Semi-Supervised Learning for Multi-View Data Classification and Visualization
Abstract
:1. Introduction
- It benefits from multiple descriptors for each sample. This can improve the unified graph estimation, which can well represent the relationship between nodes.
- It automatically fuses the information of different views.
- It not only uses the information in the feature space but also uses the available information in the label space.
- It does not require pre-constructed graphs based on individual views but can construct a single graph by considering the information from different views.
- The process of constructing the affinity graph and propagating the labels is simultaneously carried out in a single framework.
- It estimates a linear projection function to estimate the label of unseen samples.
2. Related Work
2.1. Notations
2.2. Gaussian Field and Harmonic Functions [26]
2.3. Local and Global Consistency [27]
2.4. Review on Flexible Manifold Embedding [25]
2.5. Data Smoothness Assumption
3. Proposed Method
3.1. Learning Model
3.2. Optimization
4. Experimental Results
4.1. Databases
4.2. Image Descriptors
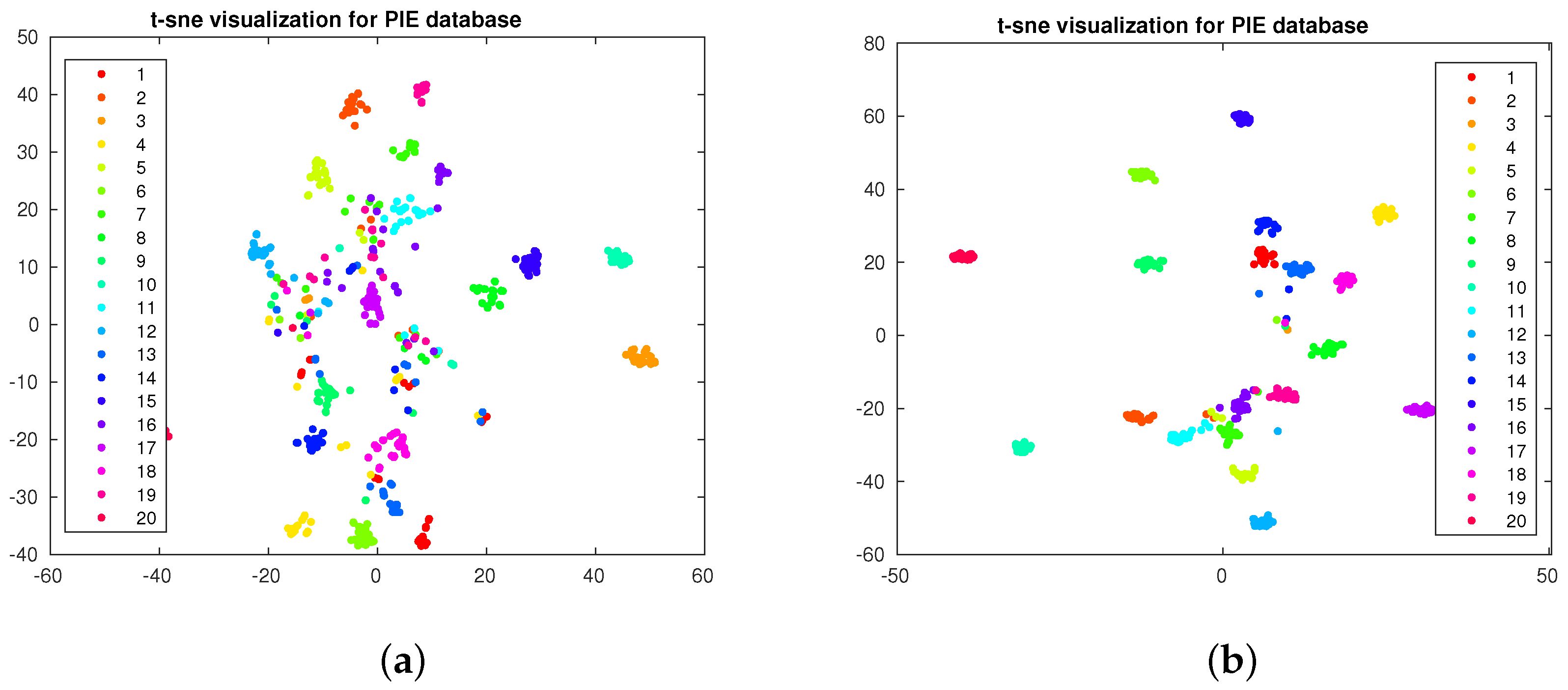
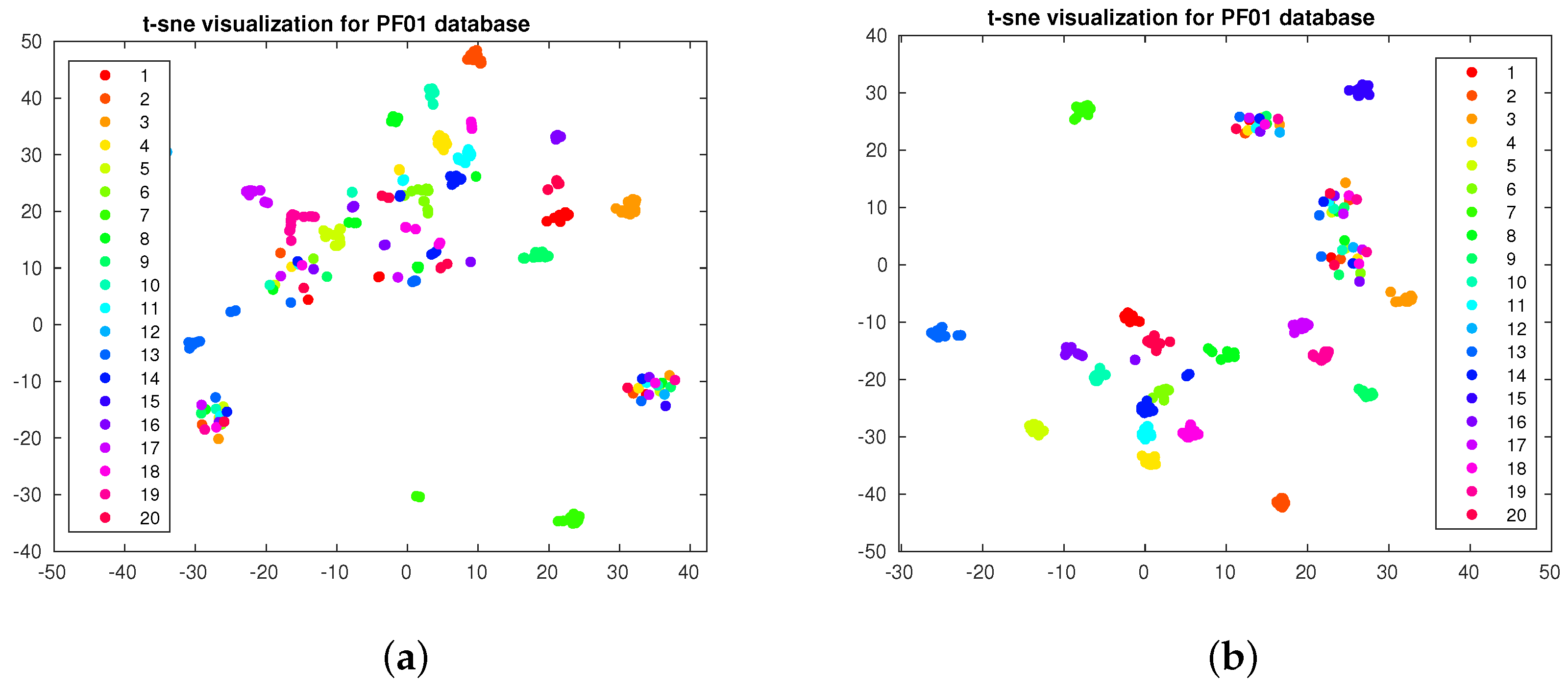
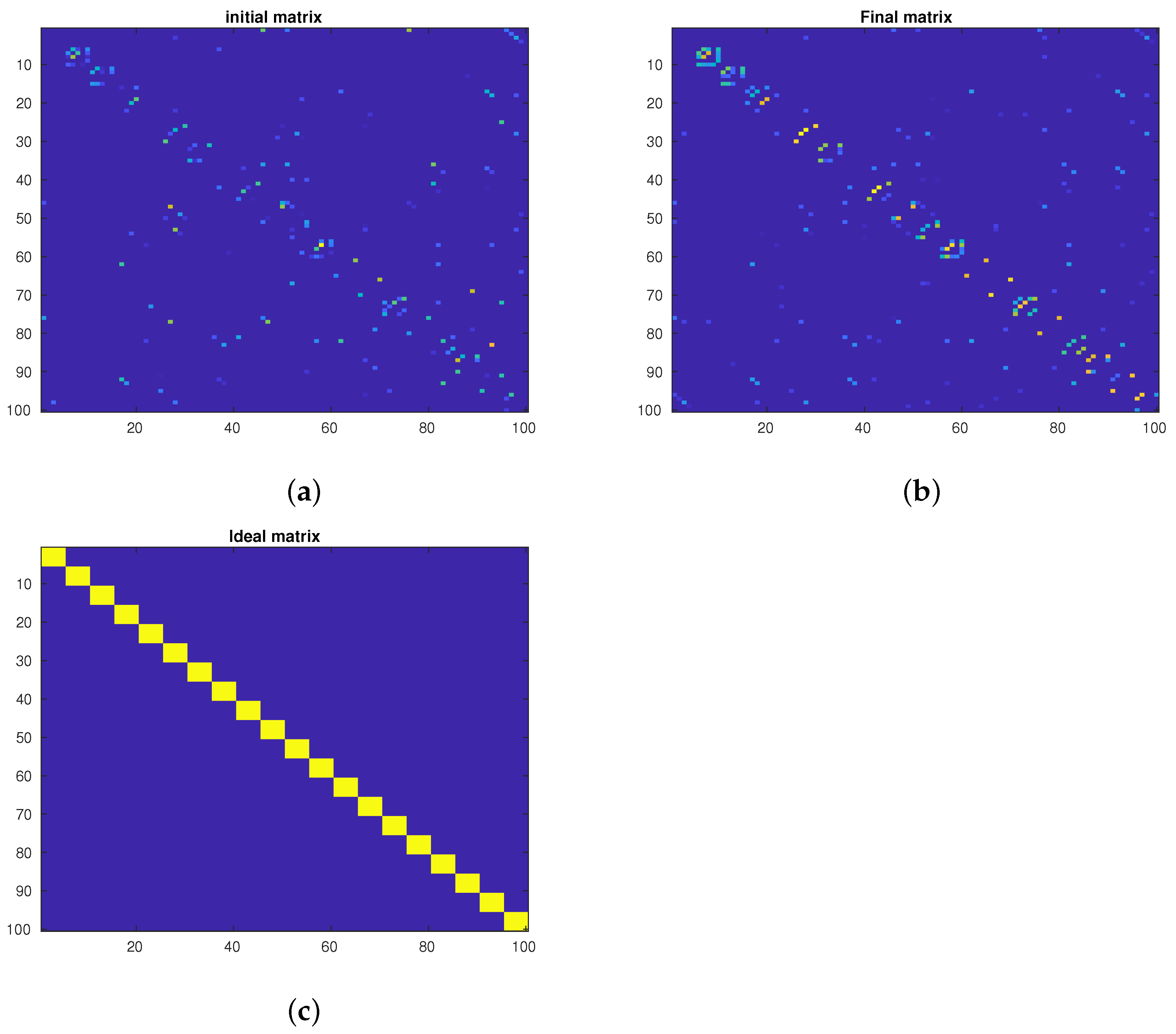
4.3. Data Visualization
4.4. Small Databases
4.5. Large Databases
4.6. Ablation Study
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, Q. Overview of Data Visualization. In Embodying Data: Chinese Aesthetics, Interactive Visualization and Gaming Technologies; Springer: Singapore, 2020; pp. 17–47. [Google Scholar] [CrossRef]
