Abstract
Since the early 19th century, linguists have collected enough linguistic data to draw a remarkably stable Uralic language family tree. However, the traditional Uralic language family tree has two main problems. First, it lacks a reliable chronology because linguistic data can suggest that some languages are closer or farther from each other, but that gives only a relative instead of a precise chronology of the branching events. Second, the extinct Mezhovskaya culture in the Ural region and the Minoan civilization on the island of Crete were not incorporated into the Uralic language family, although recent archaeogenetic and linguistic data indicate that their languages also belonged to the Uralic language family. Some recent studies took an essentially purely archaeogenetic approach to the study of the evolution of the Uralic language family. These purely archaeogenetic studies propose linguistically perplexing solutions. This is the first study of the development of the Uralic language family that fully integrates the archaeogenetic, archaeological and linguistic data and proposes a new chronology of the Uralic language family that avoids the above inconsistencies. The new chronology relies on the best current estimates of the formation of the mitochondrial DNA haplogroups that are found among present Uralic language speakers and in samples from various archaeological sites that are associated with Uralic speakers. The new chronology places the various branching events of the Uralic language family tree much earlier than usual, including the split between Proto-Finno-Permic and Proto-Ugric, which is shown to have taken place in the Mesolithic period. The new proposal makes the Bronze Age Minoan language better fit chronologically as well as linguistically into the Uralic language family.
1. Introduction
The study of the prehistory of Uralic peoples lacks a satisfying integration of archaeogenetic and linguistic data. There are not only gaps in our knowledge regarding the prehistory of Uralic peoples, but the various archaeogenetic and linguistic theories contradict each other. This paper intends to clear up the confusion and present a harmonious synthesis of the currently available archaeogenetic and linguistic data.
1.1. Why the Contradition Between Archaeogenetics and Linguistics Is Harmful?
In 1952, Michael Ventris deciphered the Mycenaeans’ Linear B script as an archaic Greek language [1]. Classics scholars then checked and accepted the decipherment. While the second step seems obvious, it may not have happened if the results of Lazaridis et al. [2] were available in 1952, for they claimed that the Mycenaeans were genetically most like Near Eastern populations who spoke Semitic languages.
Today, the situation is very different regarding the Linear A script [3], which the Minoans used on the island of Crete before the Mycenaean conquest around 1450 BCE [4]. Archaeogenetics is placed on such a pedestal that even classics scholars use it as a guide in their search for the origins of the Minoans. Classics scholars are still searching for Minoan–Near Eastern connections because Lazaridis et al. [2] said that the Minoans also were genetically most like Near Eastern populations.
As a result, the following linguistics results are ignored: (a) the Minoan language has a front–back vowel harmony [5], which is common among the Uralic languages [6] but is not a feature of Indo-European and Semitic languages, (b) a large percent of the Pre-Greek vocabulary of the ancient Greek language [7], which likely has a Minoan origin, seems cognate with the Proto-Uralic, Proto-Finno-Ugric or Proto-Ugric vocabulary [8], (c) the corresponding words show a regular sound change ([9,10], Tables 11–14), and (d) many of the Minoan texts can be translated as Proto-Ugric [11,12,13]. These results imply that the Minoan language belonged to the Uralic languages’ Ugric branch that has only three currently spoken languages—Hungarian, Khanty and Mansi—which were not spoken in the ancient Near East. If Lazaridis et al. [2] say that the Minoans came from the Near East, then why bother even reading any linguistics paper that proposes a Minoan–Ugric linguistic relationship, especially if understanding those papers would require learning those Ugric languages that are not commonly known by classics scholars?
1.2. Why Chronology and the Location of the Homeland Are Still Big Questions?
While there are well-known linguistic and genetic connections between Hungarians in Central Europe and peoples living in the Ural Mountain region, the chronology of these connections is a big question. There are two conflicting theories.
