Aquaporin 12 Is Expressed in the Stomach and Liver of the Spiny Dogfish (Squalus acanthias)
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Polymerase Chain Reactions (PCRs) and DNA Cloning
2.3. Western Blotting and Immunohistochemistry
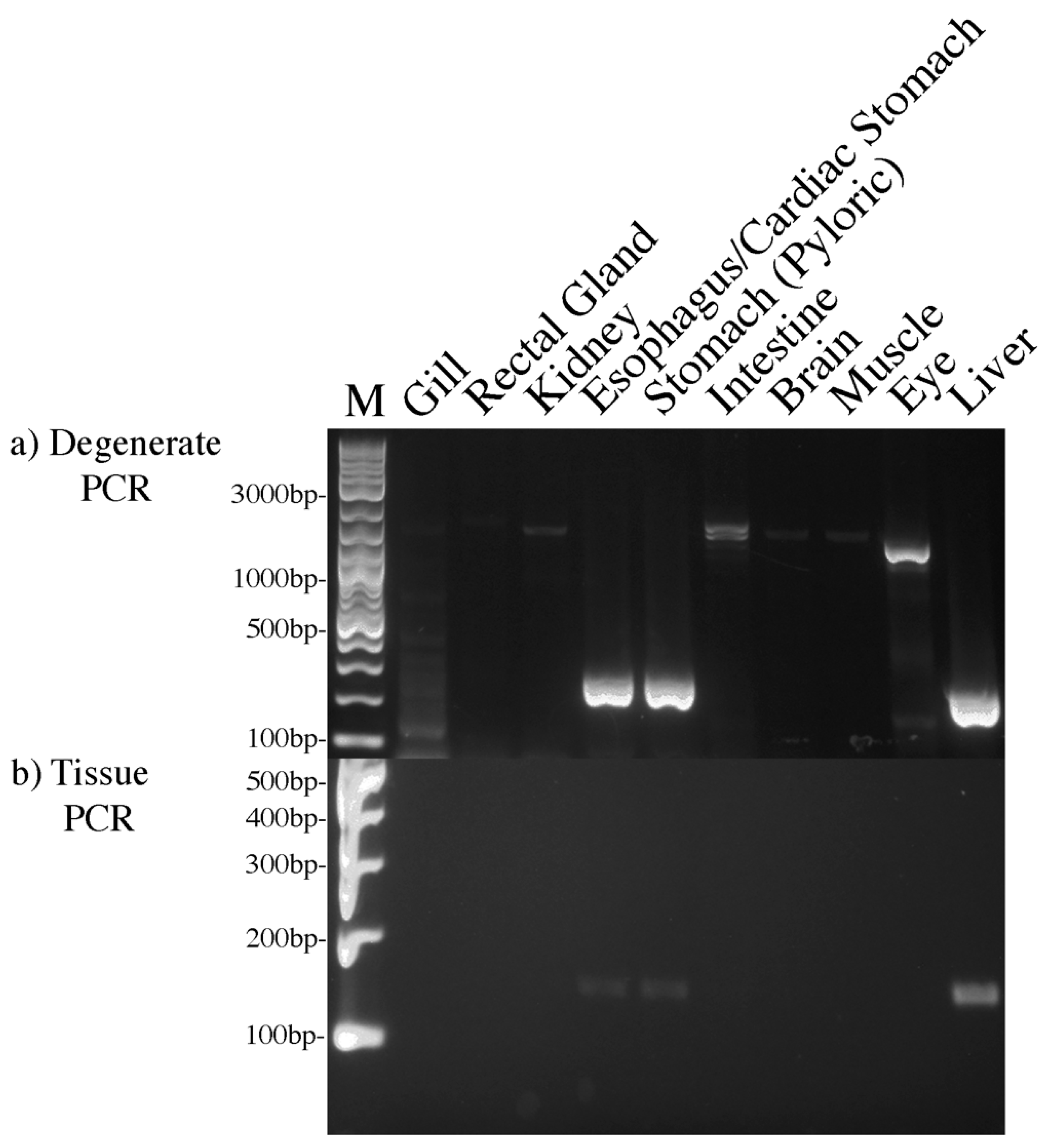
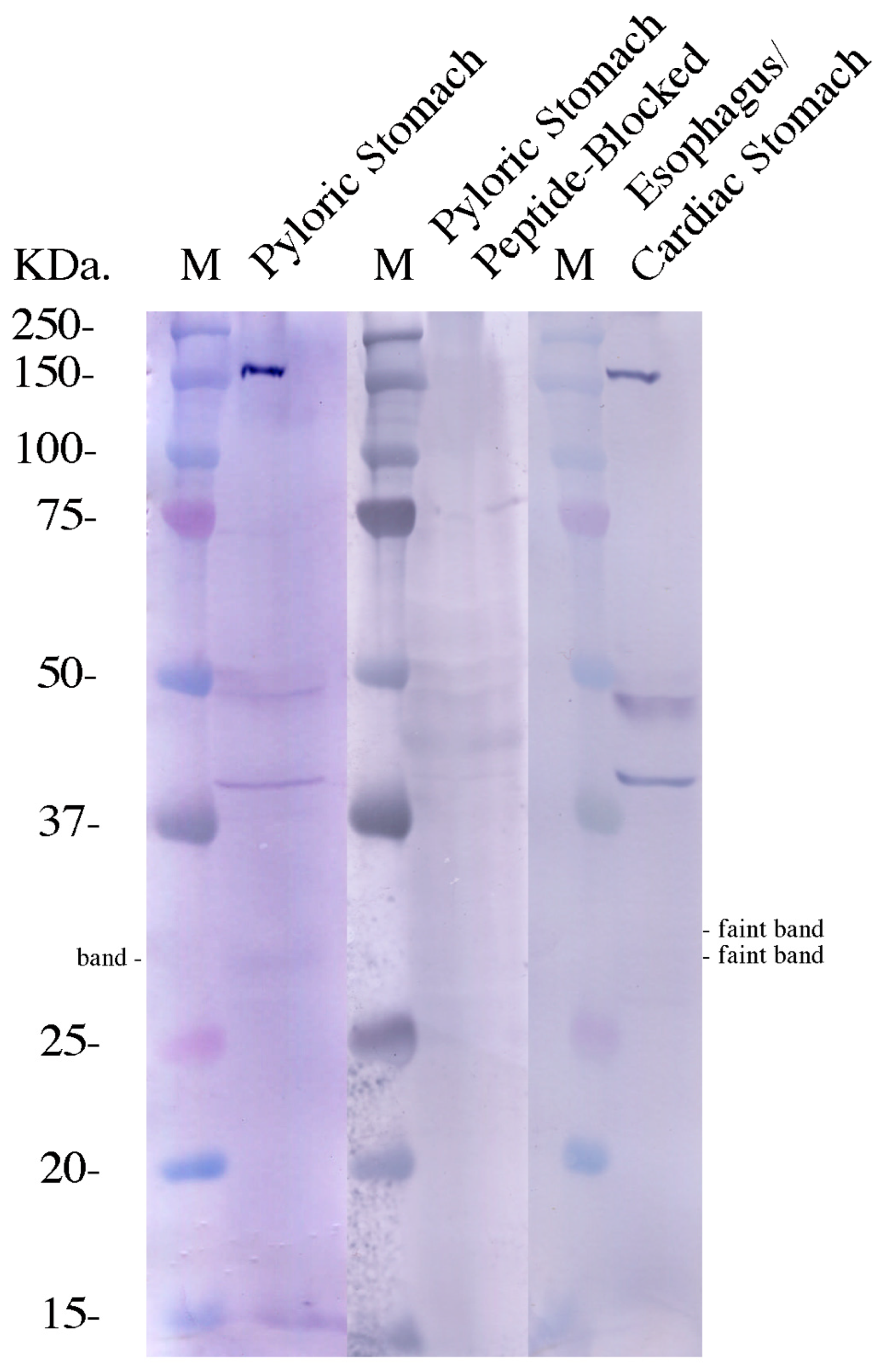
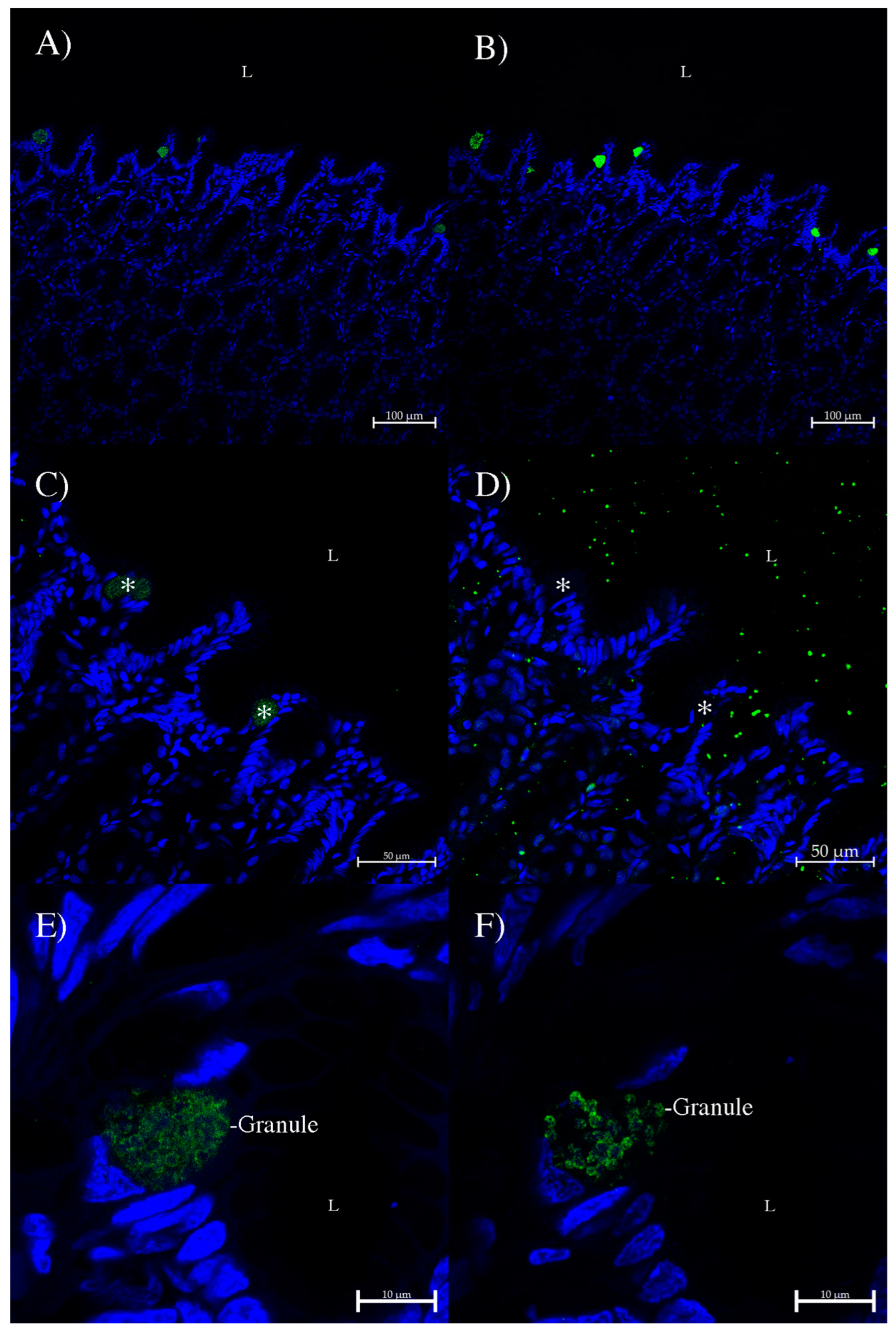
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AQP | Aquaporin |
| cDNA | Complementary DNA |
| RACE | Rapid Amplification of cDNA Ends |
| PCR | Polymerase Chain Reaction |
| Elas | Elasmobranch |
| dNTP | De-oxy Nucleotide Tri-phosphates |
| Taq | Thermus aquatus |
| ER | Endoplasmic Reticulum |
| mRNA | Messenger RNA |
| MDIBL | Mount Desert Island Biological Laboratory |
| DNA | De-oxyribo Nucleic Acid |
| IUPAC | International Union of Pure and Applied Chemistry |
| RNA | Ribo Nucleic Acid |
| KLH | Keyhole Limpet Hemocyanin |
References
- Ishibashi, K.; Hara, S.; Kondo, S. Aquaporin water channels in mammals. Clin. Exp. Nephrol. 2009, 13, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.N.; Chauvigné, F.; Hlidberg, J.B.; Cutler, C.P.; Cerdà, J. The lineage-specific evolution of aquaporin gene clusters facilitated tetrapod terrestrial adaptation. PLoS ONE 2014, 9, e113686. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Kuwahara, M.; Kageyama, Y.; Sasaki, S.; Suzuki, M.; Imai, M. Molecular cloning of a new aquaporin superfamily in mammals. In Molecular Biology and Physiology of Water and Solute Transport; Hohmann, S., Neilsen, S., Eds.; Kluwer Academic/Plenum Publishers: New York, NY, USA, 2000; pp. 123–126. [Google Scholar]
