An Attentive Multi-Modal CNN for Brain Tumor Radiogenomic Classification
Abstract
:1. Introduction
- We propose an attentive multi-modal DNN to predict the status of the MGMT promoter methylation. In addition to a multi-modal feature aggregation strategy, our proposed model integrates three performance boosters, including a lite attention mechanism to control the model size and speed up training, a separable embedding module to improve the feature representation of MRI data, and a modal-wise shortcut strategy to ensure the modal specificity. These joint efforts have improved the detection accuracy of our model by 3%, compared to the SOTA method. Experiments and results are obtained on the RSNA-MICCAI 2021 dataset [9], which is a recently released dataset with the most patients and MRI scans compared to existing datasets.
- We have made the project source code publicly available at https://github.com/ruyiq/An-Attentive-Multi-modal-CNN-for-Brain-Tumor-Radiogenomic-Classification (accessed at 26 February 2020), offering a credible benchmark for future studies.
2. Related Work
2.1. Detection of MGMT Methylation Status Based on MRI Data
2.2. Multi-Modal Learning on MRI Data
3. Materials and Methods
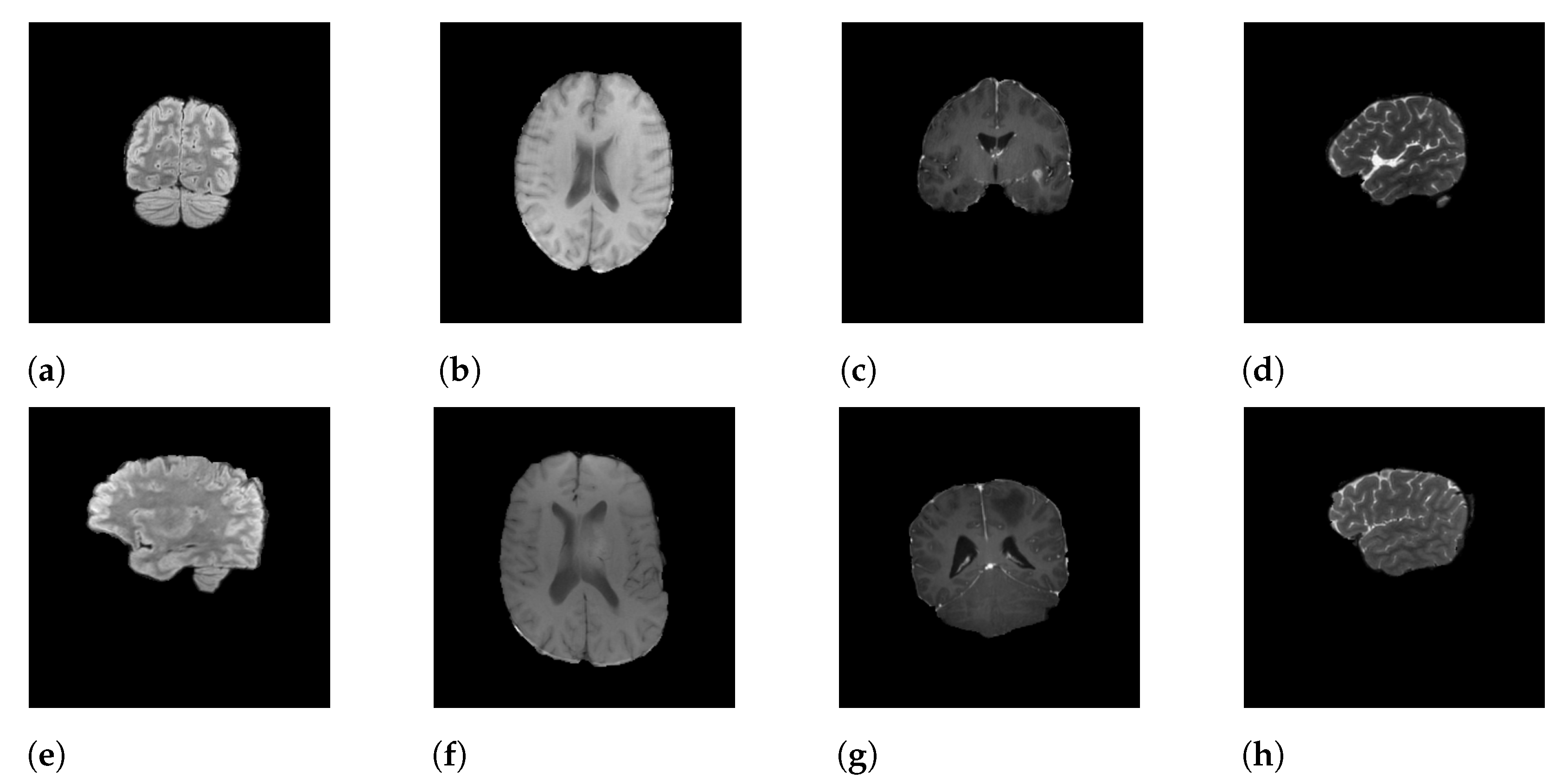
3.1. Dataset
3.2. Learning Framework
3.3. Multi-Modal Feature Fusion
3.4. Lite Attention Mechanism
- Fuse multi-modal data in the form of sequences and use recurrent neural network (RNN) models for feature extraction. This operation requires traversing the input from the first time-step to the last one, which is computationally expensive [25]. Even though improved RNN variants such as LSTM [26] and GRU [27] can effectively reduce the difficulty of parameter updates in training, the sequential arrangement of different modal data introduces unnecessary sequential priors, which can force the model to learn an unreasonable one-way information flow while understanding the inter-modal relationships to fit the main features, affecting the effectiveness of feature extraction [28,29].
- Use the attention mechanism to fuse the features extracted from different modalities. Attention can easily obtain global feature information compared to the sequential models such as LSTM and GRU mentioned above, which can better obtain contextual relationships and obtain an overall understanding of the input.
3.5. Modal-Wise Shortcut
3.6. Separable Embedding
3.7. LSTM and Detection Head
4. Experiments and Results
4.1. Evaluation Metrics
4.2. Baseline
