.; Zupanovic, Z.; Cunningham, A.; Hengstberger, S.; Whittington, R.; Kattenbelt, J.; Coupar, B. Comparative studies of piscine and amphibian iridoviruses. Arch. Virol. 2000, 145, 301–331. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.-Y.; Jia, K.-T.; Yang, B.; Huang, J. Complete genome sequence of a Megalocytivirus (family Iridoviridae) associated with turbot mortality in China. Virol. J. 2010, 7, 159. [Google Scholar] [CrossRef]
- He, J.; Zeng, K.; Weng, S.; Chan, S.-M. Experimental transmission, pathogenicity and physical–chemical properties of infectious spleen and kidney necrosis virus (ISKNV). Aquaculture 2002, 204, 11–24. [Google Scholar] [CrossRef]
- Hicks, B.D.; Worthy, G.A. Sealpox in captive grey seals (Halichoerus grypus) and their handlers. J. Wildl. Dis. 1987, 23, 1–6. [Google Scholar] [CrossRef]
- Simpson, V.; Stuart, N.; Stack, M.; Ross, H.; Head, J. Parapox infection in grey seals (Halichoerus grypus) in Cornwall. Vet. Rec. 1994, 134, 292–296. [Google Scholar] [CrossRef]
- Koonin, E.V.; Yutin, N. Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology 2010, 53, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Reavy, B.; Swanson, M.M.; Cock, P.J.; Dawson, L.; Freitag, T.E.; Singh, B.K.; Torrance, L.; Mushegian, A.R.; Taliansky, M. Distinct circular single-stranded DNA viruses exist in different soil types. Appl. Environ. Microbiol. 2015, 81, 3934–3945. [Google Scholar] [CrossRef] [PubMed]
- Cashdollar, J.; Wymer, L. Methods for primary concentration of viruses from water samples: A review and meta-analysis of recent studies. J. Appl. Microbiol. 2013, 115, 1–11. [Google Scholar] [CrossRef]
