A Comparison of Machine Learning Techniques for the Detection of Type-2 Diabetes Mellitus: Experiences from Bangladesh
Abstract
:1. Introduction
- We created a dataset of 508 study populations for diabetes.
- We applied and compared state-of-the-art clinically applicable ML models to conduct a benchmark analysis, aiming to contribute to further research in the field.
- We proposed a framework that utilizes ML techniques to detect diabetic patients.
- We ranked and identified significant features associated with diabetes mellitus.
2. Materials and Methods
2.1. Data Collection and Description
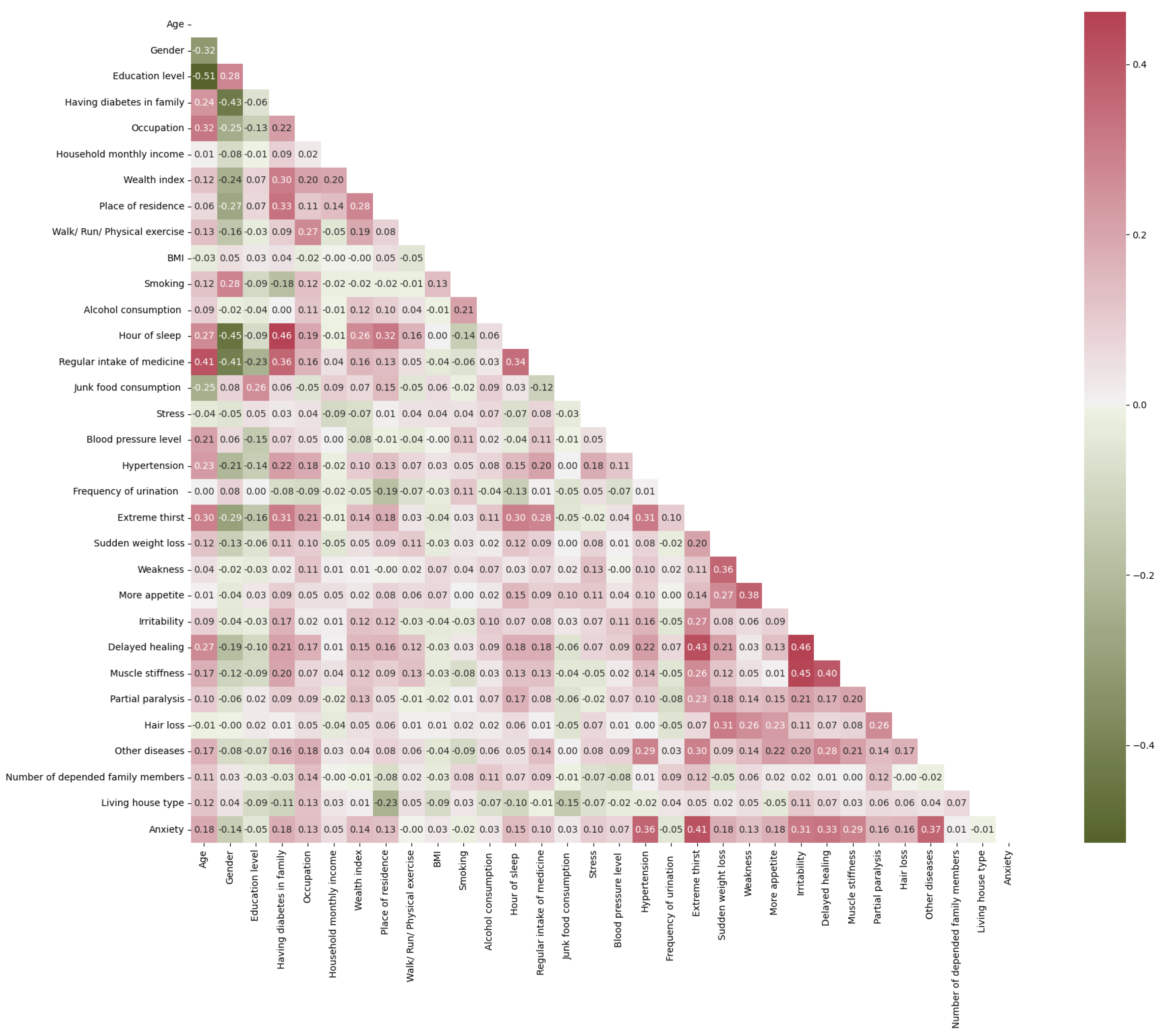
2.2. Explanatory Data Analysis
2.3. Data Preprocessing
2.4. Data Balancing Technique
2.5. Feature Transformation
2.6. Feature Selection
2.7. Statistical Methods Identifying the Most Significant and Associative Diabetes Features
2.8. Machine Learning Model
- DT is a white box concept that has an effective learning component. Numerous leaf nodes, multiple internal nodes, and a central root node constitute DT. Each leaf node is labelled according to its class and linked to the root of the tree via internal nodes. A DT’s root node serves as its beginning point, and the route from this node to its leaf nodes produces the classification rules [48].
- LR is an excellent method for predicting the probability of a result in a variety of classification situations. Commonly, the LR model is used when people can make predictions about health or illness. The LR algorithm predicts the probability of the target category dependent variable by applying the training examples to a logistic sigmoid activation function. In LR, the target attribute’s calculated probability ranges from 0 to 1. Additionally, a threshold is established to classify an event into a certain target class. The predicted probability is input into a certain target category based on the threshold value [49,50].