- ResNet by He et al. [4] is an effort to understand how deepening a neural network can increase the expressiveness and the complexity of the network. It is found that for DNN, if a newly added layer can be treated as an identity function, the deepened network is as effective as the original one. This finding drives the development of the residual block, which adds a shortcut connection to the layer output before the activation function. The simple design allows a DNN to be trained more easily and efficiently. ResNet was the winning solution for the ImageNet Large-Scale Visual Recognition Challenge in 2015 and has been applied to numerous computer vision tasks with SOTA performance. Therefore, we consider ResNet a decent baseline. Our empirical result shows that ResNet34 presents the highest accuracy. We thus use ResNet34 to represent the baseline result.
- The EfficientNet [36] paper makes two major contributions. First, a simple and mobile-size neural architecture was proposed. Second, a compound-scaling method was proposed to increase the network size to achieve maximum performance gains. It is suggested that to pursue better performance, the key is to balance all three dimensions, including network depth, width, and resolution, during ConvNet scaling. Thus, the authors of EfficientNet adopted a global scaling factor to uniformly scale the depth, width, and resolution of the network. The scaling factor makes it possible to apply grid searching to find the parameters that lead to the best performance. EfficientNet offers a generic neural architecture optimization technique applied to existing CNNs such as ResNet. It has shown superior performance in numerous tasks with SOTA results, which is why we chose it as a strong baseline.
- The gold-medal-winning strategy was developed by Firas Baba, who open-sourced the code at https://github.com/FirasBaba/rsna-resnet10 (accessed at 24 January 2020). The final model of the winning team is a 3D CNN using the ResNet10 backbone with the following design choices: BCE Loss, Adam optimizer, 15 epochs, a learning rate of 0.00001 (from epoch 1 to 10) and 0.000005 (from epoch 10–15), image size 256 by 256, batch size 8. Each epoch took around 80 s on an RTX 3090. The author also reported the best central image trick, which is a strategy to select the biggest MRI scan that contains the largest brain cutaway view for training. In this study, we refer to the model developed by Firas Baba as the SOTA since it was in first place on the contest leader board.
