Automatic Text Summarization of Biomedical Text Data: A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Selection
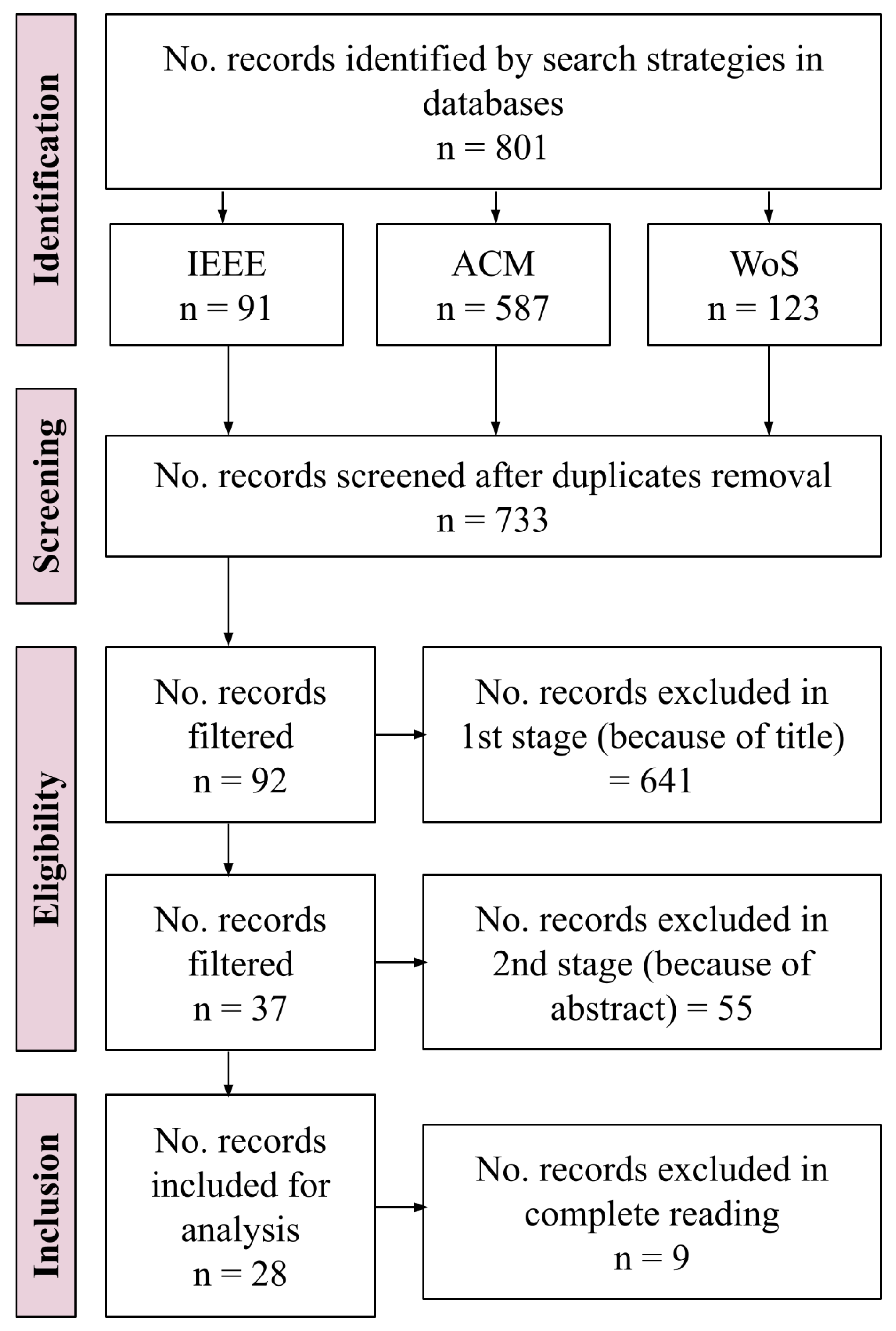
2.1.1. Identification
2.1.2. Screening
2.1.3. Eligibility
- Studies or summarization tools that describe the evaluation component (metric) and method(s) used.
- Related Natural Language Processing techniques that can be used as text summarization methods (e.g., text mining, text generation).
