Enhancing Biomedical Question Answering with Large Language Models
Abstract
:1. Introduction
2. Related Work
2.1. Biomedical Question Answering Systems
2.1.1. MedQA: A Medical Quality Assurance System
2.1.2. HONQA: Quality Assurance via Certified Websites
2.1.3. EAGLi: Extracting Answers from MEDLINE Records
2.1.4. AskHERMES: Clinical QA with Concise Summaries
2.1.5. SemBT: Biomedical QA via Semantic Relations
2.2. Text Re-Ranking
2.3. Retrieval Models
3. Methodology
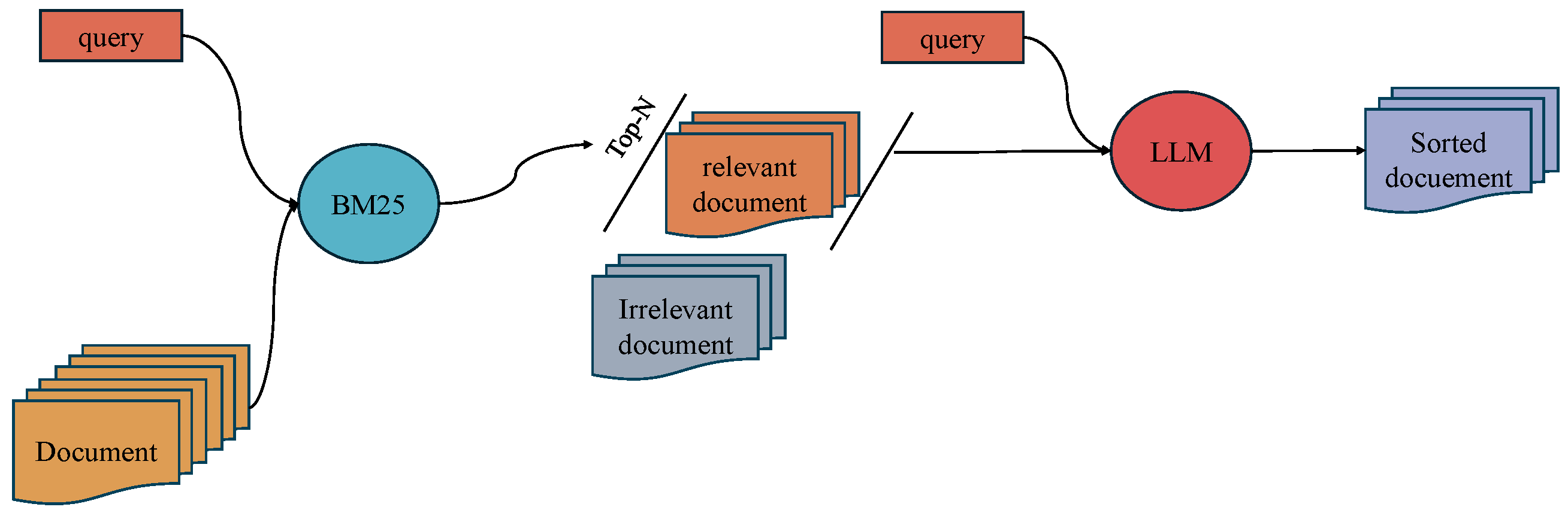
3.1. Two-Stage Retrieval
3.2. Prompt Strategy
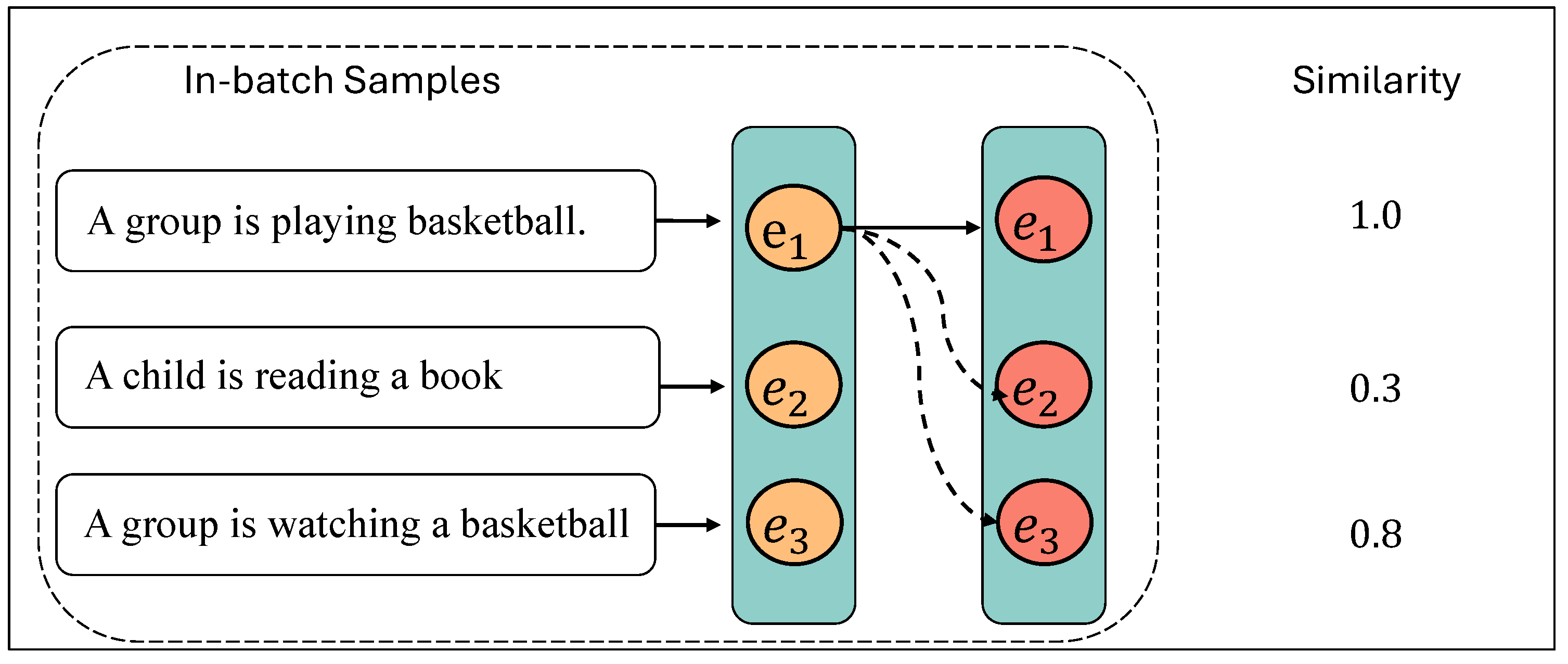