4.3. Training Setting
- Horizontal flip with a probability of 0.5;
- Random affine transformation configured as shift-limit = 0.0625, scale_limit = 0.1, rotate_limit = 10 with a probability of 0.5;
- Random contrast transformation with 0.5 probability.
4.4. Performance Evaluation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Patil, N.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2013–2017. Neuro-Oncology 2020, 22, iv1–iv96. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Ruan, S.; Canu, S. A review: Deep learning for medical image segmentation using multi-modality fusion. Array 2019, 3–4, 100004. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; Volume 30, pp. 770–778. [Google Scholar]
- Le, N.Q.K.; Do, D.T.; Chiu, F.Y.; Yapp, E.K.Y.; Yeh, H.Y.; Chen, C.Y. XGBoost Improves Classification of MGMT Promoter Methylation Status in IDH1 Wildtype Glioblastoma. J. Pers. Med. 2020, 10, 128. [Google Scholar] [CrossRef]
- Korfiatis, P.; Kline, T.L.; Lachance, D.H.; Parney, I.F.; Buckner, J.C.; Erickson, B.J. Residual Deep Convolutional Neural Network Predicts MGMT Methylation Status. J. Digit. Imaging 2017, 30, 622–628. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.C.; Bai, H.; Sun, Q.; Li, Q.; Liu, L.; Zou, Y.; Chen, Y.; Liang, C.; Zheng, H. Multiregional radiomics features from multiparametric MRI for prediction of MGMT methylation status in glioblastoma multiforme: A multicentre study. Eur. Radiol. 2018, 28, 3640–3650. [Google Scholar] [CrossRef]
- Han, L.; Kamdar, M.R. MRI to MGMT: Predicting methylation status in glioblastoma patients using convolutional recurrent neural networks. In Pacific symposium on Biocomputing 2018, Proceedings of the Pacific Symposium, Coast, HI, USA, 3–7 January 2018; World Scientific: Singapore, 2018; pp. 331–342. [Google Scholar]
- Baid, U.; Ghodasara, S.; Mohan, S.; Bilello, M.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.C.; Pati, S.; et al. The rsna-asnr-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv 2021, arXiv:2107.02314. [Google Scholar]
- Korfiatis, P.; Kline, T.L.; Coufalova, L.; Lachance, D.H.; Parney, I.F.; Carter, R.E.; Buckner, J.C.; Erickson, B.J. MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med. Phys. 2016, 43, 2835–2844. [Google Scholar] [CrossRef]
- Kanas, V.G.; Zacharaki, E.I.; Thomas, G.A.; Zinn, P.O.; Megalooikonomou, V.; Colen, R.R. Learning MRI-based classification models for MGMT methylation status prediction in glioblastoma. Comput. Methods Programs Biomed. 2017, 140, 249–257. [Google Scholar] [CrossRef]
- Chen, X.; Zeng, M.; Tong, Y.; Zhang, T.; Fu, Y.; Li, H.; Zhang, Z.; Cheng, Z.; Xu, X.; Yang, R.; et al. Automatic Prediction of MGMT Status in Glioblastoma via Deep Learning-Based MR Image Analysis. Biomed Res. Int. 2020, 2020, 9258649. [Google Scholar] [CrossRef]
- Yogananda, C.; Shah, B.R.; Nalawade, S.; Murugesan, G.; Yu, F.; Pinho, M.; Wagner, B.; Mickey, B.; Patel, T.R.; Fei, B.; et al. MRI-based deep-learning method for determining glioma MGMT promoter methylation status. Am. J. Neuroradiol. 2021, 42, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Du, C.; Xue, Z.; Chen, X.; Zhao, H.; Huang, L. What Makes Multi-modal Learning Better than Single (Provably). Adv. Neural Inf. Process. Syst. 2021, 34. [Google Scholar]