- The SVM [51,52] is a type of linear generalized classifier that sorts binary data using supervised learning. It is appropriate for small data collections with minimal outliers. The goal is to identify a hyperplane that can be used to connect data points. This hyperplane divides the space into separate domains, each of which holding different kinds of data. There are numerous hyperplanes from which to choose to split the two groups of data. Our objective was to find the plane with the largest margin. The margin is the distance between the hyperplane and two data points that are closest to it that represent two subclasses. The SVM attempts to optimize the algorithm by increasing this margin value, thereby determining the optimal superplane to divide the dataset into two layers. The nearest data points to the hyperplane are referred to as support vectors.
- GB is a prominent supervised ML method for disease forecasting since it creates an ensemble forecasting model using weak classifiers based on a DT. It constructs DTs using a gradient decent iterative optimization technique to discover the best parameter values, unlike RF. Then, we use the weighted majority votes from each DT to forecast the predicted value [53,54].
- XGBoost [55,56] constructs multiple new algorithms and merges them into a single ensemble model. First, the inaccuracy the of residuals for every observation is determined based on an established model. Based on previous errors, a revised model is developed to predict the residuals. The predictions of this model are then incorporated into the ensemble models. XGBoost is superior to GB algorithms because it finds a balance between bias and variation.
- RF is an ML algorithm that uses a random subspace approach and bagging ensemble learning. In the training stage, RF builds several DTs for arbitrarily partitioning data. For each node in the root DT, a subset of K attributes is chosen at random from the node’s attribute set. From this subset, an effective attribute is then chosen for partitioning. Each tree submits a classification as a vote for the other trees, and the RF selects the classification with the most votes [29,57].
- ET is a procedure for data mining that combines multiple methods into a single optimal predictive model to improve predictions. This technique provides superior predictive performance when compared to a single model. We combined DT, XGB, and RF to benefits from all the algorithms to improve the overall predictive performance. By uniting the strengths of multiple models, ETs can provide enhanced generalization, increased robustness, and enhanced precision. It can help reduce individual model biases and improve model performance overall [58].
2.9. Model Evaluation
3. Results and Discussion
3.1. Experimental Setup
3.2. Result Analysis
3.3. Discussion
4. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ML | Machine learning |
| DT | Decision tree |
| RF | Random forest |
| LR | Logistic regression |
| SVM | Support vector machine |
| GB | Gradient boosting |
References
- Association, A.D. Diagnosis and classification of diabetes mellitus. Diabetes Care 2014, 37, S81–S90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- IDF. Type 2 Diabetes. Available online: https://www.idf.org/aboutdiabetes/type-2-diabetes.html (accessed on 7 May 2023).
- John, J.E.; John, N.A. Imminent risk of COVID-19 in diabetes mellitus and undiagnosed diabetes mellitus patients. Pan Afr. Med. J. 2020, 32874422. [Google Scholar] [CrossRef] [PubMed]
- Gahlan, D.; Rajput, R.; Singh, V. Metabolic syndrome in north indian type 2 diabetes mellitus patients: A comparison of four different diagnostic criteria of metabolic syndrome. Diabetes Metab. Syndr. 2019, 13, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Atlas, I.D. Diabetes around the World in 2021. Available online: https://diabetesatlas.org/ (accessed on 7 May 2023).
- Williams, R.; Karuranga, S.; Malanda, B.; Saeedi, P.; Basit, A.; Besançon, S.; Bommer, C.; Esteghamati, A.; Ogurtsova, K.; Zhang, P.; et al. Global and regional estimates and projections of diabetes-related health expenditure: Results from the International Diabetes Federation Diabetes Atlas. Diabetes Res. Clin. Pract. 2020, 162, 108072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Htay, T.; Soe, K.; Lopez-Perez, A.; Doan, A.H.; Romagosa, M.A.; Aung, K. Mortality and cardiovascular disease in type 1 and type 2 diabetes. Curr. Cardiol. Rep. 2019, 21, 45. [Google Scholar] [CrossRef]