1.2.1. From the Danube Basin to the Urals?
Pamjav et al. [14] found a genetic connection between Hungarians and Mansi based on the Y-DNA haplotypes R1a-Z280 and N-M46, which the most recent common ancestor, TMRCA, of these groups estimated to have lived 9800 and 7500 YBP (years before present), respectively. Hence, according to these, Hungarian and Minoan (the West-Ugric sub-branch of Ugric [11]) could have separated by 7500 YBP from Khanty and Mansi (the Ob-Ugric sub-branch of Ugric).
Another study by Post et al. [15] found a genetic connection between Hungarians and Mansi based on the Y-DNA haplotype N-B540/L1034, which was estimated to have formed only about 4000 YBP, which is about a thousand years after the beginning of the Minoan Civilization [16] but is before the beginning of the use of the Linear A script, which shows many similarities to the script of Old European Civilization in the Danube Basin according to Gimbutas [17] (p. 319) and Haarmann [18]. Consistent with [17,18], there is some archaeogenetic evidence that the Minoans came from the Danube Basin, which includes the Danube Delta area on the western shores of the Black Sea [19,20]. It is easily imaginable that people living in the Danube Delta area could sail to the island of Crete via the Bosporus Strait. The idea of placing the Uralic homeland in the Carpathian Basin, which is included in the Danube Basin, goes back to Krantz [21]. Wiik [22] placed the Uralic homeland on the northern Black Sea coastal region, which is also near the Danube Delta area.
1.2.2. From the Urals to the Danube Basin?
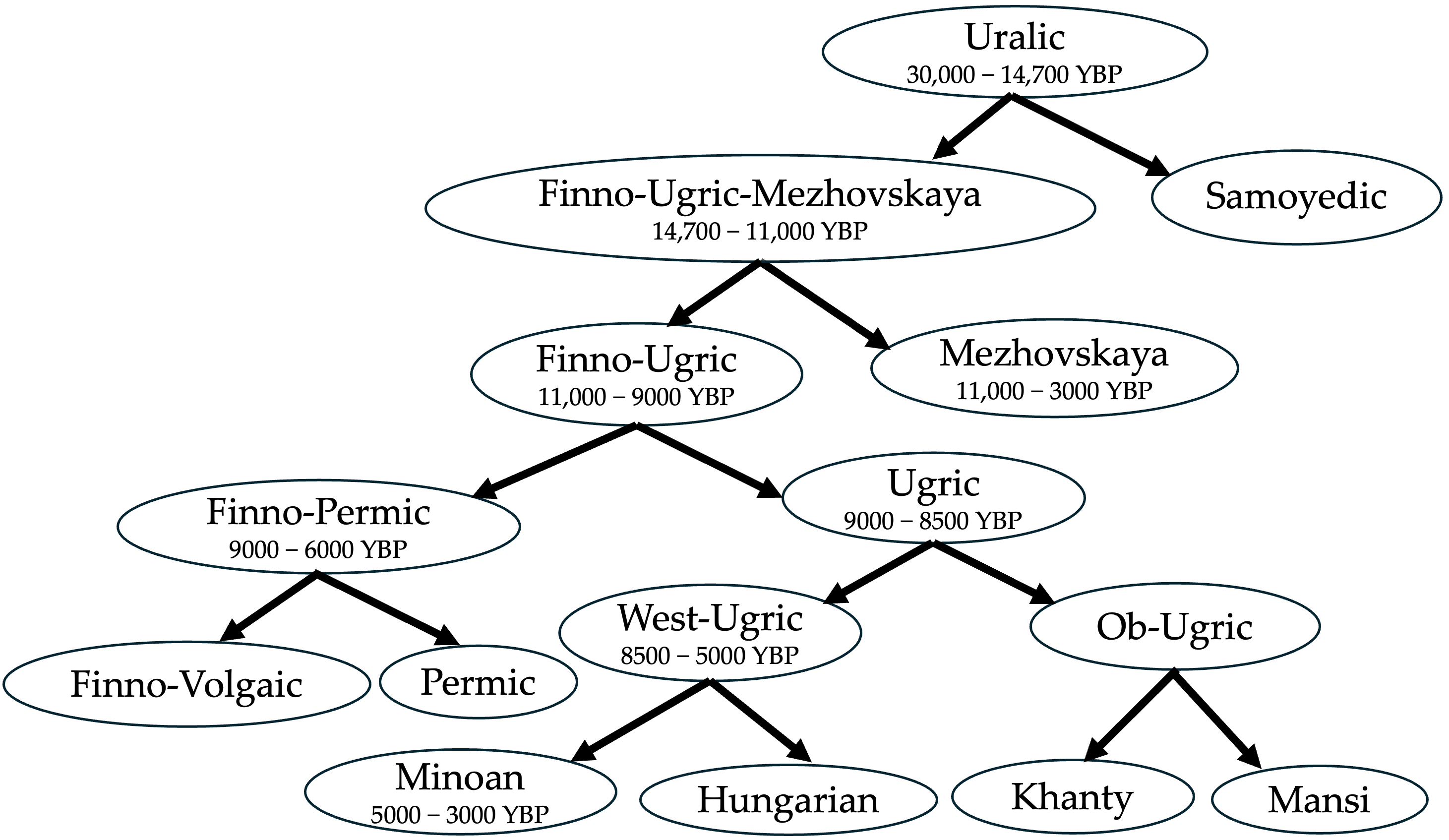
Hajdú [23] placed the Uralic homeland near the Ural Mountains based on the current habitat areas of plants and animals that have a Proto-Uralic name. He ignored the possibility that the Uralic language family originated at a time when the habitat areas were very different than today. Török [24] also placed the Proto-Ugric homeland in the southern Ural region in the Bronze Age. According to Török [24], the Proto-Ugric community formed via the admixture of the ancestors of the Nganasans and the people of the Mezhovskaya culture (denoted as Russia_Mezhovskaya). Török’s argument is based on qpAdm admixture analysis of 10–11th century Hungarian, modern Mansi, Nganasan, Bronze Age Russia_Mezhovskaya and other populations. The separation of the Hungarians and the Mansi happened sometime after 2700 YBP.