2.1.4. Included
2.2. Summarization Factors
2.2.1. Input
2.2.2. Purpose
2.2.3. Output
2.2.4. Method
2.2.5. Evaluation Metrics
3. Results
3.1. Study Frequency According Geographical Distribution, Years, and Type of Publication
3.2. Study Frequency according to Summarization Factors
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Resource | Type of Input Text | Description |
|---|---|---|
| PubMed Central (PMC) | Biomedical Literature | More than 7 million full-text records of biomedical and life sciences journal literature at the U.S. National Institutes of Health’s National Library of Medicine (NIH/NLM). Open access [74] |
| CRAFT: The Colorado Richly Annotated Full Text Corpus | Biomedical Literature | It is a manually annotated corpus consisting of 67 full-text biomedical journal articles. Each article is a member of the PMC subset. Open access [75,76] |
| BioASQ Task-6a | Biomedical Literature | Contains 13 million citations from PubMed dataset, and each citation contains the title and abstract. Open access [77] |
| PubMed | Biomedical Literature | Contains more than 34 million citations and abstracts supporting the search and retrieval of biomedical and life sciences literature. Open access [78] |
| BioMed Central (BMC) | Biomedical Literature | 300 peer-reviewed journals in science, technology, engineering, and medicine. Open access [79] |
| MEDLINE | Biomedical Literature | This database contains more than 29 million references to journal articles in life sciences with a concentration on biomedicine. The records are indexed with NLM Medical Subject Headings (MeSH). Open access [80,81] |
| MEDIQA-AnS | Biomedical Literature | The dataset includes 156 questions with related documents as the answers for each. Each answer also has an extractive and an abstractive single-answer summaries and multidocument extractive and abstractive summary considering the information presented in all of the answers. [82] |
| CORD-19: The Covid-19 Open Research Dataset | Biomedical Literature | It is a resource of scientific papers on COVID-19 and related historical coronavirus research. Open access [83] |
| Radiology Reports | EHR | 41,066 real-world radiology reports from MedStar Georgetown University Hospital. Each report describes clinical findings about a specific diagnostic case, and an impression summary [40] |
| DIAC-WoZ dataset | EHR | Clinical interviews designed to support the diagnosis of psychological distress conditions created by the Institute for Creative Technologies at the University of Southern California. Open access [84,85] |
| NTUH-iMD | EHR | The corpus contains 258,050 discharge diagnoses obtained from the National Taiwan University Hospital Integrated Medical Database and the highlighted extractive summaries written by experienced doctors [47] |
| Clinical trials | EHR | Dataset generation of 101,016 records usable for the summarization task from clinical trials. Open access [86] |
References
- Aggarwal, C.C.; Zhai, C. An Introduction to Text Mining. In Mining Text Data; Aggarwal, C.C., Zhai, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–10. [Google Scholar] [CrossRef]
- Davidoff, F.; Miglus, J. Delivering clinical evidence where it’s needed: Building an information system worthy of the profession. JAMA 2011, 305, 1906–1907. [Google Scholar] [CrossRef] [PubMed]
- Smith, R. Strategies for coping with information overload. BMJ 2010, 341, c7126. [Google Scholar] [CrossRef] [PubMed]
- Nadif, M.; Role, F. Unsupervised and self-supervised deep learning approaches for biomedical text mining. Briefings Bioinform. 2021, 22, 1592–1603. [Google Scholar] [CrossRef] [PubMed]
- Dash, S.; Acharya, B.R.; Mittal, M.; Abraham, A.; Kelemen, A. Deep Learning Techniques for Biomedical and Health Informatics; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Mallick, C.; Das, A.; Nayak, J.; Pelusi, D.; Shanmuganathan, V. Evolutionary Algorithm based Ensemble Extractive Summarization for Developing Smart Medical System. Interdiscip. Sci. Comput. Life Sci. 2021, 13, 229–259. [Google Scholar] [CrossRef] [PubMed]
- Moradi, M.; Ghadiri, N. Different approaches for identifying important concepts in probabilistic biomedical text summarization. Artif. Intell. Med. 2018, 84, 101–116. [Google Scholar] [CrossRef]
- Johnsi, R.; Kumar, G.B.; Sariki, T.P. A Concise Survey on Datasets, Tools and Methods for Biomedical Text Mining. Int. J. Appl. Eng. Res. 2022, 17, 200–217. [Google Scholar] [CrossRef]
- Menachemi, N.; Collum, T.H. Benefits and drawbacks of electronic health record systems. Risk Manag. Healthc. Policy 2011, 4, 47. [Google Scholar] [CrossRef]
- Buchan, K.; Filannino, M.; Uzuner, Ö. Automatic prediction of coronary artery disease from clinical narratives. J. Biomed. Inform. 2017, 72, 23–32. [Google Scholar] [CrossRef]
- Zhou, L.; Baughman, A.W.; Lei, V.J.; Lai, K.H.; Navathe, A.S.; Chang, F.; Sordo, M.; Topaz, M.; Zhong, F.; Murrali, M.; et al. Identifying patients with depression using free-text clinical documents. In MEDINFO 2015: eHealth-Enabled Health; IOS Press: Amsterdam, The Netherlands, 2015; pp. 629–633. [Google Scholar]