- Nipa, N.; Riyad, M.M.H.; Satu, M.S.; Walliullah, M.; Howlader, K.C.; Moni, M.A. Clinically Adaptable Machine Learning Model To Identify Early Appreciable Features of Diabetes In Bangladesh. Intell. Med. 2023. [Google Scholar] [CrossRef]
- Huang, Y.; Roy, N.; Dhar, E.; Upadhyay, U.; Kabir, M.A.; Uddin, M.; Tseng, C.L.; Syed-Abdul, S. Deep Learning Prediction Model for Patient Survival Outcomes in Palliative Care Using Actigraphy Data and Clinical Information. Cancers 2023, 15, 2232. [Google Scholar] [CrossRef]
- Panday, A.; Kabir, M.A.; Chowdhury, N.K. A survey of machine learning techniques for detecting and diagnosing COVID-19 from imaging. Quant. Biol. 2022, 10. [Google Scholar] [CrossRef]
- Uddin, M.J.; Ahamad, M.M.; Sarker, P.K.; Aktar, S.; Alotaibi, N.; Alyami, S.A.; Kabir, M.A.; Moni, M.A. An Integrated Statistical and Clinically Applicable Machine Learning Framework for the Detection of Autism Spectrum Disorder. Computers 2023, 12, 92. [Google Scholar] [CrossRef]
- Hossain, M.D.; Kabir, M.A.; Anwar, A.; Islam, M.Z. Detecting autism spectrum disorder using machine learning techniques: An experimental analysis on toddler, child, adolescent and adult datasets. Health Inf. Sci. Syst. 2021, 9, 17. [Google Scholar] [CrossRef]
- Aguilera-Venegas, G.; López-Molina, A.; Rojo-Martínez, G.; Galán-García, J.L. Comparing and tuning machine learning algorithms to predict type 2 diabetes mellitus. J. Comput. Appl. Math. 2023, 427, 115115. [Google Scholar] [CrossRef]
- Zhao, M.; Wan, J.; Qin, W.; Huang, X.; Chen, G.; Zhao, X. A machine learning-based diagnosis modelling of type 2 diabetes mellitus with environmental metal exposure. Comput. Methods Programs Biomed. 2023, 235, 107537. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Zhang, Y.; Peng, B.; Hu, X.; Zhou, L.; Chen, C.; Lu, C.; Chen, M.; Pang, C.; Dai, Y.; et al. Detection of mild cognitive impairment in type 2 diabetes mellitus based on machine learning using privileged information. Neurosci. Lett. 2022, 791, 136908. [Google Scholar] [CrossRef] [PubMed]
- Ejiyi, C.J.; Qin, Z.; Amos, J.; Ejiyi, M.B.; Nnani, A.; Ejiyi, T.U.; Agbesi, V.K.; Diokpo, C.; Okpara, C. A robust predictive diagnosis model for diabetes mellitus using Shapley-incorporated machine learning algorithms. Healthc. Anal. 2023, 3, 100166. [Google Scholar] [CrossRef]
- Hennebelle, A.; Materwala, H.; Ismail, L. HealthEdge: A Machine Learning-Based Smart Healthcare Framework for Prediction of Type 2 Diabetes in an Integrated IoT, Edge, and Cloud Computing System. Procedia Comput. Sci. 2023, 220, 331–338. [Google Scholar] [CrossRef]
- Haque, M.; Alharbi, I. A Dataset-Specific Machine Learning Study for Predicting Diabetes (Type-2) in a Developing Country Context. Indian J. Sci. Technol. 2022, 15, 1932–1940. [Google Scholar] [CrossRef]
- Tasin, I.; Nabil, T.U.; Islam, S.; Khan, R. Diabetes prediction using machine learning and explainable AI techniques. Healthc. Technol. Lett. 2022, 1684017. [Google Scholar] [CrossRef]
- Kaur, H.; Kumari, V. Predictive modelling and analytics for diabetes using a machine learning approach. Appl. Comput. Inform. 2022, 18, 90–100. [Google Scholar] [CrossRef]
- Zheng, T.; Xie, W.; Xu, L.; He, X.; Zhang, Y.; You, M.; Yang, G.; Chen, Y. A machine learning-based framework to identify type 2 diabetes through electronic health records. Int. J. Med. Inform. 2017, 97, 120–127. [Google Scholar] [CrossRef] [Green Version]
- Saha, P.K.; Patwary, N.S.; Ahmed, I. A widespread study of diabetes prediction using several machine learning techniques. In Proceedings of the 2019 22nd International Conference on Computer and Information Technology (ICCIT), Dhaka, Bangladesh, 18–20 December 2019; IEEE: Piscataway NJ, USA, 2019; pp. 1–5. [Google Scholar]
- Ali, M.S.; Islam, M.K.; Das, A.A.; Duranta, D.; Haque, M.; Rahman, M.H. A novel approach for best parameters selection and feature engineering to analyze and detect diabetes: Machine learning insights. Biomed Res. Int. 2023, 8583210. [Google Scholar] [CrossRef]
- Howlader, K.C.; Satu, M.S.; Awal, M.A.; Islam, M.R.; Islam, S.M.S.; Quinn, J.M.; Moni, M.A. Machine learning models for classification and identification of significant attributes to detect type 2 diabetes. Health Inf. Sci. Syst. 2022, 10, 2. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Zhao, X.; Miao, C. A comprehensive exploration to the machine learning techniques for diabetes identification. In Proceedings of the 2018 IEEE 4th World Forum on Internet of Things (WF-IoT), Singapore, 5–8 February 2018; IEEE: Piscataway NJ, USA, 2018; pp. 291–295. [Google Scholar]