- Borrego, J.; Morinigo, M.; Martinez-Manzanares, E.; Bosca, M.; Castro, D.; Barja, J.; Toranzo, A.E. Plasmid associated virulence properties of environmental isolates of Aeromonas hydrophila. J. Med. Microbiol. 1991, 35, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Katayama, H.; Shimasaki, A.; Ohgaki, S. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl. Environ. Microbiol. 2002, 68, 1033–1039. [Google Scholar] [CrossRef]
- Kamata, S.-I.; Suzuki, S. Concentration of marine birnavirus from seawater with a glass fiber filter precoated with bovine serum albumin. Mar. Biotechnol. 2003, 5, 157–162. [Google Scholar] [CrossRef]
- Colombet, J.; Robin, A.; Lavie, L.; Bettarel, Y.; Cauchie, H.; Sime-Ngando, T. Virioplankton ‘pegylation’: Use of PEG (polyethylene glycol) to concentrate and purify viruses in pelagic ecosystems. J. Microbiol. Methods 2007, 71, 212–219. [Google Scholar] [CrossRef]

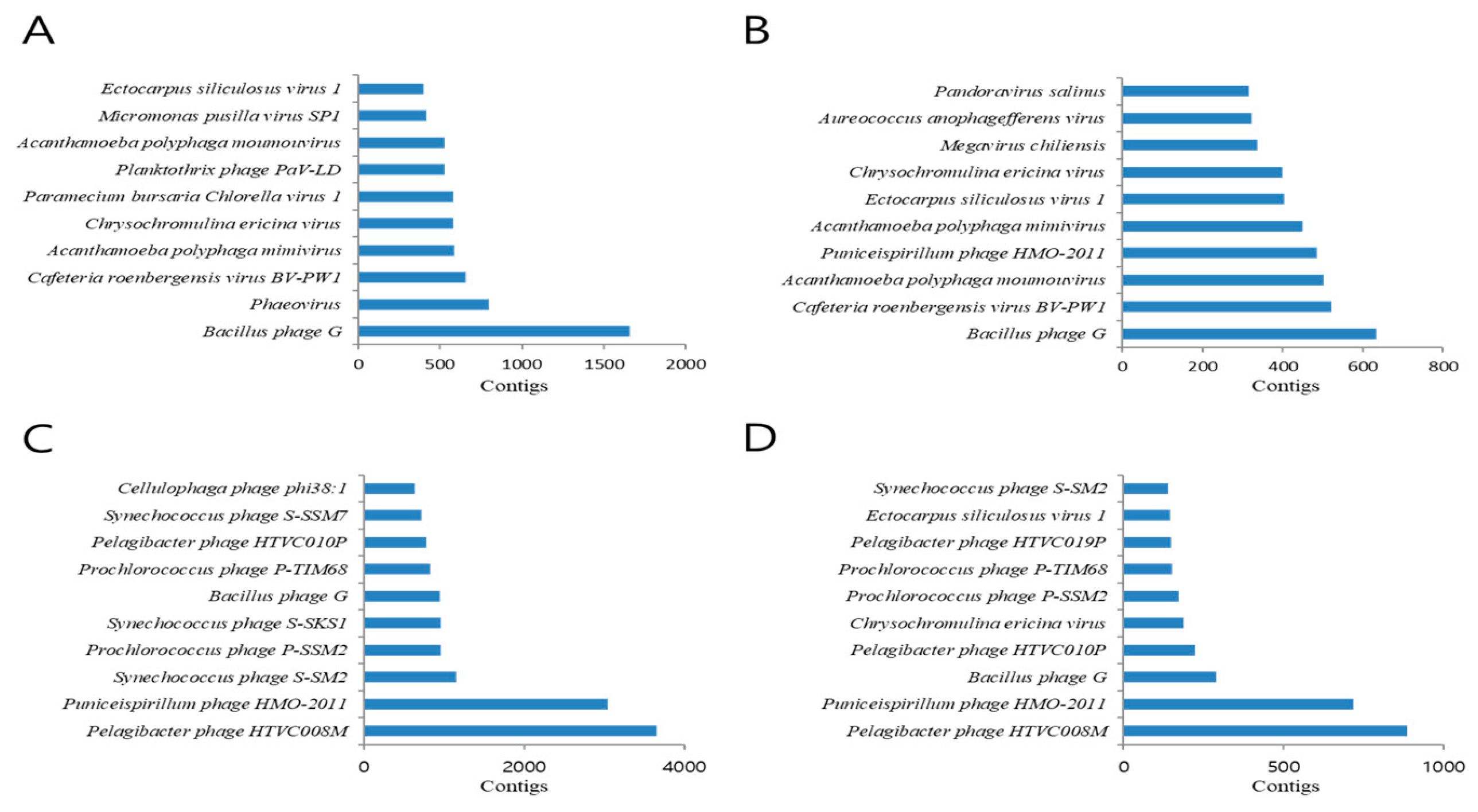
| March | June | September | December | |||||
|---|---|---|---|---|---|---|---|---|
| Index | Contigs | Virus/Phage | Contigs | Virus/Phage | Contigs | Virus/Phage | Contigs | Virus/Phage |
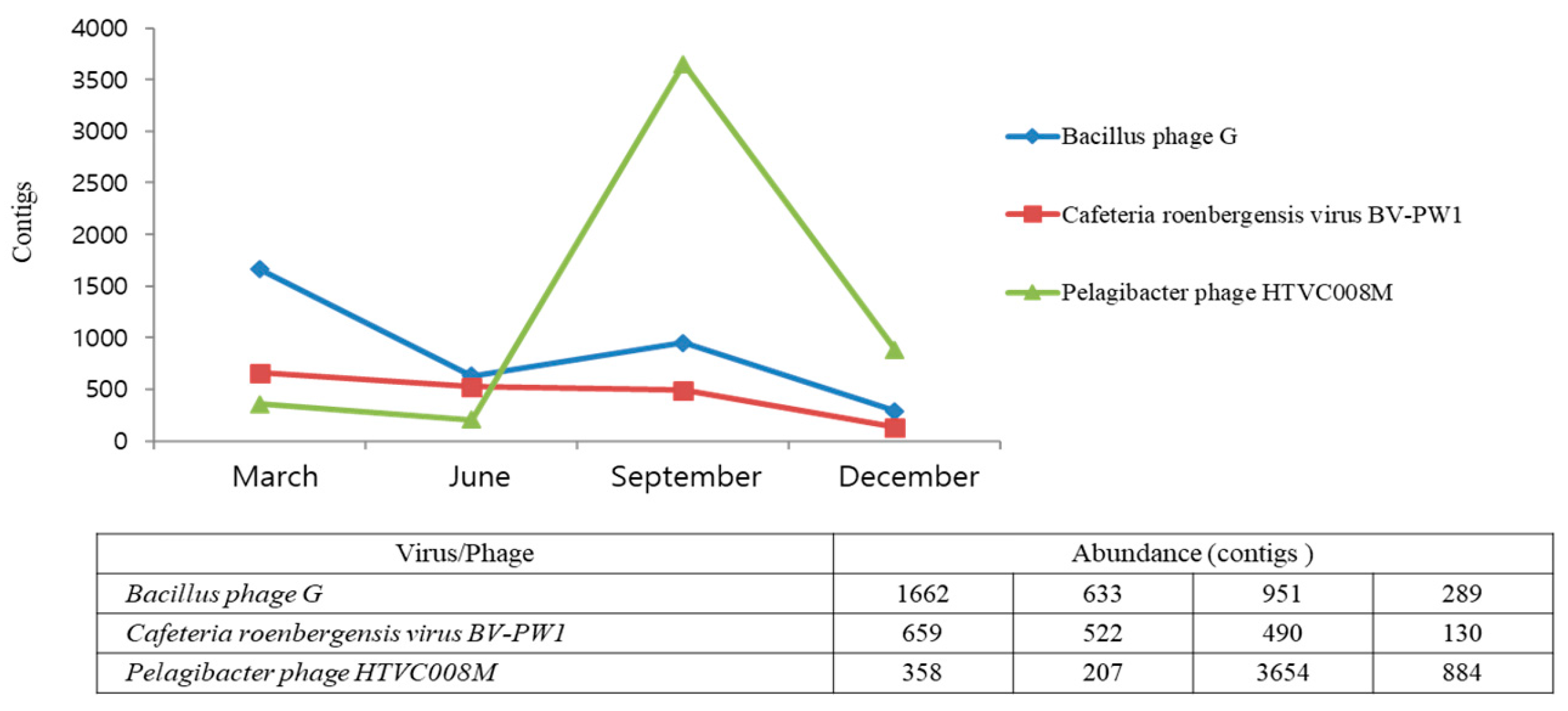
| 1 | 1662 | Bacillus phage G | 633 | Bacillus phage G | 3654 | Pelagibacter phage HTVC008M | 884 | Pelagibacter phage HTVC008M |
| 2 | 798 | Phaeovirus | 522 | Cafeteria roenbergensis virus BV-PW1 | 3047 | Puniceispirillum phage HMO-2011 | 717 | Puniceispirillum phage HMO-2011 |
| 3 | 659 | Cafeteria roenbergensis virus BV-PW1 | 502 | Acanthamoeba polyphaga moumouvirus | 1153 | Synechococcus phage S-SM2 | 289 | Bacillus phage G |
| 4 | 586 | Acanthamoeba polyphaga mimivirus | 486 | Puniceispirillum phage HMO-2011 | 964 | Prochlorococcus phage P-SSM2 | 222 | Pelagibacter phage HTVC010P |