3.3. Hard Negative Mining and Data Preparation
4. Datasets Analysis
4.1. BioASQ
4.2. TREC-COVID
5. Experiments and Results
5.1. Setup
5.2. Evaluation Metrics
5.3. Indexing
5.4. Fine-Tuned BM25 Model
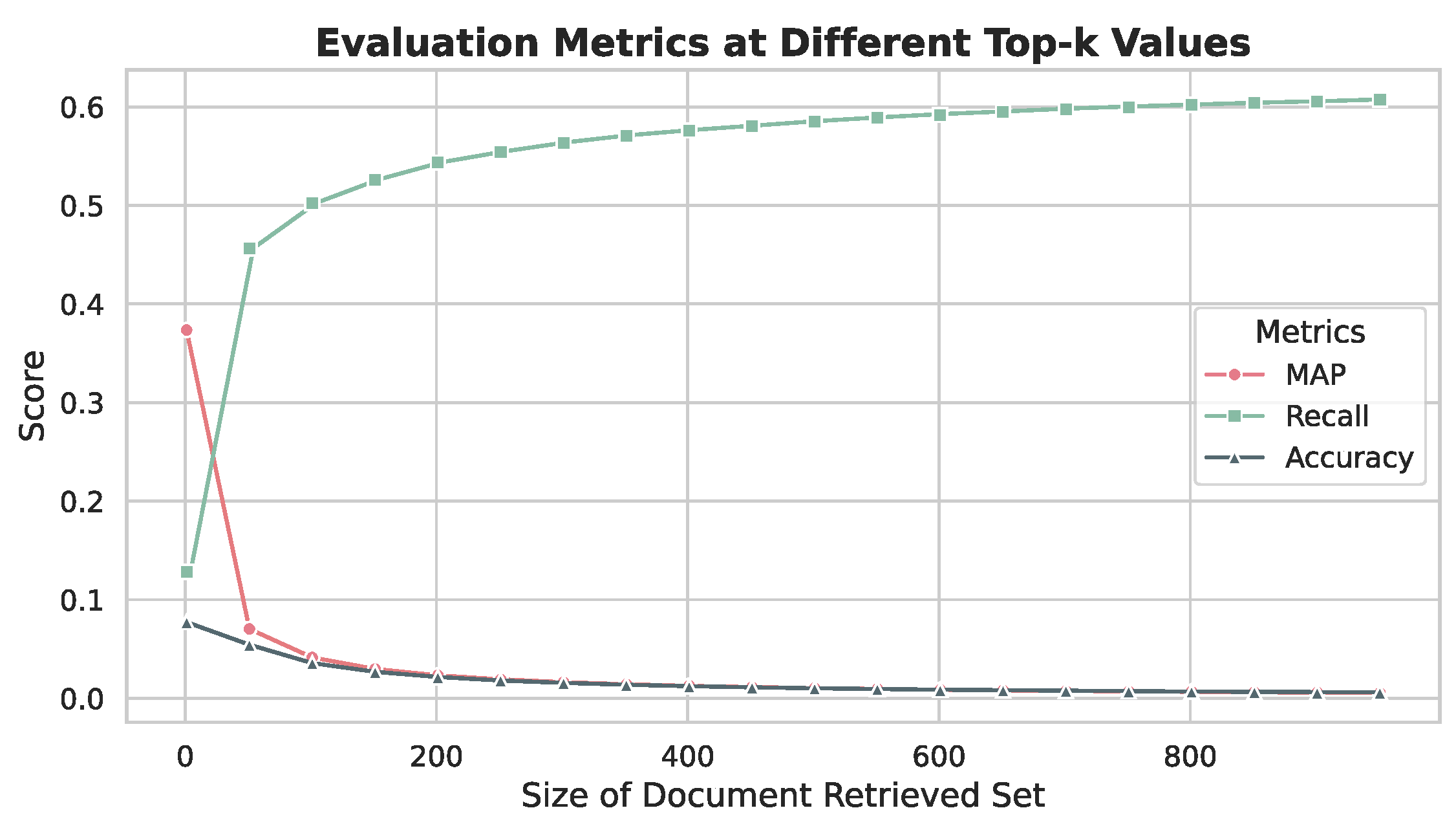
5.5. LLM Selection and k-Value Optimization
5.5.1. Experimental Setup
5.5.2. Results and Analysis
5.5.3. Optimal Configuration
5.6. BioASQ Results Analysis
5.7. Results on TREC-COVID Dataset
- BM25 [20]: A classical probabilistic retrieval model that serves as a fundamental sanity check by directly using the ranking results from the first-stage retrieval.