- Morishita, Y.; Sakube, Y.; Sasaki, S.; Ishibashi, K. Molecular mechanisms and drug development in aquaporin water channel diseases: Aquaporin superfamily (superaquaporins): Expansion of aquaporins restricted to multicellular organisms. J. Pharmacol. Sci. 2004, 96, 276–279. [Google Scholar] [CrossRef]
- Itoh, T.; Rai, T.; Kuwahara, M.; Ko, S.B.; Uchida, S.; Sasaki, S.; Ishibashi, K. Identification of a novel aquaporin, AQP12, expressed in pancreatic acinar cells. Biochem. Biophys. Res. Commun. 2005, 330, 832–838. [Google Scholar] [CrossRef]
- Ishibashi, K. Aquaporin subfamily with unusual NPA boxes. Biochim. Biophys. Acta 2006, 1758, 989–993. [Google Scholar] [CrossRef]
- Eisler, K.; Dropmann, L.M.; Bugert, P.; Ewers, M.; Witt, H. Genetic analysis of the aquaporin water channels AQP12A and AQP12B in patients with chronic pancreatitis. Pancreatology 2022, 22, 1079–1083. [Google Scholar] [CrossRef] [PubMed]
- Ohta, E.; Itoh, T.; Nemoto, T.; Kumagai, J.; Ko, S.B.; Ishibashi, K.; Ohno, M.; Uchida, K.; Ohta, A.; Sohara, E.; et al. Pancreas-specific aquaporin 12 null mice showed increased susceptibility to caerulein-induced acute pancreatitis. Am. J. Physiol. 2009, 297, C1368–C1378. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Tanaka, Y.; Morishita, Y. The role of mammalian superaquaporins inside the cell: An update. Biochim. Biophys. Acta 2021, 1863, 183617. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Kondo, S.; Hara, S.; Morishita, Y. The evolutionary aspects of aquaporin family. Am. J. Physiol. 2011, 300, R566–R576. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Kondo, S.; Hara, S.; Morishita, Y. The role of mammalian superaquaporins inside the cell. Biochim. Biophys. Acta 2014, 1840, 1507–1512. [Google Scholar] [CrossRef]