- Birjais, R.; Mourya, A.K.; Chauhan, R.; Kaur, H. Prediction and diagnosis of future diabetes risk: A machine learning approach. SN Appl. Sci. 2019, 1, 1112. [Google Scholar] [CrossRef] [Green Version]
- Yahyaoui, A.; Jamil, A.; Rasheed, J.; Yesiltepe, M. A decision support system for diabetes prediction using machine learning and deep learning techniques. In Proceedings of the 2019 1st International Informatics and Software Engineering Conference (UBMYK), Ankara, Turkey, 6–7 November 2019; IEEE: Piscataway NJ, USA, 2019; pp. 1–4. [Google Scholar]
- Abdulhadi, N.; Al-Mousa, A. Diabetes detection using machine learning classification methods. In Proceedings of the 2021 International Conference on Information Technology (ICIT), Amman, Jordan, 14–15 July 2021; IEEE: Piscataway NJ, USA, 2021; pp. 350–354. [Google Scholar]
- Battineni, G.; Sagaro, G.G.; Nalini, C.; Amenta, F.; Tayebati, S.K. Comparative machine-learning approach: A follow-up study on type 2 diabetes predictions by cross-validation methods. Machines 2019, 7, 74. [Google Scholar] [CrossRef] [Green Version]
- Tigga, N.P.; Garg, S. Prediction of Type 2 Diabetes using Machine Learning Classification Methods. Procedia Comput. Sci. 2020, 167, 706–716. [Google Scholar] [CrossRef]
- Pranto, B.; Mehnaz, S.M.; Mahid, E.B.; Sadman, I.M.; Rahman, A.; Momen, S. Evaluating machine learning methods for predicting diabetes among female patients in Bangladesh. Information 2020, 11, 374. [Google Scholar] [CrossRef]
- Sneha, N.; Gangil, T. Analysis of diabetes mellitus for early prediction using optimal features selection. J. Big Data 2019, 121, 54–64. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, Y.; Niu, M.; Wang, C.; Wang, Z. Machine learning for characterizing risk of type 2 diabetes mellitus in a rural Chinese population: The Henan Rural Cohort Study. Sci. Rep. 2020, 10, 4406. [Google Scholar] [CrossRef] [Green Version]
- Bonifazi, G.; Enrico Corradini, D.U.; Virgili, L. Defining user spectra to classify Ethereum users based on their behavior. J. Big Data 2022, 9, 37. [Google Scholar] [CrossRef]
- Le, T.M.; Vo, T.M.; Pham, T.N.; Dao, S.V.T. A novel wrapper–based feature selection for early diabetes prediction enhanced with a metaheuristic. IEEE Access 2020, 9, 7869–7884. [Google Scholar] [CrossRef]
- Islam, M.M.; Rahman, M.J.; Roy, D.C.; Maniruzzaman, M. Automated detection and classification of diabetes disease based on Bangladesh demographic and health survey data, 2011 using machine learning approach. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 217–219. [Google Scholar] [CrossRef]
- Haq, A.U.; Li, J.P.; Khan, J.; Memon, M.H.; Nazir, S.; Ahmad, S.; Khan, G.A.; Ali, A. Intelligent machine learning approach for effective recognition of diabetes in E-healthcare using clinical data. Sensors 2020, 20, 2649. [Google Scholar] [CrossRef] [PubMed]
- Shuja, M.; Mittal, S.; Zaman, M. Effective Prediction of Type II Diabetes Mellitus Using Data Mining Classifiers and SMOTE. In Proceedings of the Advances in Computing and Intelligent Systems; Sharma, H., Govindan, K., Poonia, R.C., Kumar, S., El-Medany, W.M., Eds.; Springer: Singapore, 2020; pp. 195–211. [Google Scholar]
- Chatrati, S.P.; Hossain, G.; Goyal, A.; Bhan, A.; Bhattacharya, S.; Gaurav, D.; Tiwari, S.M. Smart home health monitoring system for predicting type 2 diabetes and hypertension. J. King Saud Univ. Comput. Inf. Sci. 2022, 34, 862–870. [Google Scholar] [CrossRef]
- Islam, M.M.; Rahman, M.J.; Menhazul Abedin, M.; Ahammed, B.; Ali, M.; Ahmed, N.F.; Maniruzzaman, M. Identification of the risk factors of type 2 diabetes and its prediction using machine learning techniques. Health Syst. 2022, 12, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Islam, S.M.S.; Islam, M.T.; Uddin, R.; Tansi, T.; Talukder, S.; Sarker, F.; Mamun, K.A.A.; Adibi, S.; Rawal, L.B. Factors associated with low medication adherence in patients with Type 2 diabetes mellitus attending a tertiary hospital in Bangladesh. Lifestyle Med. 2021, 2, e47. [Google Scholar] [CrossRef]
- Nnamoko, N.; Korkontzelos, I. Efficient treatment of outliers and class imbalance for diabetes prediction. Artif. Intell. Med. 2020, 104, 101815. [Google Scholar] [CrossRef] [PubMed]