- Sentence-BERT [64]: A modification of the BERT model that uses Siamese and triplet network structures to derive semantically meaningful sentence embeddings.
- DPR [22]: A dense retrieval method that uses neural networks to encode queries and passages into a low-dimensional space.
- docT5query [43]: A document expansion technique that uses a sequence-to-sequence model to predict queries that a document might be relevant to.
- Manual Prompt: To benchmark against cutting-edge manual prompting techniques, we incorporated RankGPT [65], the current state-of-the-art approach in this domain.
- CoT [66]: This approach extends the manual prompt by appending the phrase “Let’s think step by step” to encourage more structured and detailed reasoning.
- ListT5 [67]: A re-ranking methodology leveraging the Fusion-in-Decoder architecture, which processes multiple candidate passages concurrently during both training and inference phases.
- COCO-DR [68]: A zero-shot dense retrieval method designed to enhance generalization by addressing distribution shifts between training and target scenarios.
- RaMDA [69]: A novel model addressing the domain adaptation for dense retrievers by synthesizing domain-specific data through pseudo queries.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AP | Average Precision |
| BERT | Bidirectional Encoder Representations from Transformers |
| BM25 | Best Match 25 |
| CoT | Chain of Thought |
| DCG | Discounted Cumulative Gain |
| DESM | Dual Embedding Space Model |
| DPR | Dense Passage Retrieval |
| DSSM | Deep Structured Semantic Models |
| EBAE | Embedding-Based Auto-Encoding |
| EBAR | Embedding-Based Auto-Regression |
| FN | False Negatives |
| FP | False Positives |
| GMAP | Geometric Mean Average Precision |
| IDCG | Ideal Discounted Cumulative Gain |
| IR | Information Retrieval |
| LoRA | Low-Rank Adaptation |
| MAP | Mean Average Precision |
| NDCG | Normalized Discounted Cumulative Gain |
| QA | Question Answering |
| RDF | Resource Description Framework |
| TREC-COVID | Text REtrieval Conference COVID |
| TP | True Positives |
References
- Qiu, M.; Li, F.L.; Wang, S.; Gao, X.; Chen, Y.; Zhao, W.; Chen, H.; Huang, J.; Chu, W. Alime chat: A sequence to sequence and rerank based chatbot engine. In Proceedings of the 55th Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers), Vancouver, BC, Canada, 30 July–4 August 2017; pp. 498–503. [Google Scholar]
- Yan, Z.; Duan, N.; Bao, J.; Chen, P.; Zhou, M.; Li, Z.; Zhou, J. Docchat: An information retrieval approach for chatbot engines using unstructured documents. In Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), Berlin, Germany, 7–12 August 2016; pp. 516–525. [Google Scholar]
- Amato, F.; Marrone, S.; Moscato, V.; Piantadosi, G.; Picariello, A.; Sansone, C. Chatbots Meet eHealth: Automatizing Healthcare. In Proceedings of the WAIAH@ AI* IA, Bari, Italy, 14 November 2017; pp. 40–49. [Google Scholar]
- Ram, A.; Prasad, R.; Khatri, C.; Venkatesh, A.; Gabriel, R.; Liu, Q.; Nunn, J.; Hedayatnia, B.; Cheng, M.; Nagar, A.; et al. Conversational ai: The science behind the alexa prize. arXiv 2018, arXiv:1801.03604. [Google Scholar]
- Kadam, A.D.; Joshi, S.D.; Shinde, S.V.; Medhane, S.P. Notice of Removal: Question Answering Search engine short review and road-map to future QA Search Engine. In Proceedings of the 2015 International Conference on Electrical, Electronics, Signals, Communication and Optimization (EESCO), Visakhapatnam, India, 24–25 January 2015; pp. 1–8. [Google Scholar] [CrossRef]
- Yaghoubzadeh, R.; Kopp, S. Toward a virtual assistant for vulnerable users: Designing careful interaction. In Proceedings of the 1st Workshop on Speech and Multimodal Interaction in Assistive Environments, Jeju Island, Republic of Korea, 12 July 2012; pp. 13–17. [Google Scholar]
- Austerjost, J.; Porr, M.; Riedel, N.; Geier, D.; Becker, T.; Scheper, T.; Marquard, D.; Lindner, P.; Beutel, S. Introducing a virtual assistant to the lab: A voice user interface for the intuitive control of laboratory instruments. SLAS TECHNOLOGY Transl. Life Sci. Innov. 2018, 23, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Bradley, N.C.; Fritz, T.; Holmes, R. Context-aware conversational developer assistants. In Proceedings of the 40th International Conference on Software Engineering, Gothenburg, Sweden, 27 May–3 June 2018; pp. 993–1003. [Google Scholar]