- Ishibashi, K.; Tanaka, Y.; Morishita, Y. Chapter One-Perspectives on the evolution of aquaporin superfamily. Vitam. Horm. 2020, 112, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Adeoye, A.; Odugbemi, A.; Ajewole, T. Structure and function of aquaporins: The membrane water channel protein. Biointerface Res. Appl. Chem. 2022, 12, 690–705. [Google Scholar] [CrossRef]
- Markou, A.; Ungera, L.; Abir-Awan, M.; Saadallah, A.; Halsey, A.; Balklava, Z.; Conner, M.; T¨ornroth-Horsefield, S.; Greenhill, S.D.; Conner, A.; et al. Molecular mechanisms governing aquaporin relocalisation. Biochim. Biophys. Acta 2022, 1864, 183853. [Google Scholar] [CrossRef] [PubMed]
- Isokpehi, R.D.; Rajnarayanan, R.V.; Jeffries, C.D.; Oyeleye, T.O.; Cohly, H.H.P. Integrative sequence and tissue expression profiling of chicken and mammalian aquaporins. BMC Genom. 2009, 10 (Suppl. 2), S7. [Google Scholar] [CrossRef]
- Tingaud-Sequeira, A.; Calusinska, M.; Finn, R.N.; Chauvigné, F.; Lozano, J.; Cerda, J. The zebrafish genome encodes the largest vertebrate repertoire of functional aquaporins with dual paralogy and substrate specificities similar to mammals. BMC Evol. Biol. 2010, 10, 38. [Google Scholar] [CrossRef]
- Da Silva, I.V.; Cardoso, C.; Méndez-Giménez, L.; Camoes, S.P.; Frühbeck, G.; Rodríguez, A.J.; Miranda, P.; Soveral, G. Aquaporin-7 and aquaporin-12 modulate the inflammatory phenotype of endocrine pancreatic beta-cells. Arch. Biochem. Biophys. 2020, 691, 108481. [Google Scholar] [CrossRef] [PubMed]