- Topaz, M.; Lai, K.; Dowding, D.; Lei, V.J.; Zisberg, A.; Bowles, K.H.; Zhou, L. Automated identification of wound information in clinical notes of patients with heart diseases: Developing and validating a natural language processing application. Int. J. Nurs. Stud. 2016, 64, 25–31. [Google Scholar] [CrossRef]
- Spasić, I.; Livsey, J.; Keane, J.A.; Nenadić, G. Text mining of cancer-related information: Review of current status and future directions. Int. J. Med. Inform. 2014, 83, 605–623. [Google Scholar] [CrossRef]
- Ye, Z.; Tafti, A.P.; He, K.Y.; Wang, K.; He, M.M. SparkText: Biomedical Text Mining on Big Data Framework. PLoS ONE 2016, 11, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Nenkova, A.; McKeown, K. A survey of text summarization techniques. In Mining Text Data; Springer: Berlin/Heidelberg, Germany, 2012; pp. 43–76. [Google Scholar]
- Widyassari, A.P.; Rustad, S.; Shidik, G.F.; Noersasongko, E.; Syukur, A.; Affandy, A.; Setiadi, D.R.I.M. Review of automatic text summarization techniques & methods. J. King Saud Univ.-Comput. Inf. Sci. 2022, 34, 1029–1046. [Google Scholar] [CrossRef]
- Bui, D.D.A.; Del Fiol, G.; Hurdle, J.F.; Jonnalagadda, S. Extractive text summarization system to aid data extraction from full text in systematic review development. J. Biomed. Inform. 2016, 64, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, N.; Jaiswal, A. Automatic text summarization and it’s methods—A review. In Proceedings of the 2016 6th International Conference—Cloud System and Big Data Engineering (Confluence), Noida, India, 14–15 January 2016; pp. 65–72. [Google Scholar] [CrossRef]
- Rahul; Adhikari, S.; Monika. NLP based Machine Learning Approaches for Text Summarization. In Proceedings of the 2020 Fourth International Conference on Computing Methodologies and Communication (ICCMC), Erode, India, 11–13 March 2020; pp. 535–538. [Google Scholar] [CrossRef]
- Gong, L. Application of biomedical text mining. Artif. Intell. Emerg. Trends Appl. 2018, 417. [Google Scholar]
- Mishra, R.; Bian, J.; Fiszman, M.; Weir, C.R.; Jonnalagadda, S.; Mostafa, J.; Del Fiol, G. Text summarization in the biomedical domain: A systematic review of recent research. J. Biomed. Inform. 2014, 52, 457–467. [Google Scholar] [CrossRef]
- Gulden, C.; Kirchner, M.; Schüttler, C.; Hinderer, M.; Kampf, M.; Prokosch, H.U.; Toddenroth, D. Extractive summarization of clinical trial descriptions. Int. J. Med. Inform. 2019, 129, 114–121. [Google Scholar] [CrossRef]
- Cintas, C.; Ogallo, W.; Walcott, A.; Remy, S.L.; Akinwande, V.; Osebe, S. Towards neural abstractive clinical trial text summarization with sequence to sequence models. In Proceedings of the 2019 IEEE International Conference on Healthcare Informatics (ICHI), Xi’an, China, 10–13 June 2019; pp. 1–3. [Google Scholar] [CrossRef]
- Reddy, S.M.; Miriyala, S. Exploring Multi Feature Optimization for Summarizing Clinical Trial Descriptions. In Proceedings of the 2020 IEEE Sixth International Conference on Multimedia Big Data (BigMM), New Delhi, India, 24–26 September 2020; pp. 341–345. [Google Scholar] [CrossRef]
- Afantenos, S.; Karkaletsis, V.; Stamatopoulos, P. Summarization from medical documents: A survey. Artif. Intell. Med. 2005, 33, 157–177. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: Explanation and elaboration. J. Clin. Epidemiol. 2009, 62, e1–e34. [Google Scholar] [CrossRef]
- Maybury, M. Advances in Automatic Text Summarization; MIT Press: Cambridge, MA, USA, 1999. [Google Scholar]
- Jones, K.S. Automatic summarising: Factors and directions. Adv. Autom. Text Summ. 1999. [Google Scholar] [CrossRef]
- Li, L.; Zhou, K.; Xue, G.R.; Zha, H.; Yu, Y. Enhancing Diversity, Coverage and Balance for Summarization through Structure Learning. In Proceedings of the 18th International Conference on World Wide Web, Madrid, Spain, 20–24 April 2009; Association for Computing Machinery: New York, NY, USA, 2009; pp. 71–80. [Google Scholar] [CrossRef]
- Ouyang, Y.; Li, W.; Li, S.; Lu, Q. Applying regression models to query-focused multi-document summarization. Inf. Process. Manag. 2011, 47, 227–237. [Google Scholar] [CrossRef]
- Moradi, M. CIBS: A biomedical text summarizer using topic-based sentence clustering. J. Biomed. Inform. 2018, 88, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, Q.A.; Duong, Q.H.; Nguyen, M.Q.; Nguyen, H.S.; Le, H.Q.; Can, D.C.; Thanh, T.D.; Tran, M.V. A Hybrid Multi-answer Summarization Model for the Biomedical Question-Answering System. In Proceedings of the 2021 13th International Conference on Knowledge and Systems Engineering (KSE), Bangkok, Thailand, 10–12 November 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Bertagnolli, M.M.; Anderson, B.; Quina, A.; Piantadosi, S. The electronic health record as a clinical trials tool: Opportunities and challenges. Clin. Trials 2020, 17, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Munot, N.; Govilkar, S. Comparative Study of Text Summarization Methods. Int. J. Comput. Appl. 2014, 102, 33–37. [Google Scholar] [CrossRef]
- Mani, I. Automatic Summarization; John Benjamins Publishing: Amsterdam, The Netherlands, 2001; Volume 3. [Google Scholar]