- Szubert, B.; Cole, J.E.; Monaco, C.; Drozdov, I. Structure-preserving visualisation of high dimensional single-cell datasets. Sci. Rep. 2019, 9, 8914. [Google Scholar] [CrossRef]
- van der Maaten, L.; Hinton, G. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- McInnes, L.; Healy, J.; Saul, N.; Großberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 2018, 3, 861. [Google Scholar] [CrossRef]
- Sainburg, T.; McInnes, L.; Gentner, T.Q. Parametric UMAP Embeddings for Representation and Semisupervised Learning. Neural Comput. 2021, 33, 2881–2907. [Google Scholar] [CrossRef]
- Nie, F.; Wang, Z.; Wang, R.; Li, X. Adaptive Local Embedding Learning for Semi-supervised Dimensionality Reduction. IEEE Trans. Knowl. Data Eng. 2021, 34, 4609–4621. [Google Scholar] [CrossRef]
- He, F.; Nie, F.; Wang, R.; Hu, H.; Jia, W.; Li, X. Fast Semi-Supervised Learning with Optimal Bipartite Graph. IEEE Trans. Knowl. Data Eng. 2020, 33, 3245–3257. [Google Scholar] [CrossRef]
- Dornaika, F.; Bosaghzadeh, A.; Raducanu, B. Efficient Graph Construction for Label Propagation Based Multi-observation Face Recognition. In Human Behavior Understanding; Salah, A.A., Hung, H., Aran, O., Gunes, H., Eds.; Springer: Cham, Switzerland, 2013; pp. 124–135. [Google Scholar]
- Bahrami, S.; Bosaghzadeh, A.; Dornaika, F. Multi Similarity Metric Fusion in Graph-Based Semi-Supervised Learning. Computation 2019, 7, 15. [Google Scholar] [CrossRef]