- Vicedo, J.L.; Ferrández, A. Importance of pronominal anaphora resolution in question answering systems. In Proceedings of the 38th Annual Meeting of the Association for Computational Linguistics, Hong Kong, China, 1–8 October 2000; pp. 555–562. [Google Scholar]
- Mollá, D.; Van Zaanen, M.; Smith, D. Named entity recognition for question answering. In Proceedings of the Australasian Language Technology Association Workshop. Australasian Language Technology Association, Canberra, Australia, 1–2 December 2006; pp. 51–58. [Google Scholar]
- Mao, Y.; Wei, C.H.; Lu, Z. NCBI at the 2014 BioASQ Challenge Task: Large-scale Biomedical Semantic Indexing and Question Answering. In Proceedings of the CLEF (Working Notes), Sheffield, UK, 15–18 September 2014; pp. 1319–1327. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Malik, N.; Sharan, A.; Biswas, P. Domain knowledge enriched framework for restricted domain question answering system. In Proceedings of the 2013 IEEE International Conference on Computational Intelligence and Computing Research, Madurai, India, 26–28 December 2013; IEEE: New York, NY, USA, 2013; pp. 1–7. [Google Scholar]
- Rajpurkar, P.; Zhang, J.; Lopyrev, K.; Liang, P. Squad: 100,000+ questions for machine comprehension of text. arXiv 2016, arXiv:1606.05250. [Google Scholar]
- Kočiskỳ, T.; Schwarz, J.; Blunsom, P.; Dyer, C.; Hermann, K.M.; Melis, G.; Grefenstette, E. The narrativeqa reading comprehension challenge. Trans. Assoc. Comput. Linguist. 2018, 6, 317–328. [Google Scholar] [CrossRef]
- Weissenborn, D.; Tsatsaronis, G.; Schroeder, M. Answering factoid questions in the biomedical domain. BioASQ@ CLEF 2013, 1094. Available online: https://ceur-ws.org/Vol-1094/bioasq2013_submission_5.pdf (accessed on 15 August 2024).
- Yang, H.; Gonçalves, T. Field features: The impact in learning to rank approaches. Appl. Soft Comput. 2023, 138, 110183. [Google Scholar] [CrossRef]
- Antonio, M.; Soares, C.; Parreiras, F. A literature review on question answering techniques, paradigms and systems. J. King Saud Univ. Comput. Inf. Sci. 2018, 8, 1–12. [Google Scholar]
- Russell-Rose, T.; Chamberlain, J. Expert search strategies: The information retrieval practices of healthcare information professionals. JMIR Med. Inform. 2017, 5, e7680. [Google Scholar] [CrossRef]
- Robertson, S.; Zaragoza, H. The Probabilistic Relevance Framework: BM25 and Beyond. Found. Trends Inf. Retr. 2009, 3, 333–389. [Google Scholar] [CrossRef]
- Robertson, S.E.; Walker, S. Some Simple Effective Approximations to the 2-Poisson Model for Probabilistic Weighted Retrieval. In Proceedings of the SIGIR ’94, Dublin, Ireland, 3–6 July 1994; Croft, B.W., van Rijsbergen, C.J., Eds.; Springer: London, UK, 1994; pp. 232–241. [Google Scholar]
- Karpukhin, V.; Oguz, B.; Min, S.; Lewis, P.; Wu, L.; Edunov, S.; Chen, D.; Yih, W.T. Dense Passage Retrieval for Open-Domain Question Answering. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), Online, 16–20 November 2020; pp. 6769–6781. [Google Scholar] [CrossRef]
- Berger, A.; Caruana, R.; Cohn, D.; Freitag, D.; Mittal, V. Bridging the lexical chasm: Statistical approaches to answer-finding. In Proceedings of the 23rd Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, New York, NY, USA, 24–28 July 2000; SIGIR ’00. pp. 192–199. [Google Scholar] [CrossRef]
- Fang, H.; Zhai, C. Semantic term matching in axiomatic approaches to information retrieval. In Proceedings of the 29th Annual International ACM SIGIR Conference on Research and Development in Information Retrieval, New York, NY, USA, 6 August 2006; SIGIR ’06. pp. 115–122. [Google Scholar] [CrossRef]
- Humeau, S.; Shuster, K.; Lachaux, M.A.; Weston, J. Poly-encoders: Transformer Architectures and Pre-training Strategies for Fast and Accurate Multi-sentence Scoring. arXiv 2019, arXiv:1905.01969. [Google Scholar]
- Soldaini, L.; Moschitti, A. The Cascade Transformer: An Application for Efficient Answer Sentence Selection. arXiv 2020, arXiv:2005.02534. [Google Scholar]
- Guo, J.; Fan, Y.; Ai, Q.; Croft, W.B. A Deep Relevance Matching Model for Ad-hoc Retrieval. In Proceedings of the 25th ACM International on Conference on Information and Knowledge Management, New York, NY, USA, 24–28 October 2016; CIKM ’16. pp. 55–64. [Google Scholar] [CrossRef]
- Ma, X.; Sun, K.; Pradeep, R.; Li, M.; Lin, J. Another Look at DPR: Reproduction of Training and Replication of Retrieval. In Proceedings of the Advances in Information Retrieval: 44th European Conference on IR Research, ECIR 2022, Stavanger, Norway, 10–14 April 2022; Part I. Springer: Berlin/Heidelberg, Germany, 2022; pp. 613–626. [Google Scholar] [CrossRef]