- Jones, K.S.; Galliers, J.R. Evaluating Natural Language Processing Systems: An Analysis and Review; Springer: Berlin/Heidelberg, Germany, 1996. [Google Scholar]
- Saziyabegum, S.; Sajja, P. Review on text summarization evaluation methods. Indian J. Comput. Sci. Eng. 2017, 8, 497500. [Google Scholar]
- Moradi, M.; Ghadiri, N. Quantifying the informativeness for biomedical literature summarization: An itemset mining method. Comput. Methods Programs Biomed. 2017, 146, 77–89. [Google Scholar] [CrossRef]
- Steinberger, J. Evaluation measures for text summarization. Comput. Inform. 2009, 28, 251–275. [Google Scholar]
- MacAvaney, S.; Sotudeh, S.; Cohan, A.; Goharian, N.; Talati, I.; Filice, R.W. Ontology-Aware Clinical Abstractive Summarization. In Proceedings of the 42nd International ACM SIGIR Conference on Research and Development in Information Retrieval (SIGIR’19), Paris, France, 21–25 July 2019; Association for Computing Machinery: New York, NY, USA, 2019; pp. 1013–1016. [Google Scholar] [CrossRef]
- Yongkiatpanich, C.; Wichadakul, D. Extractive Text Summarization Using Ontology and Graph-Based Method. In Proceedings of the 2019 IEEE 4th International Conference on Computer and Communication Systems (ICCCS 2019), Singapore, 23–25 February 2019; pp. 105–110. [Google Scholar]
- Gigioli, P.; Sagar, N.; Rao, A.; Voyles, J. Domain-Aware Abstractive Text Summarization for Medical Documents. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 3–6 December 2018; pp. 2338–2343. [Google Scholar] [CrossRef]
- Manas, G.; Aribandi, V.; Kursuncu, U.; Alambo, A.; Shalin, V.L.; Thirunarayan, K.; Beich, J.; Narasimhan, M.; Sheth, A. Knowledge-Infused Abstractive Summarization of Clinical Diagnostic Interviews: Framework Development Study. JMIR Ment. Health 2021, 8, e20865. [Google Scholar] [CrossRef]
- Du, Y.; Li, Q.; Wang, L.; He, Y. Biomedical-domain pre-trained language model for extractive summarization. Knowl.-Based Syst. 2020, 199, 105964. [Google Scholar] [CrossRef]
- Moradi, M.; Dorffner, G.; Samwald, M. Deep contextualized embeddings for quantifying the informative content in biomedical text summarization. Comput. Methods Programs Biomed. 2020, 184, 105117. [Google Scholar] [CrossRef]
- Lee, E.K.; Uppal, K. CERC: An interactive content extraction, recognition, and construction tool for clinical and biomedical text. BMC Med. Inform. Decis. Mak. 2020, 20-S, 306. [Google Scholar] [CrossRef]
- Chen, Y.P.; Chen, Y.Y.; Lin, J.J.; Huang, C.H.; Lai, F. Modified Bidirectional Encoder Representations From Transformers Extractive Summarization Model for Hospital Information Systems Based on Character-Level Tokens (AlphaBERT): Development and Performance Evaluation. JMIR Med. Inform. 2020, 8, e17787. [Google Scholar] [CrossRef] [PubMed]
- Moradi, M.; Dashti, M.; Samwald, M. Summarization of biomedical articles using domain-specific word embeddings and graph ranking. J. Biomed. Inform. 2020, 107, 103452. [Google Scholar] [CrossRef] [PubMed]
- Davoodijam, E.; Ghadiri, N.; Shahreza, M.L.; Rinaldi, F. MultiGBS: A multi-layer graph approach to biomedical summarization. J. Biomed. Inform. 2021, 116, 103706. [Google Scholar] [CrossRef] [PubMed]
- Moradi, M. Frequent itemsets as meaningful events in graphs for summarizing biomedical texts. In Proceedings of the 2018 8th International Conference on Computer and Knowledge Engineering (ICCKE), Mashhad, Iran, 25–26 October 2018; pp. 135–140. [Google Scholar]
- Shah, D.J.; Yu, L.; Lei, T.; Barzilay, R. Nutri-bullets: Summarizing Health Studies by Composing Segments. In Proceedings of the Thirty-Fifth AAAI Conference on Artificial Intelligence, Thirty-Third Conference on Innovative Applications of Artificial Intelligence and the Eleventh Symposium on Educational Advances in Artificial Intelligence (Assoc Advancement Artificial Intelligence), Online, 22 February–1 March 2021; Volume 35, pp. 13780–13788. [Google Scholar]
- Xie, T.; Zhen, Y.; Li, T.; Li, C.; Ge, Y. Self-supervised extractive text summarization for biomedical literatures. In Proceedings of the 2021 IEEE 9th International Conference on Healthcare Informatics (ICHI), Victoria, BC, Canada, 9–12 August 2021; pp. 503–504. [Google Scholar] [CrossRef]
- S, D.; N, L.K.; S, S. Extractive Text Summarization for COVID-19 Medical Records. In Proceedings of the 2021 Innovations in Power and Advanced Computing Technologies (i-PACT), Vellore, India, 27–29 November 2021; pp. 1–5. [Google Scholar] [CrossRef]
- Vinod, P.; Safar, S.; Mathew, D.; Venugopal, P.; Joly, L.M.; George, J. Fine-tuning the BERTSUMEXT model for Clinical Report Summarization. In Proceedings of the 2020 International Conference for Emerging Technology (INCET), Belgaum, India, 5–7 June 2020; pp. 1–7. [Google Scholar] [CrossRef]
- Nguyen, E.; Theodorakopoulos, D.; Pathak, S.; Geerdink, J.; Vijlbrief, O.; van Keulen, M.; Seifert, C. A Hybrid Text Classification and Language Generation Model for Automated Summarization of Dutch Breast Cancer Radiology Reports. In Proceedings of the 2020 IEEE Second International Conference on Cognitive Machine Intelligence (CogMI), Atlanta, GA, USA, 28–31 October 2020; pp. 72–81. [Google Scholar] [CrossRef]