- Zheng, F.; Liu, Z.; Chen, Y.; An, J.; Zhang, Y. A Novel Adaptive Multi-View Non-Negative Graph Semi-Supervised ELM. IEEE Access 2020, 8, 116350–116362. [Google Scholar] [CrossRef]
- Li, L.; He, H. Bipartite Graph based Multi-view Clustering. IEEE Trans. Knowl. Data Eng. 2020, 34, 3111–3125. [Google Scholar] [CrossRef]
- Angelou, M.; Solachidis, V.; Vretos, N.; Daras, P. Graph-based multimodal fusion with metric learning for multimodal classification. Pattern Recognit. 2019, 95, 296–307. [Google Scholar] [CrossRef]
- Nie, F.; Tian, L.; Wang, R.; Li, X. Multiview Semi-Supervised Learning Model for Image Classification. IEEE Trans. Knowl. Data Eng. 2019, 32, 2389–2400. [Google Scholar] [CrossRef]
- Wang, H.; Yang, Y.; Liu, B.; Fujita, H. A study of graph-based system for multi-view clustering. Knowl.-Based Syst. 2019, 163, 1009–1019. [Google Scholar] [CrossRef]
- Manna, S.; Khonglah, J.R.; Mukherjee, A.; Saha, G. Robust kernelized graph-based learning. Pattern Recognit. 2021, 110, 107628. [Google Scholar] [CrossRef]
- Kang, Z.; Shi, G.; Huang, S.; Chen, W.; Pu, X.; Zhou, J.T.; Xu, Z. Multi-graph fusion for multi-view spectral clustering. Knowl.-Based Syst. 2020, 189, 105102. [Google Scholar] [CrossRef]
- Karasuyama, M.; Mamitsuka, H. Multiple Graph Label Propagation by Sparse Integration. IEEE Trans. Neural Netw. Learn. Syst. 2013, 24, 1999–2012. [Google Scholar] [CrossRef]
- An, L.; Chen, X.; Yang, S. Multi-graph feature level fusion for person re-identification. Neurocomputing 2017, 259, 39–45. [Google Scholar] [CrossRef]