- Cutler, C.P.; Sanders, I.L.; Hazon, N.; Cramb, G. Primary sequence, tissue specificity and expression of the Na+, K+-ATPase β1 subunit in the European eel (Anguilla anguilla). Fish Physiol. Biochem. 1995, 14, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Cutler, C.P.; Sanders, I.L.; Cramb, G. Expression of Na+, K+-ATPase subunit isoforms in the European eel (Anguilla anguilla). Fish Physiol. Biochem. 1997, 17, 371–376. [Google Scholar] [CrossRef]
- Cutler, C.P.; Brezillon, S.; Bekir, S.; Sanders, I.L.; Hazon, N.; Cramb, G. Expression of a duplicate Na+, K+-ATPase β1 isoform in the European eel (Anguilla anguilla). Am. J. Physiol. 2000, 279, R222–R229. [Google Scholar] [CrossRef] [PubMed]
- Cutler, C.P.; Cramb, G. Branchial expression of an aquaporin 3 (AQP-3) homologue is downregulated in the European eel Anguilla anguilla following seawater acclimation. J. Exp. Biol. 2002, 205, 2643–2651. [Google Scholar] [CrossRef] [PubMed]
- Cutler, C.P.; Cramb, G. Two isoforms of the Na+/K+/2Cl− cotransporter (NKCC1) are expressed in the European eel (Anguilla anguilla). Biochim. Biophys. Acta 2002, 1566, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.-S.; Cutler, C.P.; Wilson, G.; Phillips, C.; Hazon, N.; Cramb, G. Regulation of expression of two aquaporin homologues in the intestine of the European eel: Effects of seawater acclimation and cortisol treatment. Am. J. Physiol. 2005, 288, R1733–R1743. [Google Scholar]
- Cutler, C.P.; Cramb, G. Differential expression of absorptive cation-chloride cotransporters in the intestinal and renal tissues of the European eel (Anguilla anguilla). Comp. Biochem. Physiol. 2008, 149, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Cutler, C.P.; MacIver, B.; Cramb, G.; Zeidel, M.L. Aquaporin 4 is a ubiquitously expressed isoform in the dogfish (Squalus acanthias) shark. Front. Physiol. 2012, 2, 107. [Google Scholar] [CrossRef] [PubMed]