Table 1 summarizes the various estimates about the separation of the ancestors of the Hungarian and the Mansi speakers. The difference is over 7100 years between the oldest and the youngest age estimates.

Table 1.
The estimated time when the ancestors of the Hungarians and the Mansi separated, meaning the dissolution of the Proto-Ugric homeland into an Ob-Ugric branch containing Khanty and Mansi and a proposed West-Ugric branch containing Hungarian and Minoan.
1.3. The Main Goals of This Paper
- We show that the Danube Basin to the Urals is the correct direction of the spread of the Uralic languages. We show that the interpretation of Török [24] is mathematically inconsistent with the admixture data presented in Maróti et al. [25]. We also argue against some other recent theories that place the homeland in Siberia.
- We derive archaeogenetics-based dates for the various branches of the Uralic language family. The new dates are based on the estimated formation date of the mitochondrial haplogroups found in the DNA samples from various Uralic and Minoan-related archaeological sites. The new branching dates are much earlier than usual and allow the Mezhovskaya and the Minoan languages to be placed within the Uralic language family.
The rest of this paper is organized as follows. Section 2 describes our data sources. Section 3 (referring to Appendix A) shows that the hypothesis of Török [24] cannot be true. Section 3 also gives a mitochondrial DNA (mtDNA) data analysis of the Russia_Mezhovskaya samples and their relations to Finno-Ugric peoples. Section 4 discusses the results and presents an alternative hypothesis for the relationship between the Finno-Ugric peoples and the Russia_Mezhovskaya culture. Section 5 presents some conclusions and open problems. In addition, Appendix B gives a review of the linguistic results.
2. Data Sources
The main data source that we use is the admixture results from Maróti et al. [25] (Figure 3A; Data S9A,B), which are summarized in Table 2. According to most historians, the Hungarian language came to the Carpathian Basin area in 896, led by Árpád the Conqueror, who founded the first royal dynasty. Hence, the Hungarian Conqueror population refers to 10–11th century Hungarian samples from the Carpathian Basin. The Mansi population consists of current Mansi samples near the Ob River in Western Siberia.

Table 2.
The qpAdm admixture data from Maróti et al. [25].