| 5 | 580 | Chrysochromulina ericina virus | 449 | Acanthamoeba polyphaga mimivirus | 954 | Synechococcus phage S-SKS1 | 187 | Chrysochromulina ericina virus |
| 6 | 580 | Paramecium bursaria Chlorella virus 1 | 404 | Ectocarpus siliculosus virus 1 | 951 | Bacillus phage G | 173 | Prochlorococcus phage P-SSM2 |
| 7 | 526 | Planktothrix phage PaV-LD | 398 | Chrysochromulina ericina virus | 823 | Prochlorococcus phage P-TIM68 | 152 | Prochlorococcus phage P-TIM68 |
| 8 | 526 | Acanthamoeba polyphaga moumouvirus | 336 | Megavirus chiliensis | 782 | Pelagibacter phage HTVC010P | 149 | Pelagibacter phage HTVC019P |
| 9 | 419 | Micromonas pusilla virus SP1 | 323 | Aureococcus anophagefferens virus | 716 | Synechococcus phage S-SSM7 | 145 | Ectocarpus siliculosus virus 1 |
| 10 | 401 | Ectocarpus siliculosus virus 1 | 314 | Pandoravirus salinus | 637 | Cellulophaga phage phi38:1 | 141 | Synechococcus phage S-SM2 |
| 11 | 368 | Paramecium bursaria Chlorella virus AR158 | 286 | Pandoravirus inopinatum | 618 | Chrysochromulina ericina virus | 137 | Cellulophaga phage phi38:1 |
| 12 | 361 | Puniceispirillum phage HMO-2011 | 279 | Pandoravirus dulcis | 590 | Synechococcus phage S-CBS2 | 135 | Synechococcus phage S-SKS1 |
| 13 | 358 | Trichoplusia ni ascovirus 2c | 259 | Planktothrix phage PaV-LD | 506 | Cyanophage KBS-S-2A | 130 | Cafeteria roenbergensis virus BV-PW1 |
| 14 | 358 | Pelagibacter phage HTVC008M | 227 | Phaeocystis globosa virus | 493 | Caulobacter phage Cr30 | 115 | Phaeocystis globosa virus |
| 15 | 356 | Burkholderia phage phi1026b | 221 | Feldmannia species virus | 490 | Cafeteria roenbergensis virus BV-PW1 | 105 | Caulobacter phage Cr30 |
| 16 | 335 | Pandoravirus inopinatum | 207 | Pelagibacter phage HTVC008M | 489 | Cronobacter phage vB_CsaM_GAP32 | 103 | Acanthamoeba polyphaga moumouvirus |