- Cutler, C.P.; Murray, D.; Ojo, T.; Harmon, S.; MacIver, B.; Cramb, G.; Zeidel, M.L. Aquaporin (AQP) channels in the spiny dogfish, Squalus acanthias I: Characterization of AQP3 and AQP15, and localization of their proteins in the gill and spiral valve intestine. Comp. Biochem. Physiol. 2022, 258, 110702. [Google Scholar] [CrossRef]
- Cutler, C.P.; Mainer, S.; Ojo, T. The aquaporin 8 (AQP8) membrane channel gene is present in the elasmobranch dogfish (Squalus acanthias) genome and is expressed in brain but not in gill, kidney or intestine. Comp. Biochem. Physiol. 2022, 260, 110730. [Google Scholar] [CrossRef] [PubMed]
- Cutler, C.P.; Canicatti, M.E.; Omoregie, E. Evidence that Aquaporin 11 (AQP11) in the spiny dogfish (Squalus acanthias) may represent a pseudogene. Int. J. Mol. Sci. 2024, 25, 2028. [Google Scholar] [CrossRef] [PubMed]
- Cutler, C.P.; Ojo, T. Immunohistochemical localization and expression of aquaporin 3-2 (AQP3C1) in the spiny dogfish, Squalus acanthias. Hydrobiology 2024, 3, 378–391. [Google Scholar] [CrossRef]
- Cutler, C.P.; Omoregie, E.; Ojo, T. UT-1 transporter expression in the spiny dogfish (Squalus acanthias): UT-1 protein shows a different localization in comparison to that of other sharks. Biomolecules 2024, 14, 1151. [Google Scholar] [CrossRef]
- Cutler, C.P.; Harmon, S.; Walsh, J.; Burch, K. Characterization of aquaporin 4 protein expression and localization in tissues of the dogfish (Squalus acanthias). Front. Physiol. 2012, 3, 21. [Google Scholar] [CrossRef] [PubMed]
- Cutler, C.P.; Kurt, K.; Campbell, K.E.; Ojo, T. Aquaporin (AQP) channels in the spiny dogfish, Squalus acanthias II: Localization of AQP3, AQP4 and AQP15 in the kidney. Comp. Biochem. Physiol. 2022, 258, 110701. [Google Scholar] [CrossRef] [PubMed]