According to Maróti et al. [25] (Figure 7) and Török [24], the admixture results of Table 2 imply that the Hungarian Conqueror population and the Mansi population had a common ancestor that was the mixture of Nganasan and Russia_Mezhovskaya populations. After the breakup of this common ancestor population, the ancestors of the Hungarian Conqueror had admixture from the Mongolia_MLBA population, and the ancestors of the Mansi had further admixture from the Kazakhstan_Eneolithic_Botai population.
Appendix A shows that Török’s hypothesis cannot be true. The challenge is to show that Török’s admixture data can be explained in an alternative way that is consistent with other known archaeogenetic data and with linguistic data. By other archaeogenetic data about the Russia_Mezhovskaya culture, we primarily mean the mtDNA haplogroups shown in Table 3.

Table 3.
The Russia_Mezhovskaya samples from the ancient DNA database [26].
Table 3 is based on the ancient DNA database [26], which we also searched for related haplogroup samples. We also used the YFull DNA database [27] and the Family Tree DNA (FTDNA) database [28] in our search for related haplogroup samples.
3. Data Analysis and Results
We start our data analysis by showing in Appendix A that Török’s hypothesis [24] is impossible, which means that a new theory is needed that better explains his admixture data. The following sections investigate the origin of the Russia_Mezhovskaya culture using mitochondrial DNA (mtDNA) data. We consider separately the history of the three Russia_Mezhovskaya mtDNA haplogroups in the next three sections, with Section 3.1 considering the J2b1a, Section 3.2 considering the I5, and Section 3.3 considering the M12′G mtDNA haplogroup. Finally, Section 3.4 gives a summary of the results.