- Wang, B.; Mezlini, A.M.; Demir, F.; Fiume, M.; Tu, Z.; Brudno, M.; Haibe-Kains, B.; Goldenberg, A. Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods 2014, 11, 333–337. [Google Scholar] [CrossRef]
- Bahrami, S.; Dornaika, F.; Bosaghzadeh, A. Joint auto-weighted graph fusion and scalable semi-supervised learning. Inf. Fusion 2021, 66, 213–228. [Google Scholar] [CrossRef]
- Lin, G.; Liao, K.; Sun, B.; Chen, Y.; Zhao, F. Dynamic graph fusion label propagation for semi-supervised multi-modality classification. Pattern Recognit. 2017, 68, 14–23. [Google Scholar] [CrossRef]
- Nie, F.; Cai, G.; Li, X. Multi-View Clustering and Semi-Supervised Classification with Adaptive Neighbours. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Kang, Z.; Peng, C.; Cheng, Q.; Liu, X.; Peng, X.; Xu, Z.; Tian, L. Structured graph learning for clustering and semi-supervised classification. Pattern Recognit. 2021, 110, 107627. [Google Scholar] [CrossRef]
- Deng, J.; Yu, J.G. A simple graph-based semi-supervised learning approach for imbalanced classification. Pattern Recognit. 2021, 118, 108026. [Google Scholar] [CrossRef]
- Nie, F.; Xu, D.; Tsang, I.W.; Zhang, C. Flexible Manifold Embedding: A Framework for Semi-Supervised and Unsupervised Dimension Reduction. IEEE Trans. Image Process. 2010, 19, 1921–1932. [Google Scholar] [CrossRef]
- Zhu, X.; Ghahramani, Z.; Lafferty, J.D. Semi-supervised learning using gaussian fields and harmonic functions. In Proceedings of the 20th International conference on Machine learning (ICML-03), Washington, DC, USA, 21–24 August 2003; pp. 912–919. [Google Scholar]