- Tsatsaronis, G.; Balikas, G.; Malakasiotis, P.; Partalas, I.; Zschunke, M.; Alvers, M.R.; Weissenborn, D.; Krithara, A.; Petridis, S.; Polychronopoulos, D.; et al. An overview of the BIOASQ large-scale biomedical semantic indexing and question answering competition. BMC Bioinform. 2015, 16, 138. [Google Scholar] [CrossRef]
- Voorhees, E.; Alam, T.; Bedrick, S.; Demner-Fushman, D.; Hersh, W.R.; Lo, K.; Roberts, K.; Soboroff, I.; Wang, L.L. TREC-COVID: Constructing a pandemic information retrieval test collection. In Proceedings of the ACM SIGIR Forum, New York, NY, USA, 19 February 2021; Volume 54, pp. 1–12. [Google Scholar]
- Lee, M.; Cimino, J.; Zhu, H.R.; Sable, C.; Shanker, V.; Ely, J.; Yu, H. Beyond information retrieval—Medical question answering. AMIA Annu. Symp. Proc. 2006, 2006, 469–473. [Google Scholar]
- Cruchet, S.; Gaudinat, A.; Rindflesch, T.; Boyer, C. What about trust in the question answering world. In Proceedings of the AMIA 2009 Annual Symposium, San Francisco, CA, USA, 14–18 November 2009. [Google Scholar]
- Gobeill, J.; Patsche, E.; Theodoro, D.; Veuthey, A.L.; Lovis, C.; Ruch, P. Question answering for biology and medicine. In Proceedings of the 2009 9th International Conference on Information Technology and Applications in Biomedicine, Larnaka, Cyprus, 4–7 November 2009; pp. 1–5. [Google Scholar]
- Cao, Y.; Liu, F.; Simpson, P.; Antieau, L.; Bennett, A.; Cimino, J.J.; Ely, J.; Yu, H. AskHERMES: An online question answering system for complex clinical questions. J. Biomed. Inform. 2011, 44, 277–288. [Google Scholar] [CrossRef]
- Hristovski, D.; Dinevski, D.; Kastrin, A.; Rindflesch, T.C. Biomedical question answering using semantic relations. BMC Bioinform. 2015, 16, 6. [Google Scholar] [CrossRef]
- Liu, T.Y. Learning to rank for information retrieval. Found. Trends® Inf. Retr. 2009, 3, 225–331. [Google Scholar] [CrossRef]
- Nogueira, R.; Yang, W.; Cho, K.; Lin, J. Multi-stage document ranking with BERT. arXiv 2019, arXiv:1910.14424. [Google Scholar]
- Pradeep, R.; Liu, Y.; Zhang, X.; Li, Y.; Yates, A.; Lin, J. Squeezing water from a stone: A bag of tricks for further improving cross-encoder effectiveness for reranking. In Proceedings of the European Conference on Information Retrieval, Stavanger, Norway, 10–14 April 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 655–670. [Google Scholar]
- Zhou, Y.; Shen, T.; Geng, X.; Tao, C.; Xu, C.; Long, G.; Jiao, B.; Jiang, D. Towards robust ranker for text retrieval. arXiv 2022, arXiv:2206.08063. [Google Scholar]
- Ponte, J.M.; Croft, W.B. A language modeling approach to information retrieval. In Proceedings of the ACM SIGIR Forum, New York, NY, USA, 2 August 2017; Volume 51, pp. 202–208. [Google Scholar]
- Zheng, G.; Callan, J. Learning to reweight terms with distributed representations. In Proceedings of the 38th International ACM SIGIR Conference on Research and Development in Information Retrieval, Santiago, Chile, 9–13 August 2015; pp. 575–584. [Google Scholar]
- Dai, Z.; Callan, J. Context-aware sentence/passage term importance estimation for first stage retrieval. arXiv 2019, arXiv:1910.10687. [Google Scholar]
- Nogueira, R.; Lin, J.; Epistemic, A. From doc2query to docTTTTTquery. Online Prepr. 2019, 6, 2. Available online: https://cs.uwaterloo.ca/~jimmylin/publications/Nogueira_Lin_2019_docTTTTTquery.pdf (accessed on 15 August 2024).
- Bai, Y.; Li, X.; Wang, G.; Zhang, C.; Shang, L.; Xu, J.; Wang, Z.; Wang, F.; Liu, Q. SparTerm: Learning term-based sparse representation for fast text retrieval. arXiv 2020, arXiv:2010.00768. [Google Scholar]
- Mallia, A.; Khattab, O.; Suel, T.; Tonellotto, N. Learning passage impacts for inverted indexes. In Proceedings of the 44th International ACM SIGIR Conference on Research and Development in Information Retrieval, Virtual, 11–15 July 2021; pp. 1723–1727. [Google Scholar]
- Jang, K.R.; Kang, J.; Hong, G.; Myaeng, S.H.; Park, J.; Yoon, T.; Seo, H. Ultra-high dimensional sparse representations with binarization for efficient text retrieval. arXiv 2021, arXiv:2104.07198. [Google Scholar]
- Chen, Q.; Wang, H.; Li, M.; Ren, G.; Li, S.; Zhu, J.; Li, J.; Liu, C.; Zhang, L.; Wang, J. SPTAG: A Library for Fast Approximate Nearest Neighbor Search. GitHub. 2018. Available online: https://github.com/microsoft/SPTAG (accessed on 15 August 2024).