3.1. The J2b1a mtDNA Haplogroup
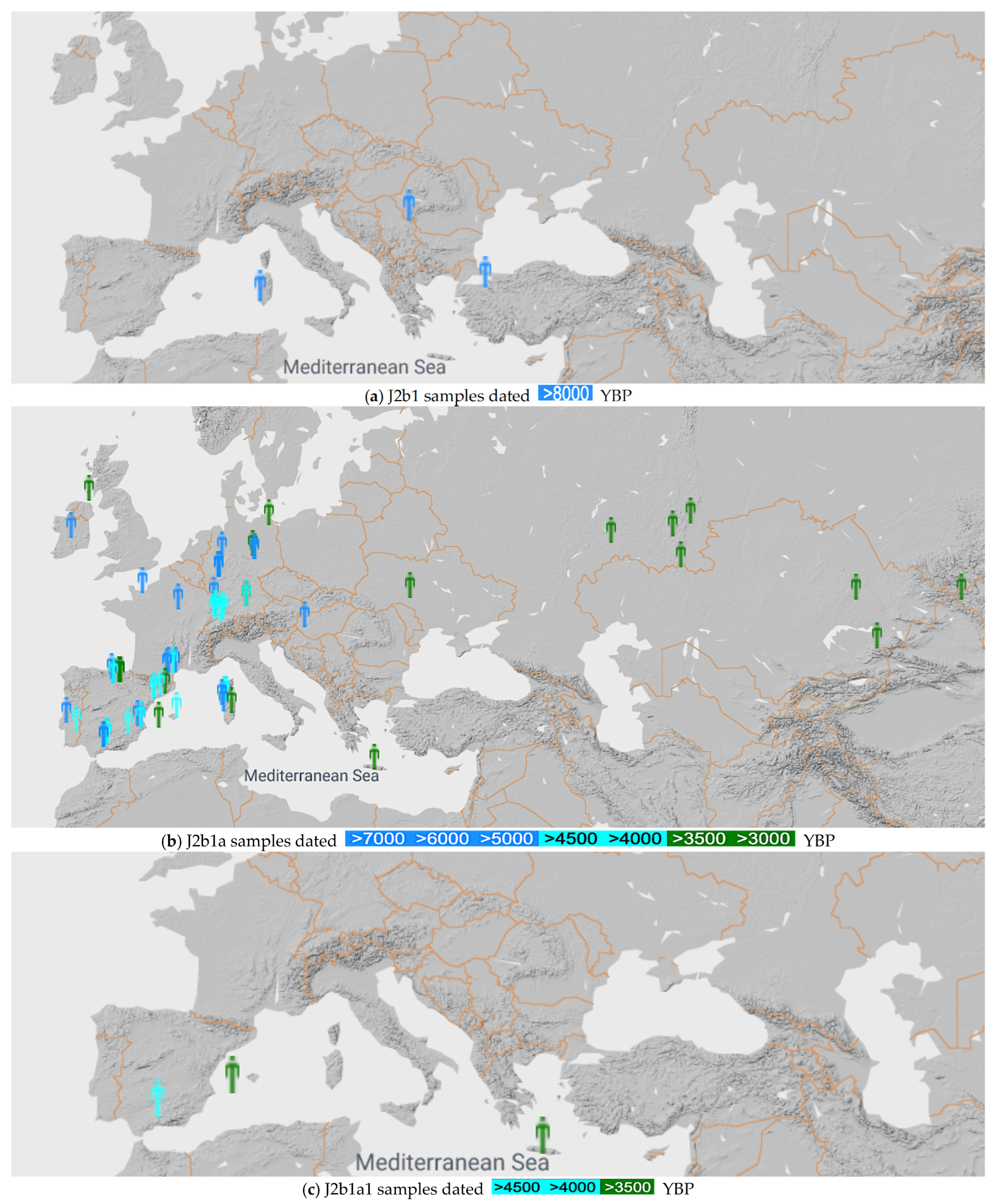
We searched and mapped the locations of the J2b1, J2b1a and J2b1a1 samples found in the ancient DNA database [26], as shown in Figure 1. Searching in this systematic way simultaneously shows the time and the locations when mutations took place that refined the haplogroup J2b1 into J2b1a and then into J2b1a1.
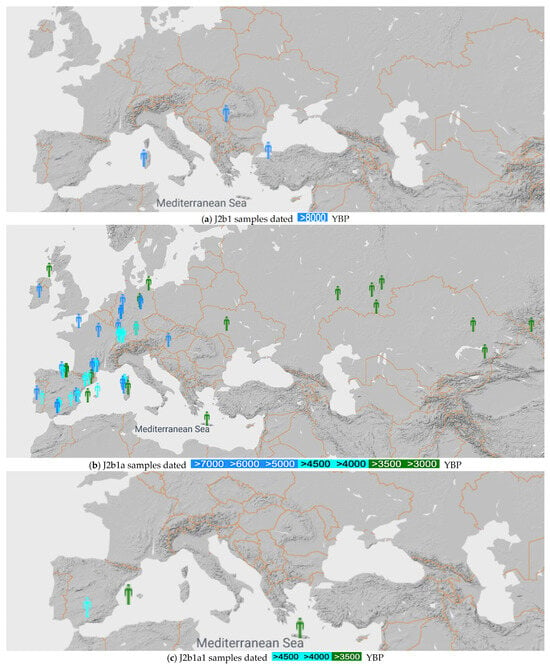
Figure 1.
mtDNA J2b1, J2b1a and J2b1a1 samples before 8000 YBP (a), 3000 YBP (b) and 3500 YBP (c), respectively [26].
Figure 1a shows that the J2b1 mtDNA haplogroup originates in Italy, with the earliest known sample from Mesolithic Sardinia. The color legend shows that all these samples are over 8000 years old.
Figure 1b shows that the J2b1a mtDNA haplogroup originates in Western Europe, with the earliest known sample from Spain. The mtDNA J2b1a expanded from Western Europe to Central Europe, where the earliest sample (I1908) is at Keszthely-Fenékpuszta, Hungary from 6500 YBP. It continued to spread east to Ukraine, the southern Ural Mountains area and the Altai Mountains by 3000 YBP.
Figure 1c shows that the earliest J2b1a1 occurs near Córdoba, Spain at 4650 YBP. The J2b1a1 sample from Crete (I9127) is a Minoan sample that was first reported by Lazaridis et al. [2]. In addition, a J2b1a1b mtDNA haplogroup sample (I23210) was reported from Vojvodina, Serbia from 3900 YBP by the YFull database [27].