- Zhou, D.; Bousquet, O.; Lal, T.N.; Weston, J.; Schölkopf, B. Learning with Local and Global Consistency. In Advances in Neural Information Processing Systems 16; Thrun, S., Saul, L.K., Schölkopf, B., Eds.; MIT Press: Cambridge, MA, USA, 2004; pp. 321–328. [Google Scholar]
- Budd, S.; Robinson, E.C.; Kainz, B. A survey on active learning and human-in-the-loop deep learning for medical image analysis. Med. Image Anal. 2021, 71, 102062. [Google Scholar] [CrossRef]
- Tharwat, A.; Schenck, W. A Survey on Active Learning: State-of-the-Art, Practical Challenges and Research Directions. Mathematics 2023, 11, 820. [Google Scholar] [CrossRef]
- Ferdinands, G.; Schram, R.; de Bruin, J.; Bagheri, A.; Oberski, D.L.; Tummers, L.; Teijema, J.J.; van de Schoot, R. Performance of active learning models for screening prioritization in systematic reviews: A simulation study into the Average Time to Discover relevant records. Syst. Rev. 2023, 12, 100. [Google Scholar] [CrossRef]
- Nie, F.; Li, J.; Li, X. Parameter-free Auto-weighted Multiple Graph Learning: A Framework for Multiview Clustering and Semi-supervised Classification. In Proceedings of the Twenty-Fifth International Joint Conference on Artificial Intelligence IJCAI’16, New York, NY, USA, 9–15 July 2016; AAAI Press: Palo Alto, CA, USA, 2016; pp. 1881–1887. [Google Scholar]
- Dong, H.; Gu, N. Asian face image database PF01. 2001. Available online: http://imlab.postech.ac.kr/databases.htm (accessed on 5 January 2006).