- Mitra, B.; Nalisnick, E.; Craswell, N.; Caruana, R. A dual embedding space model for document ranking. arXiv 2016, arXiv:1602.01137. [Google Scholar]
- Gao, L.; Dai, Z.; Callan, J. COIL: Revisit exact lexical match in information retrieval with contextualized inverted list. arXiv 2021, arXiv:2104.07186. [Google Scholar]
- Huang, P.S.; He, X.; Gao, J.; Deng, L.; Acero, A.; Heck, L. Learning deep structured semantic models for web search using clickthrough data. In Proceedings of the 22nd ACM international Conference on Information & Knowledge Management, San Francisco, CA, USA, 27 October–1 November 2013; pp. 2333–2338. [Google Scholar]
- Hu, B.; Lu, Z.; Li, H.; Chen, Q. Convolutional neural network architectures for matching natural language sentences. Adv. Neural Inf. Process. Syst. 2014, 27, 2042–2050. [Google Scholar]
- Li, C.; Liu, Z.; Xiao, S.; Shao, Y. Making Large Language Models A Better Foundation For Dense Retrieval. arXiv 2023, arXiv:2312.15503. [Google Scholar]
- Chen, J.; Xiao, S.; Zhang, P.; Luo, K.; Lian, D.; Liu, Z. BGE M3-Embedding: Multi-Lingual, Multi-Functionality, Multi-Granularity Text Embeddings through Self-Knowledge Distillation. arXiv 2024, arXiv:2402.03216. [Google Scholar]
- Team, G.; Mesnard, T.; Hardin, C.; Dadashi, R.; Bhupatiraju, S.; Pathak, S.; Sifre, L.; Rivière, M.; Kale, M.S.; Love, J.; et al. Gemma: Open models based on gemini research and technology. arXiv 2024, arXiv:2403.08295. [Google Scholar]
- Douze, M.; Guzhva, A.; Deng, C.; Johnson, J.; Szilvasy, G.; Mazaré, P.E.; Lomeli, M.; Hosseini, L.; Jégou, H. The Faiss library. arXiv 2024, arXiv:2401.08281. [Google Scholar]
- Doms, A.; Schroeder, M. GoPubMed: Exploring PubMed with the gene ontology. Nucleic Acids Res. 2005, 33, W783–W786. [Google Scholar] [CrossRef]
- Wang, L.L.; Lo, K.; Chandrasekhar, Y.; Reas, R.; Yang, J.; Burdick, D.; Eide, D.; Funk, K.; Katsis, Y.; Kinney, R.M.; et al. CORD-19: The COVID-19 Open Research Dataset. In Proceedings of the 1st Workshop on NLP for COVID-19 at ACL 2020, Online, 5–10 July 2020. [Google Scholar]
- Lin, J.; Ma, X.; Lin, S.C.; Yang, J.H.; Pradeep, R.; Nogueira, R. Pyserini: A Python toolkit for reproducible information retrieval research with sparse and dense representations. In Proceedings of the 44th International ACM SIGIR Conference on Research and Development in Information Retrieval, Virtual, 11–15 July 2021; pp. 2356–2362. [Google Scholar]
- Dubey, A.; Jauhri, A.; Pandey, A.; Kadian, A.; Al-Dahle, A.; Letman, A.; Mathur, A.; Schelten, A.; Yang, A.; Fan, A.; et al. The Llama 3 Herd of Models. arXiv 2024, arXiv:2407.21783. [Google Scholar]
- Yang, A.; Yang, B.; Hui, B.; Zheng, B.; Yu, B.; Zhou, C.; Li, C.; Li, C.; Liu, D.; Fan, Z.; et al. Qwen2 Technical Report. arXiv 2024, arXiv:2407.10671. [Google Scholar]
- Shin, A.; Jin, Q.; Lu, Z. Multi-stage literature retrieval system trained by PubMed search logs for biomedical question answering. In Proceedings of the Conference and Labs of the Evaluation Forum (CLEF), Thessaloniki, Greece, 18–21 September 2023. [Google Scholar]
- Rosso-Mateus, A.; Muñoz-Serna, L.A.; Montes-y Gómez, M.; González, F.A. Deep Metric Learning for Effective Passage Retrieval in the BioASQ Challenge. In Proceedings of the CLEF 2023: Conference and Labs of the Evaluation Forum, Thessaloniki, Greece, 18–21 September 2023. [Google Scholar]