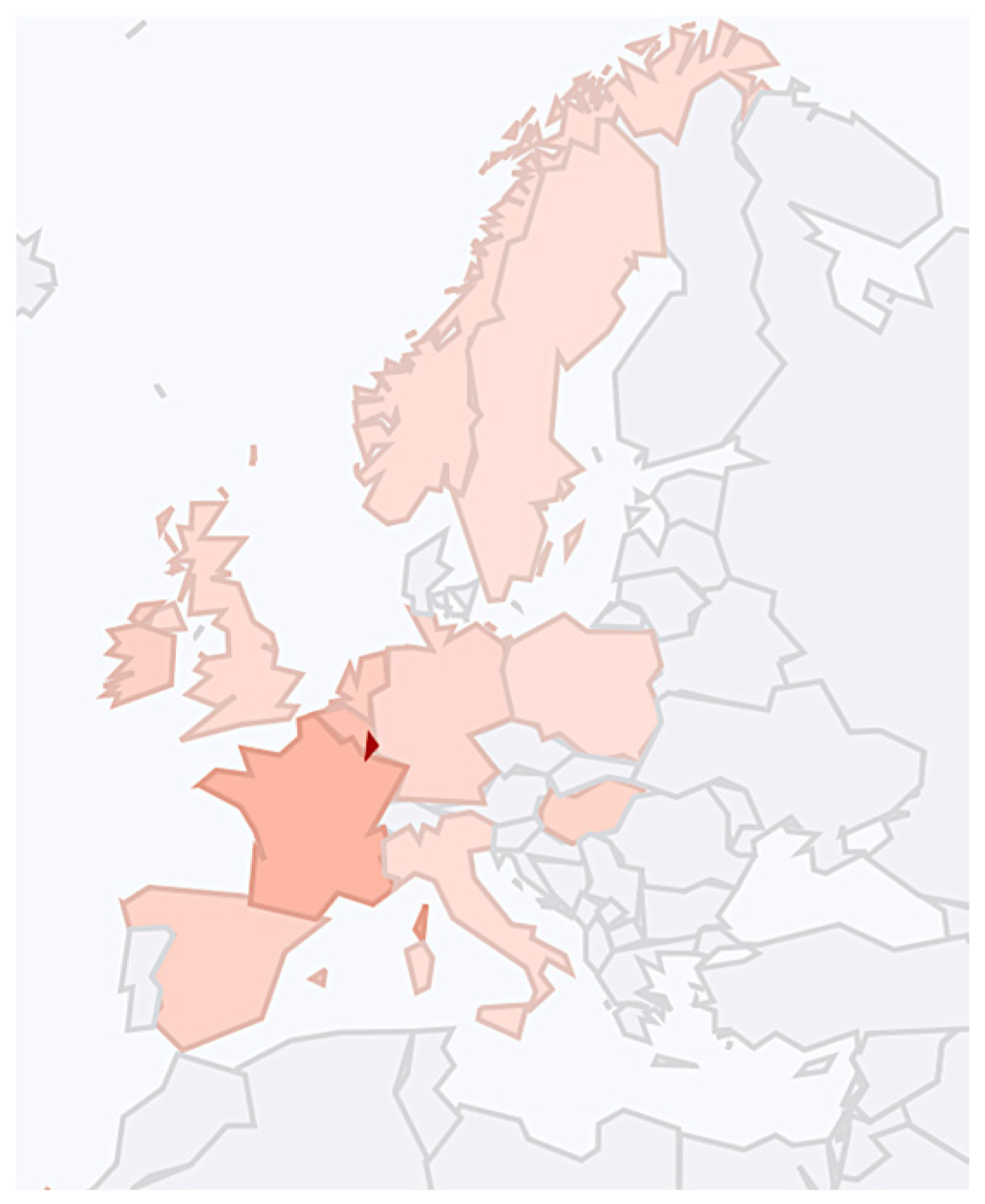
We also searched for J2b1 and J2b1a mtDNA haplogroups among Finno-Ugric peoples. The YFull database contained a J2b1a20b Hungarian sample (SHper175) from 975 YBP and several modern Finnish samples. No J2b1a1 mtDNA haplogroup samples were found in Estonia, Finland and Hungary in the YFull database. However, the Family Tree DNA database reported a J2b1a1 mtDNA haplogroup sample from Hungary, as shown in Figure 2.