- Rai, A.; Sangwan, S.; Goel, T.; Verma, I.; Dey, L. Query Specific Focused Summarization of Biomedical Journal Articles. In Proceedings of the 2021 16th Conference on Computer Science and Intelligence Systems (FedCSIS), Online, 2–5 September 2021; pp. 91–100. [Google Scholar] [CrossRef]
- Purbawa, D.P.; Malikhah; Esti Anggraini, R.N.; Sarno, R. Automatic Text Summarization using Maximum Marginal Relevance for Health Ethics Protocol Document in Bahasa. In Proceedings of the 2021 13th International Conference on Information Communication Technology and System (ICTS), Surabaya, Indonesia, 20–21 October 2021; pp. 324–329. [Google Scholar] [CrossRef]
- Sibunruang, C.; Polpinij, J. Finding Clinical Knowledge from MEDLINE Abstracts by Text Summarization Technique. In Proceedings of the 2018 International Conference on Information Technology (InCIT), Khon Kaen, Thailand, 24–25 October 2018; pp. 1–6. [Google Scholar] [CrossRef]
- Rouane, O.; Belhadef, H.; Bouakkaz, M. Combine clustering and frequent itemsets mining to enhance biomedical text summarization. Expert Syst. Appl. 2019, 135, 362–373. [Google Scholar] [CrossRef]
- Allahyari, M.; Pouriyeh, S.; Assefi, M.; Safaei, S.; Trippe, E.D.; Gutierrez, J.B.; Kochut, K. Text summarization techniques: A brief survey. arXiv 2017, arXiv:1707.02268. [Google Scholar] [CrossRef]
- Wang, M.; Wang, M.; Yu, F.; Yang, Y.; Walker, J.; Mostafa, J. A systematic review of automatic text summarization for biomedical literature and EHRs. J. Am. Med. Inform. Assoc. 2021, 28, 2287–2297. [Google Scholar] [CrossRef]
- Li, W. Abstractive multi-document summarization with semantic information extraction. In Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing, Lisbon, Portugal, 17–21 September 2015; pp. 1908–1913. [Google Scholar]
- Chu, E.; Liu, P. Meansum: A neural model for unsupervised multi-document abstractive summarization. In Proceedings of the International Conference on Machine Learning, PMLR, Long Beach, CA, USA, 9–15 June 2019; pp. 1223–1232. [Google Scholar]
- Banerjee, S.; Mitra, P.; Sugiyama, K. Multi-document abstractive summarization using ilp based multi-sentence compression. In Proceedings of the Twenty-Fourth International Joint Conference on Artificial Intelligence, Buenos Aires, Argentina, 25–31 July 2015. [Google Scholar]
- Pasunuru, R.; Celikyilmaz, A.; Galley, M.; Xiong, C.; Zhang, Y.; Bansal, M.; Gao, J. Data augmentation for abstractive query-focused multi-document summarization. In Proceedings of the AAAI Conference on Artificial Intelligence (AAAI 2021), Online, 2–9 February 2021; pp. 13666–13674. [Google Scholar]
- Lin, C.Y. ROUGE: A Package for Automatic Evaluation of Summaries. In Text Summarization Branches Out; Association for Computational Linguistics: Barcelona, Spain, 2004; pp. 74–81. [Google Scholar]
- Amer, E.; Fouad, K.M. Keyphrase Extraction methodology from short abstracts of Medical Documents. In Proceedings of the 8th Cairo International Biomedical Engineering Conference (CIBEC), Cairo, Egypt, 15–17 December 2016; pp. 23–26. [Google Scholar]
- Olaronke, I.; Olaleke, J. A Systematic Review of Natural Language Processing in Healthcare. Int. J. Inf. Technol. Comput. Sci. 2015, 08, 44–50. [Google Scholar] [CrossRef]
- Deaton, J. Transformers and Pointer-Generator Networks for Abstractive Summarization. 2019. Available online: https://www.semanticscholar.org/paper/Transformers-and-Pointer-Generator-Networks-for-Deaton/46adc063c1c46e02f6457e45503cbb65495f6494 (accessed on 29 June 2022).
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. In Proceedings of the Advances in Neural Information Processing Systems 30 (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Gambhir, M.; Gupta, V. Recent Automatic Text Summarization Techniques: A Survey. Artif. Intell. Rev. 2017, 47, 1–66. [Google Scholar] [CrossRef]
- Zhang, T.; Kishore, V.; Wu, F.; Weinberger, K.Q.; Artzi, Y. BERTScore: Evaluating Text Generation with BERT. arXiv 2019, arXiv:1904.09675. [Google Scholar]
- Jurafsky, D.; Martin, J.H. Speech and Language Processing: An Introduction to Natural Language Processing, Computational Linguistics, and Speech Recognition; Pearson/Prentice Hall: Hoboken, NJ, USA, 2009. [Google Scholar]
- About PMC. Available online: https://www.ncbi.nlm.nih.gov/pmc/about/intro/ (accessed on 19 July 2022).
- Bada, M.; Eckert, M.; Evans, D.; Garcia, K.; Shipley, K.; Sitnikov, D.; Baumgartner, W., Jr.; Cohen, K.; Verspoor, K.; Blake, J.; et al. Concept annotation in the CRAFT corpus. BMC Bioinform. 2012, 13, 161. [Google Scholar] [CrossRef]
- Craft: The Colorado Richly Annotated Full Text Corpus. Available online: http://bionlp-corpora.sourceforge.net/CRAFT/ (accessed on 19 July 2022).
- Tsatsaronis, G.; Balikas, G.; Malakasiotis, P.; Partalas, I.; Zschunke, M.; Alvers, M.R.; Weissenborn, D.; Krithara, A.; Petridis, S.; Polychronopoulos, D.; et al. An overview of the BIOASQ large-scale biomedical semantic indexing and question answering competition. BMC Bioinform. 2015, 16, 138. [Google Scholar] [CrossRef] [PubMed]
- PubMed.gov. Available online: https://pubmed.ncbi.nlm.nih.gov/ (accessed on 19 July 2022).