| Degenerate PCR (expected product 305 bp):- I* = deoxy-inosine IUPAC Codes: Y = C/T R = A/G H = A/C/T | |
| Elas AQP12 Sense 2 | ACI* GAY YTI* CAY YTI* ATH CAR AAY ATG ATG GC |
| Elas AQP12 Anti 2 | GG RCT I*AR CCA RTA I*AC I*AI* CAT RTA YTC |
| 5′ (5R) and 3′ (3R) RACE | |
| Squal AQP12 5R 1 | CAGCTACGTG CACCAGGGCA GT |
| Squal AQP12 5R 2 | AGGGCTTTGG ACAGGATCCT GTATCCA |
| Squal AQP12 3R 1 | CCTAGTTGTG ATGAAGTTTG AGGGAGCAGC T |
| Squal AQP12 3R 2 | GATACAGGAT CCTGTCCAAA GCCCTAACCA |
| Tissue PCR (expected product 152 bp) | |
| AQP12 sense | GGTGCACGTA GCTGGCCCAT AT |
| AQP12 anti | ACAGAAAGAT GGCCAGAATT GTTGCAATGA ATGG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cutler, C.P.; Bender, J.; Conner, S.; Omoregie, E. Aquaporin 12 Is Expressed in the Stomach and Liver of the Spiny Dogfish (Squalus acanthias). J. Mar. Sci. Eng. 2025, 13, 161. https://doi.org/10.3390/jmse13010161
Cutler CP, Bender J, Conner S, Omoregie E. Aquaporin 12 Is Expressed in the Stomach and Liver of the Spiny Dogfish (Squalus acanthias). Journal of Marine Science and Engineering. 2025; 13(1):161. https://doi.org/10.3390/jmse13010161
Chicago/Turabian StyleCutler, Christopher P., Jade Bender, Sarah Conner, and Esosa Omoregie. 2025. "Aquaporin 12 Is Expressed in the Stomach and Liver of the Spiny Dogfish (Squalus acanthias)" Journal of Marine Science and Engineering 13, no. 1: 161. https://doi.org/10.3390/jmse13010161
APA StyleCutler, C. P., Bender, J., Conner, S., & Omoregie, E. (2025). Aquaporin 12 Is Expressed in the Stomach and Liver of the Spiny Dogfish (Squalus acanthias). Journal of Marine Science and Engineering, 13(1), 161. https://doi.org/10.3390/jmse13010161