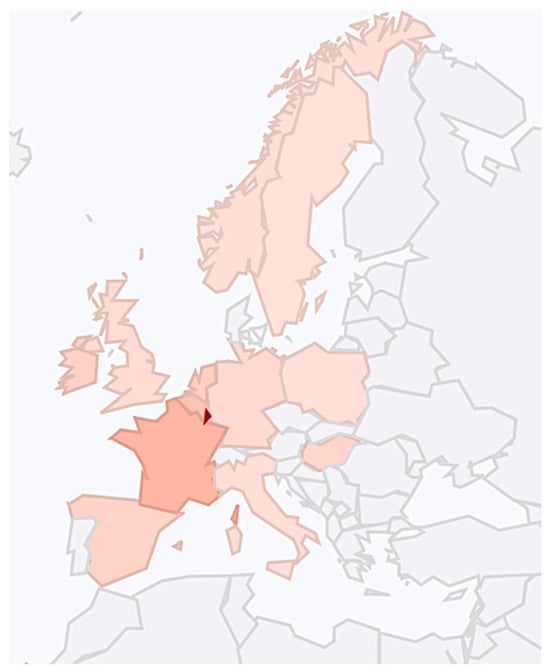
Figure 2.
The Family Tree DNA database summary regarding mtDNA J2b1a1 [28].
Table 4 gives a comprehensive list of all the ancient J2b1a mtDNA haplogroup samples from Eastern Europe and the Black Sea region. The Black Sea region is meant broadly and includes those regions of Central Europe that are near the Danube and can connect to the Black Sea. For example, Vojvodina, Serbia in the third data row in Table 4 was classified as belonging to the broader Black Sea region.

Table 4.
J2b1a and J2b1a1 samples from Eastern Europe and the Black Sea region [26,27,28].
3.2. The I5 mtDNA Haplogroup
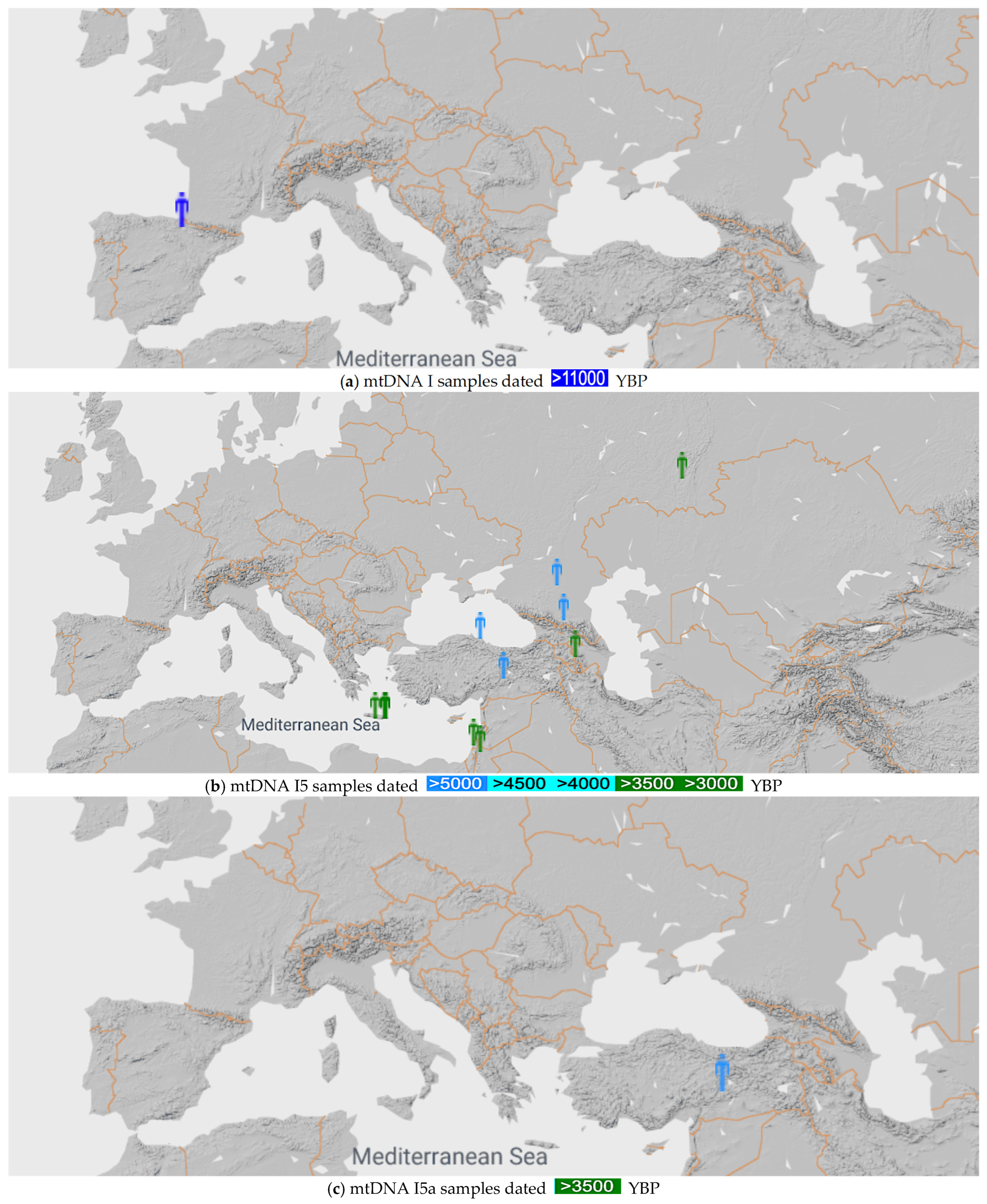