- Myronenko, A. 3D MRI Brain Tumor Segmentation Using Autoencoder Regularization. In Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention Workshop(MICCAI), Shenzhen, China, 13–17 October 2019; pp. 311–320. [Google Scholar] [CrossRef]
- Tseng, K.L.; Lin, Y.L.; Hsu, W.; Huang, C.Y. Joint Sequence Learning and Cross-Modality Convolution for 3D Biomedical Segmentation. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 311–320. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.; Lu, J.; Cai, J.; Cham, T.J.; Wang, G. Large-margin multi-modal deep learning for RGB-D object recognition. IEEE Trans. Multimed. 2015, 17, 1887–1898. [Google Scholar] [CrossRef]
- Liu, W.; Luo, Z.; Cai, Y.; Yu, Y.; Ke, Y.; Junior, J.M.; Gonçalves, W.N.; Li, J. Adversarial unsupervised domain adaptation for 3D semantic segmentation with multi-modal learning. ISPRS J. Photogramm. Remote Sens. 2021, 176, 211–221. [Google Scholar] [CrossRef]
- Wang, Z.; She, Q.; Smolic, A. TEAM-Net: Multi-modal Learning for Video Action Recognition with Partial Decoding. arXiv 2021, arXiv:2110.08814. [Google Scholar]
- Ning, Z.; Xiao, Q.; Feng, Q.; Chen, W.; Zhang, Y. Relation-induced multi-modal shared representation learning for Alzheimer’s disease diagnosis. IEEE Trans. Med. Imaging 2021, 40, 1632–1645. [Google Scholar] [CrossRef]
- Rani, G.; Oza, M.G.; Dhaka, V.S.; Pradhan, N.; Verma, S.; Rodrigues, J.J. Applying deep learning-based multi-modal for detection of coronavirus. Multimed. Syst. 2021, 1–12. [Google Scholar] [CrossRef]
- Shachor, Y.; Greenspan, H.; Goldberger, J. A mixture of views network with applications to multi-view medical imaging. IEEE Trans. Med. Imaging 2020, 374, 1–9. [Google Scholar] [CrossRef]
- Nie, D.; Wang, L.; Gao, Y.; Shen, D. Fully convolutional networks for multi-modality isointense infant brain image segmentation. In Proceedings of the 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), Prague, Czech Republic, 13–16 April 2016; pp. 1342–1345. [Google Scholar] [CrossRef] [Green Version]
- Kamnitsas, K.; Ledig, C.; Newcombe, V.F.; Simpson, J.P.; Kane, A.D.; Menon, D.K.; Rueckert, D.; Glocker, B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med. Image Anal. 2017, 36, 61–78. [Google Scholar] [CrossRef]
- Cho, K.; Merrienboer, B.V.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning Phrase Representations using RNN Encoder-Decoder for Statistical Machine Translationn. Comput. Sci. Comput. Lang. 2014, 36, 61–78. [Google Scholar] [CrossRef]
- Sainath, T.N.; Vinyals, O.; Senior, A.; Sak, H. Convolutional, Long Short-Term Memory, fully connected Deep Neural Networks. In Proceedings of the 2015 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), South Brisbane, QLD, Australia, 19–24 April 2015. [Google Scholar] [CrossRef]
- Zaremba, W.; Sutskever, I.; Vinyals, O. Recurrent Neural Network Regularization. Neural Evol. Comput. 2014, arXiv:1409.2329. [Google Scholar]
- Bahdanau, D.; Cho, K.; Bengio, Y. Neural Machine Translation by Jointly Learning to Align and Translate. Comput. Sci. Comput. Lang. 2014, arXiv:1409.0473. [Google Scholar]