- Ganie, S.M.; Malik, M.B. An ensemble Machine Learning approach for predicting Type-II diabetes mellitus based on lifestyle indicators. Healthc. Anal. 2022, 2, 100092. [Google Scholar] [CrossRef]
- Petmezas, G.; Haris, K.; Stefanopoulos, L.; Kilintzis, V.; Tzavelis, A.; Rogers, J.A.; Katsaggelos, A.K.; Maglaveras, N. Automated atrial fibrillation detection using a hybrid CNN-LSTM network on imbalanced ECG datasets. Biomed. Signal Process. Control 2021, 63, 102194. [Google Scholar] [CrossRef]
- Mehedi Hassan, M.; Mollick, S.; Yasmin, F. An unsupervised cluster-based feature grouping model for early diabetes detection. Healthc. Anal. 2022, 2, 100112. [Google Scholar] [CrossRef]
- Deberneh, H.M.; Kim, I. Prediction of Type 2 Diabetes Based on Machine Learning Algorithm. Int. J. Environ. Res. Public Health 2021, 18, 3317. [Google Scholar] [CrossRef]
- Aktar, S.; Ahamad, M.M.; Rashed-Al-Mahfuz, M.; Azad, A.; Uddin, S.; Kamal, A.; Alyami, S.A.; Lin, P.I.; Islam, S.M.S.; Quinn, J.M.; et al. Machine learning approach to predicting COVID-19 disease severity based on clinical blood test data: Statistical analysis and model development. JMIR Med. Inform. 2021, 9, e25884. [Google Scholar] [CrossRef]
- Azad, C.; Bhushan, B.; Sharma, R.; Shankar, A.; Singh, K.K.; Khamparia, A. Prediction model using SMOTE, genetic algorithm and decision tree (PMSGD) for classification of diabetes mellitus. Multimed. Syst. 2022, 28, 1289–1307. [Google Scholar] [CrossRef]
- Maniruzzaman, M.; Rahman, M.; Al-MehediHasan, M.; Suri, H.S.; Abedin, M.; El-Baz, A.; Suri, J.S. Accurate diabetes risk stratification using machine learning: Role of missing value and outliers. J. Med. Syst. 2018, 42, 92. [Google Scholar] [CrossRef] [Green Version]
- Ahlqvist, E.; Storm, P.; Käräjämäki, A.; Martinell, M.; Dorkhan, M.; Carlsson, A.; Vikman, P.; Prasad, R.B.; Aly, D.M.; Almgren, P.; et al. Novel subgroups of adult-onset diabetes and their association with outcomes: A data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 2018, 6, 361–369. [Google Scholar] [CrossRef] [Green Version]
- Boubin, M.; Shrestha, S. Microcontroller implementation of support vector machine for detecting blood glucose levels using breath volatile organic compounds. Sensors 2019, 19, 2283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muhammad, L.; Algehyne, E.A.; Usman, S.S. Predictive supervised machine learning models for diabetes mellitus. SN Comput. Sci. 2020, 1, 240. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Rahman, M.J.; Roy, D.C.; Tawabunnahar, M.; Jahan, R.; Ahmed, N.F.; Maniruzzaman, M. Machine learning algorithm for characterizing risks of hypertension, at an early stage in Bangladesh. Diabetes Metab. Syndr. Clin. Res. Rev. 2021, 15, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Ahamad, M.M.; Aktar, S.; Rashed-Al-Mahfuz, M.; Uddin, S.; Liò, P.; Xu, H.; Summers, M.A.; Quinn, J.M.; Moni, M.A. A machine learning model to identify early stage symptoms of SARS-Cov-2 infected patients. Expert Syst. Appl. 2020, 160, 113661. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Hasan, M.K.; Ahmad, M.; Awal, M.A.; Islam, M.A.; Masud, M.; Meshref, H. Early prediction of diabetes using an ensemble of machine learning models. Int. J. Environ. Res. Public Health 2022, 19, 12378. [Google Scholar] [CrossRef]
- Kibria, H.B.; Nahiduzzaman, M.; Goni, M.O.F.; Ahsan, M.; Haider, J. An ensemble approach for the prediction of diabetes mellitus using a soft voting classifier with an explainable AI. Sensors 2022, 22, 7268. [Google Scholar] [CrossRef] [PubMed]
- Ijaz, M.F.; Alfian, G.; Syafrudin, M.; Rhee, J. Hybrid prediction model for type 2 diabetes and hypertension using DBSCAN-based outlier detection, synthetic minority over sampling technique (SMOTE), and random forest. Appl. Sci. 2018, 8, 1325. [Google Scholar] [CrossRef] [Green Version]
- Amelio, A.; Bonifazi, G.; Corradini, E.; Di Saverio, S.; Marchetti, M.; Ursino, D.; Virgili, L. Defining a deep neural network ensemble for identifying fabric colors. Appl. Soft Comput. 2022, 130, 109687. [Google Scholar] [CrossRef]
- Islam, S.M.S.; Talukder, A.; Awal, M.A.; Siddiqui, M.M.U.; Ahamad, M.M.; Ahammed, B.; Rawal, L.B.; Alizadehsani, R.; Abawajy, J.; Laranjo, L.; et al. Machine Learning Approaches for Predicting Hypertension and Its Associated Factors Using Population-Level Data From Three South Asian Countries. Front. Cardiovasc. Med. 2022, 9, 839379. [Google Scholar] [CrossRef] [PubMed]