| 17 | 308 | Bathycoccus sp. RCC1105 virus BpV | 206 | Pithovirus sibericum | 487 | Pelagibacter phage HTVC019P | 97 | Cyanophage KBS-S-2A |
| 18 | 307 | Pandoravirus dulcis | 193 | Paramecium bursaria Chlorella virus AR158 | 473 | Phaeocystis globosa virus | 93 | Planktothrix phage PaV-LD |
| 19 | 293 | Phaeocystis globosa virus | 178 | Bathycoccus sp. RCC1105 virus BpV | 443 | Synechococcus phage ACG-2014f | 93 | Pelagibacter phage HTVC011P |
| 20 | 280 | Chlorovirus | 169 | Megavirus lba | 437 | Rhodothermus phage RM378 | 93 | Bathycoccus sp. RCC1105 virus BpV1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, J.; Oh, E.G.; Jin, Y. Metagenomic Analysis of Seasonal Variations in Viral Dynamics and Diversity in Seawater of Jeju Island, Republic of Korea. J. Mar. Sci. Eng. 2024, 12, 1480. https://doi.org/10.3390/jmse12091480
Hwang J, Oh EG, Jin Y. Metagenomic Analysis of Seasonal Variations in Viral Dynamics and Diversity in Seawater of Jeju Island, Republic of Korea. Journal of Marine Science and Engineering. 2024; 12(9):1480. https://doi.org/10.3390/jmse12091480
Chicago/Turabian StyleHwang, Jinik, Eun Gyoung Oh, and Youngguk Jin. 2024. "Metagenomic Analysis of Seasonal Variations in Viral Dynamics and Diversity in Seawater of Jeju Island, Republic of Korea" Journal of Marine Science and Engineering 12, no. 9: 1480. https://doi.org/10.3390/jmse12091480
APA StyleHwang, J., Oh, E. G., & Jin, Y. (2024). Metagenomic Analysis of Seasonal Variations in Viral Dynamics and Diversity in Seawater of Jeju Island, Republic of Korea. Journal of Marine Science and Engineering, 12(9), 1480. https://doi.org/10.3390/jmse12091480