- Almeida, T.; Jonker, R.A.A.; Poudel, R.; Silva, J.M.; Matos, S. BIT.UA at BioASQ 11B: Two-Stage IR with Synthetic Training and Zero-Shot Answer Generation. In Proceedings of the Conference and Labs of the Evaluation Forum, Bologna, Italy, 5–8 September 2022. [Google Scholar]
- Reimers, N.; Gurevych, I. Sentence-bert: Sentence embeddings using siamese bert-networks. arXiv 2019, arXiv:1908.10084. [Google Scholar]
- Sun, W.; Yan, L.; Ma, X.; Ren, P.; Yin, D.; Ren, Z. Is ChatGPT Good at Search? Investigating Large Language Models as Re-Ranking Agent. arXiv 2023, arXiv:2304.09542. [Google Scholar]
- Wei, J.; Wang, X.; Schuurmans, D.; Bosma, M.; Xia, F.; Chi, E.; Le, Q.V.; Zhou, D. Chain-of-thought prompting elicits reasoning in large language models. Adv. Neural Inf. Process. Syst. 2022, 35, 24824–24837. [Google Scholar]
- Yoon, S.; Choi, E.; Kim, J.; Yun, H.; Kim, Y.; won Hwang, S. ListT5: Listwise Reranking with Fusion-in-Decoder Improves Zero-shot Retrieval. arXiv 2024, arXiv:2402.15838. [Google Scholar]
- Yu, Y.; Xiong, C.; Sun, S.; Zhang, C.; Overwijk, A. Coco-dr: Combating distribution shifts in zero-shot dense retrieval with contrastive and distributionally robust learning. arXiv 2022, arXiv:2210.15212. [Google Scholar]
- Kim, J.; Kim, M.; Park, J.; Hwang, S.w. Relevance-assisted Generation for Robust Zero-shot Retrieval. In Proceedings of the 2023 Conference on Empirical Methods in Natural Language Processing: Industry Track, Singapore, 6–10 December 2023; pp. 723–731. [Google Scholar]
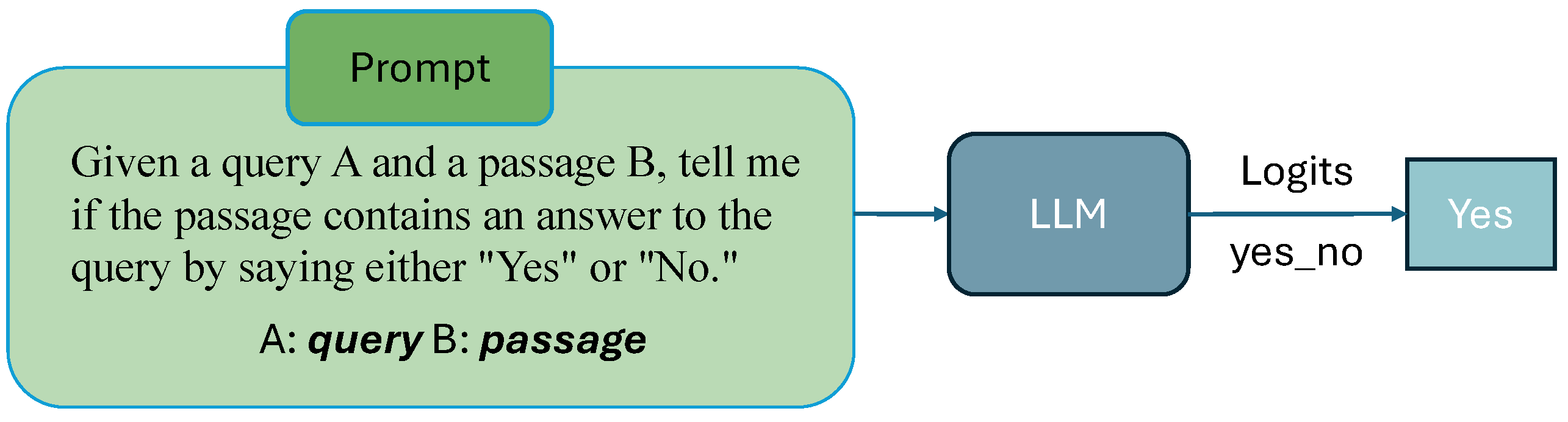





| Question | Answer | |
|---|---|---|
| 1 | What is CHARMS with respect to medical review of predictive modeling? | Checklist for critical Appraisal and data extraction for systematic Reviews of predictive Modelling Studies (CHARMS) |
| 2 | What is AUROC in the context of predictive modeling? | Area under the receiver operator characteristics curve |
| 3 | Is casimersen effective for the treatment of Duchenne muscular dystrophy? | Yes |
| Partition | Yes/No | Factoid | List | Summary | Documents | Snippets | Total |
|---|---|---|---|---|---|---|---|
| Training | 1271 | 1417 | 901 | 1130 | 9.01 | 12.03 | 4719 |
| Test 1 | 24 | 19 | 12 | 20 | 2.48 | 3.28 | 75 |
| Test 2 | 24 | 22 | 12 | 17 | 2.95 | 4.29 | 75 |
| Test 3 | 24 | 26 | 18 | 22 | 2.66 | 3.77 | 90 |
| Test 4 | 14 | 31 | 24 | 21 | 2.80 | 3.91 | 90 |
| Question | Answer | |
|---|---|---|
| 1 | What is the origin of COVID-19? | Although primary genomic analysis has revealed that severe acute respiratory syndrome coronavirus (SARS CoV) is a new type of coronavirus… |
| 2 | How does the coronavirus respond to changes in the weather? | Abstract: In this study, we aimed at analyzing the associations between transmission of and deaths caused by SARS-CoV-2 and meteorological variables… |
| 3 | Will SARS-CoV2 infected people develop immunity? Is cross protection possible? | Of the seven coronaviruses associated with disease in humans, SARS-CoV, MERS-CoV and SARS-CoV-2… |
| Test Batch-1 | |||||
| System | Mean Precision | Recall | F-Measure | MAP | GMAP |
| A&Q [61] | 0.1427 | 0.4814 | 0.1733 | 0.2931 | 0.0115 |
| A&Q2 [61] | 0.1747 | 0.5378 | 0.2069 | 0.3995 | 0.0465 |
| MindLab QA System [62] | 0.2820 | 0.3369 | 0.2692 | 0.2631 | 0.0039 |
| bioinfo-0 [63] | 0.3052 | 0.6100 | 0.3381 | 0.5053 | 0.1014 |