- BioMed Central. Available online: https://www.biomedcentral.com/ (accessed on 19 July 2022).
- MEDLINE. Available online: https://www.nlm.nih.gov/medline/index.html (accessed on 19 July 2022).
- Download MEDLINE/PubMed Data. Available online: https://www.nlm.nih.gov/databases/download/pubmed_medline.html (accessed on 19 July 2022).
- Savery, M.; Ben Abacha, A.; Gayen, S.; Demner-Fushman, D. Question-Driven Summarization of Answers to Consumer Health Questions. Sci. Data 2020, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.L.; Lo, K.; Chandrasekhar, Y.; Reas, R.; Yang, J.; Burdick, D.; Eide, D.; Funk, K.; Katsis, Y.; Kinney, R.M.; et al. CORD-19: The COVID-19 Open Research Dataset. In Proceedings of the 1st Workshop on NLP for COVID-19 at ACL 2020, Online; Association for Computational Linguistics: Stroudsburg, PA, USA, 2020. [Google Scholar]
- DAIC-WOZ Database & Extended DAIC Database. Available online: https://dcapswoz.ict.usc.edu/ (accessed on 19 July 2022).
- Gratch, J.; Artstein, R.; Lucas, G.; Stratou, G.; Scherer, S.; Nazarian, A.; Wood, R.; Boberg, J.; DeVault, D.; Marsella, S.; et al. The Distress Analysis Interview Corpus of Human and Computer Interviews; Technical Report; University of Southern California Los Angeles: Los Angeles, CA, USA, 2014. [Google Scholar]
- ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ (accessed on 19 July 2022).



| Question | Purpose | |
|---|---|---|
| Q1 | What are the most prevalent methods used for text summarization in the biomedical domain? | To determine which techniques have been applied in text summarization in the biomedical domain. |
| Q2 | What data types are used in text summarization in the biomedical domain? | To identify which types of text are most common, either single or multiple document. This will also allow us to assess the most frequently used application in biomedical literature or EHR. |
| Q3 | Which areas in the biomedical field have applied text summarization techniques? | To find out which medical areas have implemented summarization methods. |
| Q4 | What are the most common evaluation metrics of text summarization in the biomedical field? | To assess and identify suitable evaluation metrics to use when comparative studies are carried out on text summarization, mainly in the field of health care. |
| Medical Keywords | Technical Keywords | Search Strategy |
|---|---|---|
| Biomedical OR biomedicine OR medical OR medicine OR healthcare OR health OR “patient care” OR clinical OR disease OR diseases OR therapy OR therapies OR treatment OR treatment OR diagnosis OR diagnoses OR diagnostic OR etiology | “Text summarization” OR “abstractive summarization” OR “extractive summarization” OR “abstractive text summarization” OR “extractive text summarization” OR “single document summarization” OR “multi-document summarization” OR “query-based summarization” OR “generic summarization” OR “hugging face” | All fields (Medical Keywords) AND All fields (Technical Keywords) AND (1 January 2014: 15 March 2022) |
| Criteria | |
|---|---|
| Inclusion | Exclusion |
| Complete records. Studies published in journals or at conferences, where the words obtained from the search strategy appear in the title and abstract. Studies that describe the evaluation component (metric) and method(s) used. | Studies not written in English, editorials, or opinion papers. Studies based on summarization techniques in fields other than the biomedical domain. Unavailable records. |
| Parameters | Category | Frequency | |
|---|---|---|---|
| No. Studies | % | ||
| Location | Eastern Africa | 1 | 1.09% |
| Northern Africa | 3 | 3.26% | |
| Africa | 4 | 4.35% | |
| Eastern Asia | 22 | 23.91% | |
| Southern Asia | 30 | 32.61% | |
| Western Asia | 1 | 1.09% | |
| Asia | 53 | 57.61% | |
| Northern Europe | 1 | 1.09% | |
| Southern Europe | 4 | 4.35% | |
| Western Europe | 9 | 9.78% | |
| Europe | 14 | 15.22% | |
| North America | 16 | 17.39% | |
| South America | 3 | 3.26% | |
| America | 19 | 20.65% | |
| Australia/Oceania | 2 | 2.17% | |
| Year | 2014 | 3 | 3.26% |
| 2015 | 6 | 6.52% | |
| 2016 | 5 | 5.43% | |
| 2017 | 5 | 5.43% | |
| 2018 | 12 | 13.04% | |
| 2019 | 15 | 16.30% | |
| 2020 | 20 | 21.74% | |
| 2021 | 22 | 23.91% | |
| 2022 | 4 | 4.35% | |
| Type of publication | Conference | 49 | 53.26% |
| Journal | 43 | 46.74% | |
| Parameters | Category | Frequency | |
|---|---|---|---|
| No. Studies | % | ||
| Input | Single-document (SD) | 25 | 89.29% |
| Multiple-document (MD) | 1 | 3.57% | |
| Single-multiple-document (SMD) | 2 | 7.14% | |
| Biomedical literature (BL) | 20 | 71.43% | |