- Akter, T.; Ali, M.H.; Khan, M.I.; Satu, M.S.; Uddin, M.J.; Alyami, S.A.; Ali, S.; Azad, A.; Moni, M.A. Improved transfer-learning-based facial recognition framework to detect autistic children at an early stage. Brain Sci. 2021, 11, 734. [Google Scholar] [CrossRef] [PubMed]
- Ahamad, M.M.; Aktar, S.; Uddin, M.J.; Rahman, T.; Alyami, S.A.; Al-Ashhab, S.; Akhdar, H.F.; Azad, A.; Moni, M.A. Early-Stage Detection of Ovarian Cancer Based on Clinical Data Using Machine Learning Approaches. J. Pers. Med. 2022, 12, 1211. [Google Scholar] [CrossRef]
- Ahamad, M.M.; Aktar, S.; Uddin, M.J.; Rashed-Al-Mahfuz, M.; Azad, A.; Uddin, S.; Alyami, S.A.; Sarker, I.H.; Khan, A.; Liò, P.; et al. Adverse effects of COVID-19 vaccination: Machine learning and statistical approach to identify and classify incidences of morbidity and postvaccination reactogenicity. Healthcare 2022, 11, 31. [Google Scholar] [CrossRef]
- Akter, T.; Khan, M.I.; Ali, M.H.; Satu, M.S.; Uddin, M.J.; Moni, M.A. Improved machine learning based classification model for early autism detection. In Proceedings of the 2021 2nd International Conference on Robotics, Electrical and Signal Processing Techniques (ICREST), Dhaka, Bangladesh, 5–7 January 2021; IEEE: Piscataway, NJ, USA, 2021; pp. 742–747. [Google Scholar]
- Akter, T.; Satu, M.S.; Khan, M.I.; Ali, M.H.; Uddin, S.; Lio, P.; Quinn, J.M.; Moni, M.A. Machine learning-based models for early stage detection of autism spectrum disorders. IEEE Access 2019, 7, 166509–166527. [Google Scholar] [CrossRef]
- Xiong, Y.; Lin, L.; Chen, Y.; Salerno, S.; Li, Y.; Zeng, X.; Li, H. Prediction of gestational diabetes mellitus in the first 19 weeks of pregnancy using machine learning techniques. J. Matern. Fetal Neonatal Med. 2022, 35, 2457–2463. [Google Scholar] [CrossRef]
- Olisah, C.C.; Smith, L.; Smith, M. Diabetes mellitus prediction and diagnosis from a data preprocessing and machine learning perspective. Comput. Methods Programs Biomed. 2022, 220, 106773. [Google Scholar] [CrossRef]
- Wei, H.; Sun, J.; Shan, W.; Xiao, W.; Wang, B.; Ma, X.; Hu, W.; Wang, X.; Xia, Y. Environmental chemical exposure dynamics and machine learning-based prediction of diabetes mellitus. Sci. Total Environ. 2022, 806, 150674. [Google Scholar] [CrossRef]
- Rawat, V.; Joshi, S.; Gupta, S.; Singh, D.P.; Singh, N. Machine learning algorithms for early diagnosis of diabetes mellitus: A comparative study. Mater. Today Proc. 2022, 56, 502–506. [Google Scholar] [CrossRef]
- Syed, A.H.; Khan, T. Machine learning-based application for predicting risk of type 2 diabetes mellitus (T2DM) in Saudi Arabia: A retrospective cross-sectional study. IEEE Access 2020, 8, 199539–199561. [Google Scholar] [CrossRef]
- Chou, C.Y.; Hsu, D.Y.; Chou, C.H. Predicting the Onset of Diabetes with Machine Learning Methods. J. Pers. Med. 2023, 13, 406. [Google Scholar] [CrossRef] [PubMed]
- Laila, U.E.; Mahboob, K.; Khan, A.W.; Khan, F.; Taekeun, W. An ensemble approach to predict early-stage diabetes risk using machine learning: An empirical study. Sensors 2022, 22, 5247. [Google Scholar] [CrossRef] [PubMed]




| ID | Attribute Name | Feature Description | Count (N = 508), n (%) or Avg., Min, Max, Median |
|---|---|---|---|
| 1 | Age | Identifying age groups | 1–18: 244 (48.03%) 19–40: 95 (18.7%) 40–65: 99 (19.49%) 65 or more: 70 (13.78%) |
| 2 | Gender | Gender of the respondent | Male: 249 (49.01%) Female: 259 (50.99 %) |
| 3 | Education Level | Level of education can be no education, primary, secondary or higher | Primary: 52 (10.36%) Secondary: 95 (18.7%) Higher: 305 (60.03%) No education: 51 (10.03%) |
| 4 | Diabetes in the family | Respondent’s ancestors or parents suffered from the disease or not | Yes: 298 (58.66%) No: 210 (41.34%) |
| 5 | Occupation | Respondent can be unemployed, employed or find job | Looking work: 56 (11.02%) Not working: 189 (37.20%) Working: 260 (51.18%) |
| 6 | Household monthly income | Household monthly income of the respondent | Avg.: 30,477 Max: 300,000 Min: 1000 Median: 30,000 |
| 7 | Wealth index | Wealth index can be low, middle or upper class | Poor: 46 (0.09%) Middle: 406 (79.91%) Rich: 58 (11%) |
| 8 | Place of residence | Place of residence indicates in which area the respondent usually lives | Urban: 233 (45.86%) Rural: 276 (54.14%) |
| 9 | Walk/ Run/ Physical exercise | How much jogging or walking the respondent did | None: 43 (8.46%) Less half: 90 (17.71%) More half: 151 (29.72%) One hour or More: 224 (44.09%) |