- Sim, T.; Baker, S.; Bsat, M. The CMU Pose, Illumination, and Expression (PIE) database. In Proceedings of the Fifth IEEE International Conference on Automatic Face Gesture Recognition, Washington, DC, USA, 21 May 2002; pp. 46–51. [Google Scholar] [CrossRef]
- Phillips, P.J.; Moon, H.; Rizvi, S.A.; Rauss, P.J. The FERET Evaluation Methodology for Face-Recognition Algorithms. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 1090–1104. [Google Scholar] [CrossRef]
- Lecun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Ahonen, T.; Hadid, A.; Pietikainen, M. Face Description with Local Binary Patterns: Application to Face Recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2006, 28, 2037–2041. [Google Scholar] [CrossRef]
- Shen, L.; Bai, L. A review on Gabor wavelets for face recognition. Pattern Anal. Appl. 2006, 9, 273–292. [Google Scholar] [CrossRef]
- Tuzel, O.; Porikli, F.; Meer, P. Region Covariance: A Fast Descriptor for Detection and Classification. In Proceedings of the Computer Vision—ECCV 2006, Graz, Austria, 7–13 May 2006; Leonardis, A., Bischof, H., Pinz, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 589–600. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Sedrakyan, G.; Mannens, E.; Verbert, K. Guiding the choice of learning dashboard visualizations: Linking dashboard design and data visualization concepts. J. Comput. Lang. 2019, 50, 19–38. [Google Scholar] [CrossRef]
- Gong, C.; Tao, D.; Maybank, S.J.; Liu, W.; Kang, G.; Yang, J. Multi-Modal Curriculum Learning for Semi-Supervised Image Classification. IEEE Trans. Image Process. 2016, 25, 3249–3260. [Google Scholar] [CrossRef] [PubMed]
| Dataset | Num. of Classes | Num. of Samples | Num. of Samples in Each Class | Extracted Descriptors | Dimension |
|---|---|---|---|---|---|
| PF01 | 107 | 1819 | 17 | Cov [38] | 405 |
| Gabor [37] | 2560 | ||||
| LBP [36] | 900 | ||||
| PIE | 68 | 1926 | 27∼29 | Cov [38] | 405 |
| Gabor [37] | 2560 | ||||
| LBP [36] | 900 | ||||
| FERET | 200 | 1400 | 7 | Cov [38] | 405 |
| Gabor [37] | 2560 | ||||
| LBP [36] | 900 | ||||
| MNIST | 10 | 60,000 | 5421∼6742 | VGG16 [39] | 4096 |
| VGG19 [39] | 4096 |
| PIE | ||||||
|---|---|---|---|---|---|---|
| Method | Cov | Gabor | LBP | Concat. | JGC-MFME | |
| Num. of Lab. Samples/Class | ||||||
| 15 | 80.93 ± 3.71 | 91.79 ± 3.32 | 87.75 ± 3.90 | 94.31 ± 2.88 | 94.44 ± 2.76 | |
| 17 | 81.93 ± 4.06 | 91.09 ± 3.08 | 88.11 ± 2.99 | 94.54 ± 2.51 | 94.75 ± 2.58 | |
| FERET | ||||||
| Method | Cov | Gabor | LBP | Concat. | JGC-MFME | |
| Num. of Lab. Samples/Class | ||||||
| 3 | 59.17 ± 9.53 | 79.80 ± 8.96 | 72.01 ± 10.3 | 83.98 ± 8.06 | 85.50 ± 7.64 | |