| EHR (EHR) | 8 | 28.57% | |
| Purpose | Query-based (QB) | 3 | 10.71% |
| Generic (Ge) | 25 | 89.29% | |
| Output | Extractive (Ex) | 21 | 75.00% |
| Abstractive (Ab) | 6 | 21.43% | |
| Extractive and abstractive (EA) | 1 | 3.57% | |
| Method | Mathematical/Statistical (M/S) | 8 | 28.57% |
| Machine Learning (ML) | 16 | 57.14% | |
| Hybrid (Hy) | 4 | 14.29% | |
| Evaluation Metric | Rouge (Rg) | 24 | 85.71% |
| Rouge and others (R/O) | 2 | 7.14% | |
| Other (O) | 2 | 7.14% | |
| Human Evaluation | Human evaluation (HE) | 7 | 25.00% |
| No human evaluation (NHE) | 21 | 75.00% | |
| Title | C/J | Loc. | Year | Input | Purpose | Out | Method (Best) | Metric (Best) | H. Evaluation |
|---|---|---|---|---|---|---|---|---|---|
| Ontology-Aware Clinical Abstractive Summarization [40] | C | USA | 2019 | SD, EHR: Radiology Reports | Ge | Ab | ML: pointer–generator based on Seq2Seq model | Rg 1:38.42 2:23.29 L:37.02 | HE: Radiologist (Readability, Accuracy, Completeness) |
| Extractive Text Summarization using Ontology and Graph-Based Method [41] | C | Singapore | 2019 | SD, BL: Review papers | Ge | Ex | M/S: Graph-based method (PageRank) | Rg-P 1:25.46 L:23.61 | NHE |
| Domain-Aware Abstractive Text Summarization for Medical Documents [42] | C | Spain | 2019 | SD, BL: abstracts from PubMed dataset | Ge | Ab | ML: deep-reinforced pointer–generator network | R/O- 1:42.43 2:21.59 L:36.89 TFIDF UMLS MeSH | NHE |
| Knowledge-Infused Abstractive Summarization of Clinical Diagnostic Interviews: Framework Development Study [43] | J | USA | 2021 | SD, EHR: Diagnostic interviews by mental health professionals | QB: clinical diagnostic interviews | Ab | M/S: knowledge-infused abstractive summarization (KiAS) | Rg-L R:24.46 F1:32.57 | HE: Mental Health professionals (GQCC, GQUC, meaningful responses) |
| Extractive summarization of clinical trial descriptions [22] | J | Germany | 2019 | SD, EHR: clinical trial descriptions from clinicaltrials.gov | Ge | Ex | ML: TextRank | Rg-L P:30.95 R:33.86 F1:30.03 | HE: Human reviewers (Contains all information, Helpfulness) |
| Biomedical-domain pretrained language model for extractive summarization [44] | J | China | 2020 | SD, BL: titles and abstracts from PubMed dataset (Task 6a) | Ge | Ex | ML: domain-aware bidirectional language model (BioBERTSum) | Rg-F1 1:37.45 2:17.59 L:29.58 | NHE |
| Deep contextualized embeddings for quantifying the informative content in biomedical text summarization [45] | J | Austria | 2020 | SD, BL: articles from BioMed Central database | Ge | Ex | Hy: deep bidirectional language model and clustering method (BERT-based, BERT-large) | Rg 1:75.04 2:33.12 | NHE |
| CERC: an interactive content extraction, recognition, and construction tool for clinical and biomedical text [46] | J | England | 2020 | SD, BL: abstracts from Medline | Ge | Ex | ML: multistage algorithm (MINTS) | Rg 1:41.4 2:13.6 SU4:17.1 | NHE |
| Evolutionary Algorithm based Ensemble Extractive Summarization for Developing Smart Medical System [6] | J | India | 2021 | SD, BL: PubMed and MEDLINE journal citations | Ge | Ex | Hy: Multiobjective Evolutionary Algorithm based on Decomposition (MOEAD) | Rg-F1 1:70.7 2:65.5 SU:47.9 | NHE |
| Different approaches for identifying important concepts in probabilistic biomedical text summarization [7] | J | Iran | 2018 | SD, BL: Biomedical articles | Ge | Ex | M/S: Bayesian method | Rg 1:78.86 2:35.29 SU4:41.04 | NHE |
| CIBS: A biomedical text summarizer using topic-based sentence clustering [31] | J | Iran | 2018 | SMD, BL: abstracts from PubMed and BioMed | Ge | Ex | ML: Clustering and Itemset mining (CIBs) | Rg 2:34.75 SU4:39.78 | NHE |
| Modified Bidirectional Encoder Representations From Transformers Extractive Summarization Model for Hospital Information Systems Based on Character-Level Tokens (AlphaBERT): Development and Performance Evaluation [47] | J | Taiwan | 2020 | SD, EHR: diagnoses from National Taiwan University Hospital | Ge | Ex | ML: BERT-based structure with a two-stage training method (AlphaBERT) | Rg 1:76.9 2:61.0 L:75.1 | HE: Doctor feedback (Score) |
| Summarization of biomedical articles using domain-specific word embeddings and graph ranking [48] | J | Austria | 2020 | SD, BL: articles from PubMed | Ge | Ex | Hy: domain-specific word embedding and graph-based model | Rg 1:76.87 2:34.91 | NHE |
| MultiGBS: A multilayer graph approach to biomedical summarization [49] | J | Iran | 2021 | SD, BL: articles from BioMed Central | Ge | Ex | M/S: graph-based creation and sentence selection model (MultiGBS) | Rg/O Rg-F1 1:16.4 2:05.2 L:14.6 SU4:07.5 Bertscore F1:80.6 | NHE |