| 10 | BMI | Calculated from Height and Weight | Avg.: 23.2 Max: 58.44 Min: 12 Median: 23.12 |
| 11 | Smoking | Whether the respondent smokes or not | Yes: 53 (10.44%) No: 455 (89.56%) |
| 12 | Alcohol consumption | Whether or not the respondent drinks alcohol | Yes: 22 (4.33%) No: 486 (95.67%) |
| 13 | Hours of sleep | Total hour of sleep each day | Avg.: 7.77 Max: 12 Min: 1 Median: 8 |
| 14 | Regular intake of medicine (Except insulin) | Regular medication use, excluding insulin | Yes: 233 (45.86%) No: 275 (54.14%) |
| 15 | Junk food consumption | Prevalence of junk food consumption | Yes: 194 (38.18%) No: 314 (61.82%) |
| 16 | Stress | Stress level of respondent | Not at all: 66 (13%) Sometimes: 329 (64.76%) Often: 60 (11.8%) Always: 55 (10.82%) |
| 17 | Blood pressure level | Average level of blood pressure | High: 83 (16.33%) Normal: 383 (75.4%) Low: 43 (8.26%) |
| 18 | Hypertension | Whether or not the respondent has hypertension | Yes: 269 (52.95%) No: 239 (47.05%) |
| 19 | Frequency of urination | Frequency of urination each day | Not much: 360 (70.86%) Quite much: 148 (29.14%) |
| 20 | Extreme thirst | Whether or not the respondent is extremely thirsty | Yes: 253 (49.8%) No: 255 (50.2%) |
| 21 | Sudden weight loss | Whether the respondent ever noticed sudden weight loss | Yes: 110 (21.65%) No: 334 (65.74%) May be: 64 (12.6%) |
| 22 | Weakness | Whether the respondent feels more vulnerable | Yes: 225 (44.29%) No: 187 (36.81%) May be: 96 (18.9%) |
| 23 | More appetite | Whether the respondent feels more hungry | Yes: 151 (29.72%) No: 250 (49.21%) May be: 106 (20.86%) |
| 24 | Irritability | Whether the respondent feels irritability | Yes: 250 (49.2%) No: 258 (50.8%) |
| 25 | Delayed healing | Whether the respondent notices delayed healing of the wound | Yes: 213 (41.92%) No: 295 (58.08%) |
| 26 | Muscle stiffness | Whether the respondent notices muscle stiffness of their body | Yes: 234 (46.06%) No: 274 (53.94%) |
| 27 | Partial paralysis. | Partial paralysis of any part of the body is noticed or not | Yes: 69 (13.58%) No: 396 (77.95%) May be: 33 (8.46%) |
| 28 | Hair loss | Whether the respondent notices gradually losing hair or not | Yes: 231 (45.47%) No: 203 (39.76%) May be: 70 (14.76%) |
| 29 | Other diseases | Whether the respondent has other serious diseases | Yes: 260 (51.18%) No: 248 (48.82%) |
| 30 | Number of dependent family members | Total number of dependent family members | Avg.: 3.43 Max: 15 Min: 0 Median: 3 |
| 31 | Living house type | Respondent home can be rental or owned | Owned: 409 (80.51%) Rented: 99 (19.49%) |
| 32 | Anxiety | Whether the respondent has extreme anxiety or not | Yes: 293 (57.67%) No: 215 (42.33%) |
| 33 | Diabetes (output factor) | Diabetes, non-diabetes | Yes: 275 (54.14%) No: 233 (45.86%) |
| Classifier | DT | LR | SVM | XGB | GB | RF | ET |
|---|---|---|---|---|---|---|---|
| Accuracy | 0.813 | 0.8386 | 0.8602 | 0.8661 | 0.8681 | 0.874 | 0.876 |
| Precision | 0.8409 | 0.8459 | 0.8864 | 0.8791 | 0.8741 | 0.8809 | 0.8926 |
| Recall | 0.8073 | 0.8582 | 0.8509 | 0.8727 | 0.8836 | 0.8873 | 0.8764 |
| ROC-AUC | 0.8135 | 0.8368 | 0.8611 | 0.8655 | 0.8667 | 0.8728 | 0.8760 |
| F1-Score | 0.8237 | 0.852 | 0.8683 | 0.8759 | 0.8788 | 0.8841 | 0.8844 |
| Geometric Mean | 0.8135 | 0.8365 | 0.861 | 0.8655 | 0.8665 | 0.8727 | 0.8759 |
| Log-Loss | 6.7404 | 5.8181 | 5.0376 | 4.8247 | 4.7538 | 4.5409 | 4.47 |
| Classifier | DT | LR | SVM | XGB | GB | RF | ET |
|---|---|---|---|---|---|---|---|
| Accuracy | 0.8018 | 0.8509 | 0.8655 | 0.8691 | 0.8673 | 0.8636 | 0.8745 |
| Precision | 0.8029 | 0.8561 | 0.8708 | 0.8664 | 0.8686 | 0.8676 | 0.8705 |
| Recall | 0.8 | 0.8436 | 0.8582 | 0.8727 | 0.8655 | 0.8582 | 0.88 |
| ROC-AUC | 0.8018 | 0.8509 | 0.8655 | 0.8691 | 0.8673 | 0.8636 | 0.8745 |
| F1-Score | 0.8015 | 0.8498 | 0.8645 | 0.8696 | 0.867 | 0.8629 | 0.8752 |
| Geometric Mean | 0.8018 | 0.8509 | 0.8654 | 0.8691 | 0.8673 | 0.8636 | 0.8745 |
| Log-Loss | 7.1432 | 5.3738 | 4.8495 | 4.7184 | 4.784 | 4.915 | 4.5218 |
| Classifier | DT | LR | SVM | XGB | GB | RF | ET |
|---|---|---|---|---|---|---|---|
| Accuracy | 0.8473 | 0.8527 | 0.8691 | 0.8873 | 0.8764 | 0.8891 | 0.8927 |
| Precision | 0.8577 | 0.8514 | 0.883 | 0.8959 | 0.8906 | 0.8934 | 0.8971 |
| Recall | 0.8327 | 0.8545 | 0.8509 | 0.8764 | 0.8582 | 0.8836 | 0.8873 |
| ROC-AUC | 0.8473 | 0.8527 | 0.8691 | 0.8873 | 0.8764 | 0.8891 | 0.8927 |
| F1-Score | 0.845 | 0.853 | 0.8667 | 0.886 | 0.8741 | 0.8885 | 0.8921 |