| Our system | 0.2653 | 0.6753 | 0.2915 | 0.6292 | 0.1735 |
| Test Batch-2 | |||||
| System | Mean Precision | Recall | F-Measure | MAP | GMAP |
| A&Q [61] | 0.1987 | 0.4428 | 0.2079 | 0.3494 | 0.0255 |
| A&Q2 [61] | 0.1747 | 0.3728 | 0.1761 | 0.2339 | 0.0111 |
| MindLab QA System [62] | 0.2283 | 0.2835 | 0.1876 | 0.1661 | 0.0063 |
| bioinfo-0 [63] | 0.2841 | 0.4993 | 0.2913 | 0.4244 | 0.0858 |
| Our system | 0.3267 | 0.4942 | 0.3843 | 0.4597 | 0.0609 |
| Test Batch-3 | |||||
| System | Mean Precision | Recall | F-Measure | MAP | GMAP |
| A&Q [61] | 0.2144 | 0.4502 | 0.2120 | 0.3603 | 0.0439 |
| A&Q2 [61] | 0.2289 | 0.4574 | 0.2236 | 0.3775 | 0.0525 |
| MindLab QA System [62] | 0.1600 | 0.2100 | 0.1440 | 0.1312 | 0.0025 |
| bioinfo-0 [63] | 0.2823 | 0.4794 | 0.2808 | 0.3568 | 0.0646 |
| Our system | 0.3879 | 0.4521 | 0.3405 | 0.4192 | 0.0451 |
| Test Batch-4 | |||||
| System | Mean Precision | Recall | F-Measure | MAP | GMAP |
| A&Q [61] | - | - | - | - | - |
| A&Q2 [61] | - | - | - | - | - |
| MindLab QA System [62] | - | - | - | - | - |
| bioinfo-0 [63] | 0.3327 | 0.4323 | 0.3066 | 0.3751 | 0.0595 |
| Our system | 0.4447 | 0.3443 | 0.3092 | 0.3623 | 0.0282 |
| Test Batch-1 | ||||
| Type | Precision | Recall | F-Measure | MAP |
| Factoid | 0.2105 | 0.6451 | 0.2415 | 0.5491 |
| List | 0.2667 | 0.5464 | 0.3050 | 0.3470 |
| Yes/No | 0.2458 | 0.6578 | 0.2590 | 0.5248 |
| Summary | 0.1850 | 0.6208 | 0.2329 | 0.4658 |
| Test Batch-2 | ||||
| Type | Precision | Recall | F-Measure | MAP |
| Factoid | 0.2955 | 0.5426 | 0.3108 | 0.4383 |
| List | 0.3250 | 0.3723 | 0.2226 | 0.2831 |
| Yes/No | 0.2708 | 0.6397 | 0.2955 | 0.4875 |
| Summary | 0.2706 | 0.6110 | 0.3025 | 0.4558 |
| Test Batch-3 | ||||
| Type | Precision | Recall | F-Measure | MAP |
| Factoid | 0.2154 | 0.5137 | 0.2166 | 0.3093 |
| List | 0.3833 | 0.3985 | 0.3150 | 0.3294 |
| Yes/No | 0.2000 | 0.5706 | 0.2188 | 0.4455 |
| Summary | 0.2545 | 0.5378 | 0.2661 | 0.3386 |
| Test Batch-4 | ||||
| Type | Precision | Recall | F-Measure | MAP |
| Factoid | 0.1774 | 0.5288 | 0.2287 | 0.3930 |
| List | 0.3083 | 0.3665 | 0.2631 | 0.3027 |
| Yes/No | 0.2643 | 0.4406 | 0.2173 | 0.3812 |
| Summary | 0.3524 | 0.3284 | 0.2796 | 0.2494 |
| System | NDCG@10 |
|---|---|
| Our system | 0.8326 |
| BM25 | 0.4812 |
| Sentence-BERT | 0.3334 |
| DPR | 0.6420 |
| docT5query | 0.7420 |
| GPT-3.5-Manual | 0.7667 |
| GPT-3.5-CoT | 0.8213 |
| LLaMA3-Manual | 0.7746 |
| LLaMA3-CoT | 0.7454 |
| Qwen2-Manual | 0.8145 |
| Qwen2-CoT | 0.7972 |
| ListT5-base | 0.7830 |
| ListT5-3B | 0.8440 |
| PE-Rank | 0.7772 |
| COCO-DR | 0.7890 |
| RaMDA | 0.8143 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Li, S.; Gonçalves, T. Enhancing Biomedical Question Answering with Large Language Models. Information 2024, 15, 494. https://doi.org/10.3390/info15080494
Yang H, Li S, Gonçalves T. Enhancing Biomedical Question Answering with Large Language Models. Information. 2024; 15(8):494. https://doi.org/10.3390/info15080494
Chicago/Turabian StyleYang, Hua, Shilong Li, and Teresa Gonçalves. 2024. "Enhancing Biomedical Question Answering with Large Language Models" Information 15, no. 8: 494. https://doi.org/10.3390/info15080494
APA StyleYang, H., Li, S., & Gonçalves, T. (2024). Enhancing Biomedical Question Answering with Large Language Models. Information, 15(8), 494. https://doi.org/10.3390/info15080494