| Quantifying the informativeness for biomedical literature summarization: An itemset mining method [38] | J | Iran | 2017 | SD, BL: Scientific papers | Ge | Ex | M/S: Itemset mining | Rg 1:75.83 2:33.81 SU4:38.89 | NHE |
| Frequent itemsets as meaningful events in graphs for summarizing biomedical texts [50] | C | Iran | 2018 | SD, BL: scientific articles | Ge | Ex | M/S: Graph-based method | Rg 2:34.03 SU4:38.51 | NHE |
| Nutri-bullets: Summarizing Health Studies by Composing Segments [51] | C | USA | 2021 | MD, BL: scientific abstracts from PubMed and ScienceDirect | Ge | Ab | ML: reinforcement learning (Blank Language Model—BLM) | O-Meteor Me:15.0 | HE: (Faithfulness, Relevance, Fluency) |
| Self-supervised extractive text summarization for biomedical literature [52] | C | USA | 2021 | SD, BL: Radiation Therapy scientific articles from PubMed | Ge | Ex | ML: BERT | Rg-R 1:71.00 2:59.00 | NHE |
| A Hybrid Multianswer Summarization Model for the Biomedical Question-Answering System [32] | C | Vietnam | 2021 | SMD, BL: Medical Question-Answer Summarization dataset (MEDIQA-AnS) | QB: Question-driven filtering phase | EA | ML: Denoising autoencoder and BART (Extractive Abstractive hybrid model - EAHS) | Rg-F1 1:30.00 2:22.00 L:25.00 | NHE |
| Towards neural abstractive clinical trial text summarization with sequence to sequence models [23] | C | Kenya | 2019 | SD, EHR: clinical trial descriptions from clinical trials.gov | Ge | Ab | ML: Seq2Seq model with attention | Rg-F1 1:40.4 2:15.0 L:33.8 | NHE |
| Extractive Text Summarization for COVID-19 Medical Records [53] | C | India | 2021 | SD, BL: COVID-19 research articles from PubMed, Microsoft Academic and WHO COVID-19 | Ge | Ex | ML: Generative Pre-Trained Transformer 2 (GPT-2) | Rg-F1 1:78.22 2:71.17 L:78.22 | NHE |
| Fine-tuning the BERTSUMEXT model for Clinical Report Summarization [54] | C | India | 2020 | SD, EHR: clinical report summarization dataset | Ge | Ex | ML: Fine-tuned BERTSUMTEXT | Rg-F1 1:50.07 2:39.85 L:49.59 | HE: Doctor’s opinion |
| A Hybrid Text Classification and Language Generation Model for Automated Summarization of Dutch Breast Cancer Radiology Reports [55] | C | Netherlands | 2020 | SD, EHR: Dutch breast cancer radiology reports | Ge | Ab | ML: encoder–decoder attention model (EDA) | Rg-F1 1:54.0 2:38.8 L:51.5 | HE: Radiologists (correctness, relevance, comprehensible) |
| Query Specific Focused Summarization of Biomedical Journal Articles [56] | C | India | 2021 | SD, BL: articles from COVID-19 Open Research Dataset (CORD-19) | QB: User required information | Ex | M/S: Optimization and contextual method | Rg 1:47.61 2:19.62 L:44.74 | NHE |
| Exploring Multi-Feature Optimization for Summarizing Clinical Trial Descriptions [24] | C | India | 2020 | SD, EHR: Clinical Trial Descriptions from Mendeley datasets | Ge | Ex | M/S: Multi Feature Optimization (MFO) | Rg-R 1:70.0 2:39.0 L:50.0 | NHE |
| Automatic Text Summarization using Maximum Marginal Relevance for Health Ethics Protocol Document in Bahasa [57] | C | Indonesia | 2021 | SD, BL: Health research ethics protocol | Ge | Ex | M/S: Maximum Marginal Relevance (MMR) | Rg-4 P:34.0 R:71.0 F1:46.0 | NHE |
| Finding Clinical Knowledge from MEDLINE Abstracts by Text Summarization Technique [58] | C | Thailand | 2018 | SD, BL: cervical cancer in clinical trials from MEDLINE abstracts | Ge | Ex | ML: BM25 term-weighting and text filtering techniques | O P:100.0 R:84.0 F1:91.0 | NHE |
| Combining clustering and frequent item set mining to enhance biomedical text summarization [59] | J | USA | 2019 | SD, BL: articles from BioMed central database | Ge | Ex | Hy: clustering and frequent itemset meaning | Rg 1:23.84 2:08.71 SU4:11.45 | NHE |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chaves, A.; Kesiku, C.; Garcia-Zapirain, B. Automatic Text Summarization of Biomedical Text Data: A Systematic Review. Information 2022, 13, 393. https://doi.org/10.3390/info13080393
Chaves A, Kesiku C, Garcia-Zapirain B. Automatic Text Summarization of Biomedical Text Data: A Systematic Review. Information. 2022; 13(8):393. https://doi.org/10.3390/info13080393
Chicago/Turabian StyleChaves, Andrea, Cyrille Kesiku, and Begonya Garcia-Zapirain. 2022. "Automatic Text Summarization of Biomedical Text Data: A Systematic Review" Information 13, no. 8: 393. https://doi.org/10.3390/info13080393
APA StyleChaves, A., Kesiku, C., & Garcia-Zapirain, B. (2022). Automatic Text Summarization of Biomedical Text Data: A Systematic Review. Information, 13(8), 393. https://doi.org/10.3390/info13080393