| Geometric Mean | 0.8471 | 0.8527 | 0.8689 | 0.8872 | 0.8762 | 0.8891 | 0.8927 |
| Log Loss | 5.5048 | 5.3082 | 4.7184 | 4.0631 | 4.4563 | 3.9976 | 3.8665 |
| Classifier | DT | LR | SVM | XGB | GB | RF | ET | ET (Tuning) |
|---|---|---|---|---|---|---|---|---|
| Accuracy | 0.8364 | 0.8545 | 0.8673 | 0.8782 | 0.8655 | 0.8764 | 0.8818 | 0.9927 |
| Precision | 0.8627 | 0.847 | 0.8633 | 0.891 | 0.8764 | 0.8848 | 0.8977 | 1 |
| Recall | 0.8 | 0.8655 | 0.8727 | 0.8618 | 0.8509 | 0.8655 | 0.8618 | 0.9855 |
| ROC-AUC | 0.8364 | 0.8545 | 0.8673 | 0.8782 | 0.8655 | 0.8764 | 0.8818 | 0.9927 |
| F1-Score | 0.8302 | 0.8561 | 0.868 | 0.8762 | 0.8635 | 0.875 | 0.8794 | 0.9927 |
| Geometric Mean | 0.8356 | 0.8545 | 0.8673 | 0.878 | 0.8653 | 0.8763 | 0.8816 | 0.9927 |
| Log-Loss | 5.8981 | 5.2427 | 4.784 | 4.3908 | 4.8495 | 4.4563 | 4.2597 | 0.2621 |
| Feature Name | DT | SVM | LR | RF | XGB | GB | ET | Avg. | Rank |
|---|---|---|---|---|---|---|---|---|---|
| Age | 1 | 0.95 | 1 | 1 | 1 | 1 | 1 | 0.99 | 1 |
| Having diabetes in family | 0.26 | 1 | 0.94 | 0.45 | 0.34 | 0.23 | 0.23 | 0.49 | 2 |
| Regular intake of medicine | 0.08 | 0.89 | 0.79 | 0.48 | 0.13 | 0.2 | 0.19 | 0.39 | 3 |
| Extreme thirst | 0.08 | 0.82 | 0.75 | 0.39 | 0.15 | 0.13 | 0.19 | 0.36 | 4 |
| Occupation | 0.06 | 0.77 | 0.67 | 0.17 | 0.03 | 0.04 | 0.06 | 0.26 | 5 |
| Frequency of urination | 0.04 | 0.66 | 0.57 | 0.06 | 0.05 | 0.04 | 0.04 | 0.21 | 6 |
| Walk/ Run/ Physically exercise | 0.06 | 0.62 | 0.56 | 0.13 | 0.01 | 0.02 | 0.03 | 0.21 | 6 |
| Weakness | 0.06 | 0.57 | 0.58 | 0.11 | 0.01 | 0.02 | 0.05 | 0.2 | 7 |
| Smoking | 0.05 | 0.67 | 0.66 | 0 | 0.01 | 0 | 0.01 | 0.2 | 7 |
| Junk food consumption | 0.05 | 0.64 | 0.6 | 0.02 | 0.03 | 0 | 0.01 | 0.19 | 8 |
| Partial paralysis | 0 | 0.6 | 0.58 | 0.06 | 0.05 | 0.02 | 0.03 | 0.19 | 8 |
| Education level | 0.08 | 0.36 | 0.37 | 0.22 | 0.02 | 0.04 | 0.09 | 0.17 | 9 |
| Hypertension | 0.01 | 0.51 | 0.53 | 0.09 | 0 | 0.01 | 0.02 | 0.17 | 9 |
| Gender | 0.18 | 0 | 0 | 0.48 | 0.21 | 0.14 | 0.21 | 0.17 | 9 |
| Hair loss | 0.05 | 0.43 | 0.38 | 0.12 | 0.01 | 0.01 | 0.04 | 0.15 | 10 |
| Anxiety | 0.02 | 0.43 | 0.37 | 0.05 | 0.01 | 0 | 0.02 | 0.13 | 11 |
| Wealth index | 0.01 | 0.34 | 0.33 | 0.04 | 0.02 | 0 | 0.02 | 0.11 | 12 |
| Stress | 0.04 | 0.25 | 0.25 | 0.1 | 0.03 | 0.02 | 0.04 | 0.11 | 12 |
| Living house type | 0.02 | 0.38 | 0.34 | 0 | 0 | 0 | 0 | 0.11 | 12 |
| Muscle stiffness | 0.04 | 0.26 | 0.27 | 0.06 | 0.01 | 0.01 | 0.05 | 0.1 | 13 |
| Reference | Dataset | Accuracy | Precision | Recall | ROC-AUC | F1-Score | Geometric Mean | Log-Loss |
|---|---|---|---|---|---|---|---|---|
| Pranto et al. [31] | PIMA, Kurmitola Hospital, Dhaka | 0.8120 | 0.8 | 1 | 0.84 | 0.88 | – | – |
| Syed and Khan [69] | Western Region of Saudi Arabia | 0.821 | 0.776 | 0.89 | 0.867 | 0.829 | – | – |
| Chou et al. [70] | Taipei Municipal Medical Center | 0.953 | 0.927 | 0.931 | 0.991 | 0.929 | – | – |
| Laila et al. [71] | UCI Repository | 0.9711 | 0.971 | 0.971 | – | 0.971 | – | – |
| This study | Bangladesh, 2022 | 0.9927 | 1 | 0.9855 | 0.9927 | 0.9927 | 0.9927 | 0.2621 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uddin, M.J.; Ahamad, M.M.; Hoque, M.N.; Walid, M.A.A.; Aktar, S.; Alotaibi, N.; Alyami, S.A.; Kabir, M.A.; Moni, M.A. A Comparison of Machine Learning Techniques for the Detection of Type-2 Diabetes Mellitus: Experiences from Bangladesh. Information 2023, 14, 376. https://doi.org/10.3390/info14070376
Uddin MJ, Ahamad MM, Hoque MN, Walid MAA, Aktar S, Alotaibi N, Alyami SA, Kabir MA, Moni MA. A Comparison of Machine Learning Techniques for the Detection of Type-2 Diabetes Mellitus: Experiences from Bangladesh. Information. 2023; 14(7):376. https://doi.org/10.3390/info14070376
Chicago/Turabian StyleUddin, Md. Jamal, Md. Martuza Ahamad, Md. Nesarul Hoque, Md. Abul Ala Walid, Sakifa Aktar, Naif Alotaibi, Salem A. Alyami, Muhammad Ashad Kabir, and Mohammad Ali Moni. 2023. "A Comparison of Machine Learning Techniques for the Detection of Type-2 Diabetes Mellitus: Experiences from Bangladesh" Information 14, no. 7: 376. https://doi.org/10.3390/info14070376
APA StyleUddin, M. J., Ahamad, M. M., Hoque, M. N., Walid, M. A. A., Aktar, S., Alotaibi, N., Alyami, S. A., Kabir, M. A., & Moni, M. A. (2023). A Comparison of Machine Learning Techniques for the Detection of Type-2 Diabetes Mellitus: Experiences from Bangladesh. Information, 14(7), 376. https://doi.org/10.3390/info14070376