| 5 | 67.27 ± 18.67 | 86.00 ± 15.38 | 85.15 ± 17.66 | 90.72 ± 12.98 | 91.77 ± 12.98 | |
| PF01 | ||||||
| Method | Cov | Gabor | LBP | Concat. | JGC-MFME | |
| Num. of Lab. Samples/Class | ||||||
| 5 | 70.24 ± 3.34 | 84.26 ± 3.76 | 79.57 ± 2.55 | 91.30 ± 2.03 | 92.28 ± 1.61 | |
| 7 | 71.82 ± 5.93 | 85.81 ± 4.93 | 80.71 ± 4.51 | 93.22 ± 2.39 | 94.13 ± 2.01 | |
| MNIST (Accuracy on Unlabeled Data) | ||||
|---|---|---|---|---|
| Lab./class | 20 | 30 | 40 | |
| Method | ||||
| Single descriptor (VGG16) | 94.53 ± 0.3 | 95.17 ± 0.3 | 95.59 ± 0.3 | |
| Single descriptor (VGG19) | 94.35 ± 0.5 | 94.99 ± 0.5 | 95.35 ± 0.4 | |
| Feature Concat. | 95.87 ± 0.4 | 96.28 ± 0.4 | 96.66 ± 0.3 | |
| SNF [19] | 94.53 ± 0.6 | 95.81 ± 0.4 | 96.49 ± 0.3 | |
| SMGI [17] | 88.40 ± 0.9 | 89.66 ± 0.7 | 89.84 ± 0.7 | |
| DGFLP [21] | 92.14 ± 0.5 | 92.70 ± 0.5 | 93.06 ± 0.3 | |
| MLGC [18] | 88.20 ± 1.4 | 89.45 ± 1.0 | 90.27 ± 0.7 | |
| AMGL [31] | 94.99 ± 0.5 | 95.39 ± 0.3 | 95.67 ± 0.3 | |
| MMCL [41] | 52.45 ± 14.5 | 49.98 ± 13.9 | 49.22 ± 13 | |
| JGC-MFME | 95.91 ± 0.4 | 96.34 ± 0.4 | 96.68 ± 0.3 | |
| MNIST (Accuracy on Test Data) | ||||
| Lab./class | 20 | 30 | 40 | |
| Method | ||||
| Single descriptor (VGG16) | 94.98 ± 0.5 | 95.39 ± 0.3 | 95.81 ± 0.3 | |
| Single descriptor (VGG19) | 95.05 ± 0.5 | 95.64 ± 0.4 | 95.97 ± 0.4 | |
| Feature Concat. | 95.47 ± 0.5 | 96.79 ± 0.5 | 96.21 ± 0.4 | |
| SNF [19] | 94.79 ± 0.6 | 95.97 ± 0.3 | 96.64 ± 0.2 | |
| JGC-MFME | 95.92 ± 0.4 | 95.36 ± 0.3 | 96.76 ± 0.3 | |
| Database | Evaluation Metric | Prop. Method | Prop. Method (Without PCA) |
|---|---|---|---|
| PF01 7 labeled | Acc (%) | 95.14 | 93.36 |
| CPU time (Sec) | 23 | 65 | |
| PIE 17 labeled | Acc (%) | 90.12 | 89.61 |
| CPU time (Sec) | 20 | 47 |
| Database | Prop. Method | |||||
|---|---|---|---|---|---|---|
| PF01 7 labeled | 95.14 | 94.95 | 60.84 | 60.84 | 0.93 | 0.93 |
| PIE 17 labeled | 90.12 | 90 | 66.1 | 66.1 | 1.55 | 1.55 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ziraki, N.; Bosaghzadeh, A.; Dornaika, F. Semi-Supervised Learning for Multi-View Data Classification and Visualization. Information 2024, 15, 421. https://doi.org/10.3390/info15070421
Ziraki N, Bosaghzadeh A, Dornaika F. Semi-Supervised Learning for Multi-View Data Classification and Visualization. Information. 2024; 15(7):421. https://doi.org/10.3390/info15070421
Chicago/Turabian StyleZiraki, Najmeh, Alireza Bosaghzadeh, and Fadi Dornaika. 2024. "Semi-Supervised Learning for Multi-View Data Classification and Visualization" Information 15, no. 7: 421. https://doi.org/10.3390/info15070421