- Cho, K.; van Merrienboer, B.; Bahdanau, D.; Bengio, Y. On the properties of neural machine translation: Encoder-decoder approaches. Comput. Sci. Comput. Lang. 2014, arXiv:1409.1259. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is All you Need. In Proceedings of the Conference on Neural Information Processing Systems (NeurIPS), Red Hook, NY, USA, 4–9 December 2017; Volume 30, pp. 1–12. [Google Scholar]
- Devlin, J.; Chang, M.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Lan, Z.; Chen, M.; Goodman, S.; Gimpel, K.; Sharma, P.; Soricut, R. ALBERT: A Lite BERT for Self-supervised Learning of Language Representations. Neural Evol. Comput. 2019, arXiv:1909.11942. [Google Scholar]
- Zhang, Z.; Hanand, X.; Liu, Z.; Jiang, X.; Sun, M.; Liu, Q. ERNIE: Enhanced Language Representation with Informative Entities. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics, Florence, Italy, 28 July–2 August 2019. [Google Scholar]
- Liu, Y.; Ott, M.; Goyal, N.; Du, J.; Joshi, M.; Chen, D.; Levy, O.; Lewis, M.; Zettlemoyer, L.; Stoyanov, V. RoBERTa: A Robustly Optimized BERT Pretraining Approach. arXiv 2019, arXiv:1907.11692. [Google Scholar]
- Sainath, R.Z.C.T.; Parada, C. Feature Learning with Raw-Waveform CLDNNs for Voice Activity Detection; Interspeech: Baixas, France, 2016. [Google Scholar]
- Tan, M.; Le, Q.V. Efficientnet: Rethinking model scaling for convolutional neural networks. arXiv 2019, arXiv:1905.11946. [Google Scholar]
- Dietterich, T. Overfitting and undercomputing in machine learning. Acm Comput. Surv. (CSUR) 1995, 27, 326–327. [Google Scholar] [CrossRef]




| Reference | Year | Model | D.S. | M.M. | A.M. | M.W.S. | S.E. |
|---|---|---|---|---|---|---|---|
| [10] | 2016 | SVM, RF | 155 | ✗ | ✗ | ✗ | ✗ |
| [11] | 2017 | KNN, RF, J48, NB | 86 | ✗ | ✗ | ✗ | ✗ |
| [6] | 2017 | ResNet | 155 | ✓ | ✗ | ✗ | ✗ |
| [7] | 2018 | CNN+RF | 133 | ✓ | ✗ | ✗ | ✗ |
| [8] | 2018 | CRNN | 262 | ✓ | ✗ | ✗ | ✗ |
| [5] | 2020 | XGBoost | 53 | ✓ | ✗ | ✗ | ✗ |
| [12] | 2020 | Custom CNN | 153 | ✗ | ✗ | ✗ | ✗ |
| [13] | 2021 | MGMT-Net | 247 | ✗ | ✗ | ✗ | ✗ |
| Our work | 2022 | Custom DNN | 585 | ✓ | ✓ | ✓ | ✓ |
| Scan Type | FLAIR | T1w | T1wCE | T2w |
|---|---|---|---|---|
| # files | 74,248 | 77,627 | 96,766 | 100,000 |
| Avg. # files per case | 127 | 133 | 165 | 171 |
| Method | S.E | Acc | T.D.P.E (s) |
|---|---|---|---|
| ResNet34 | NA | 53.12% | 65.4 |
| EfficientNet | NA | 54.80% | 52.3 |
| 3D CNN + ResNet10 (SOTA) | NA | 60.74% | 73.1 |
| multi-modality + attention | NA | 56.74% | 67.3 |
| multi-modality + attention | EfficientNet | 59.03% | 72.2 |
| multi-modality + attention | Resnet34 | 61.09% | 78.8 |
| multi-modality + attention + shortcut | Resnet34 | 63.71% | 79.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, R.; Xiao, Z. An Attentive Multi-Modal CNN for Brain Tumor Radiogenomic Classification. Information 2022, 13, 124. https://doi.org/10.3390/info13030124
Qu R, Xiao Z. An Attentive Multi-Modal CNN for Brain Tumor Radiogenomic Classification. Information. 2022; 13(3):124. https://doi.org/10.3390/info13030124
Chicago/Turabian StyleQu, Ruyi, and Zhifeng Xiao. 2022. "An Attentive Multi-Modal CNN for Brain Tumor Radiogenomic Classification" Information 13, no. 3: 124. https://doi.org/10.3390/info13030124
APA StyleQu, R., & Xiao, Z. (2022). An Attentive Multi-Modal CNN for Brain Tumor Radiogenomic Classification. Information, 13(3), 124. https://doi.org/10.3390/info13030124