Figure 3 shows the I, I5 and I5a mtDNA haplogroups found in [26].
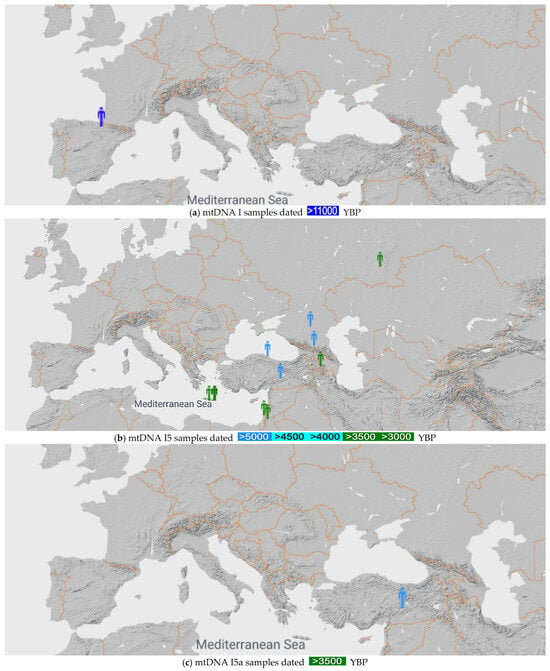
Figure 3.
mtDNA I, I5 and I5a samples before 10,000 YBP (a), 3000 YBP (b) and 3500 YBP (c), respectively [26].
Figure 3a shows that the I5 mtDNA haplogroup originated in Western Europe, with the earliest known sample from Paleolithic Spain.
Figure 3b shows that the I5 mtDNA haplogroup originated around the Black Sea.
Figure 3c shows that the I5a mtDNA haplogroup also originated around the Black Sea. In addition, an I5a mtDNA haplogroup Minoan sample (HGC024) was reported from the Charalambos Cave near the Lasithi Plateau, Crete from 3700 YBP by Skourtanioti [29].
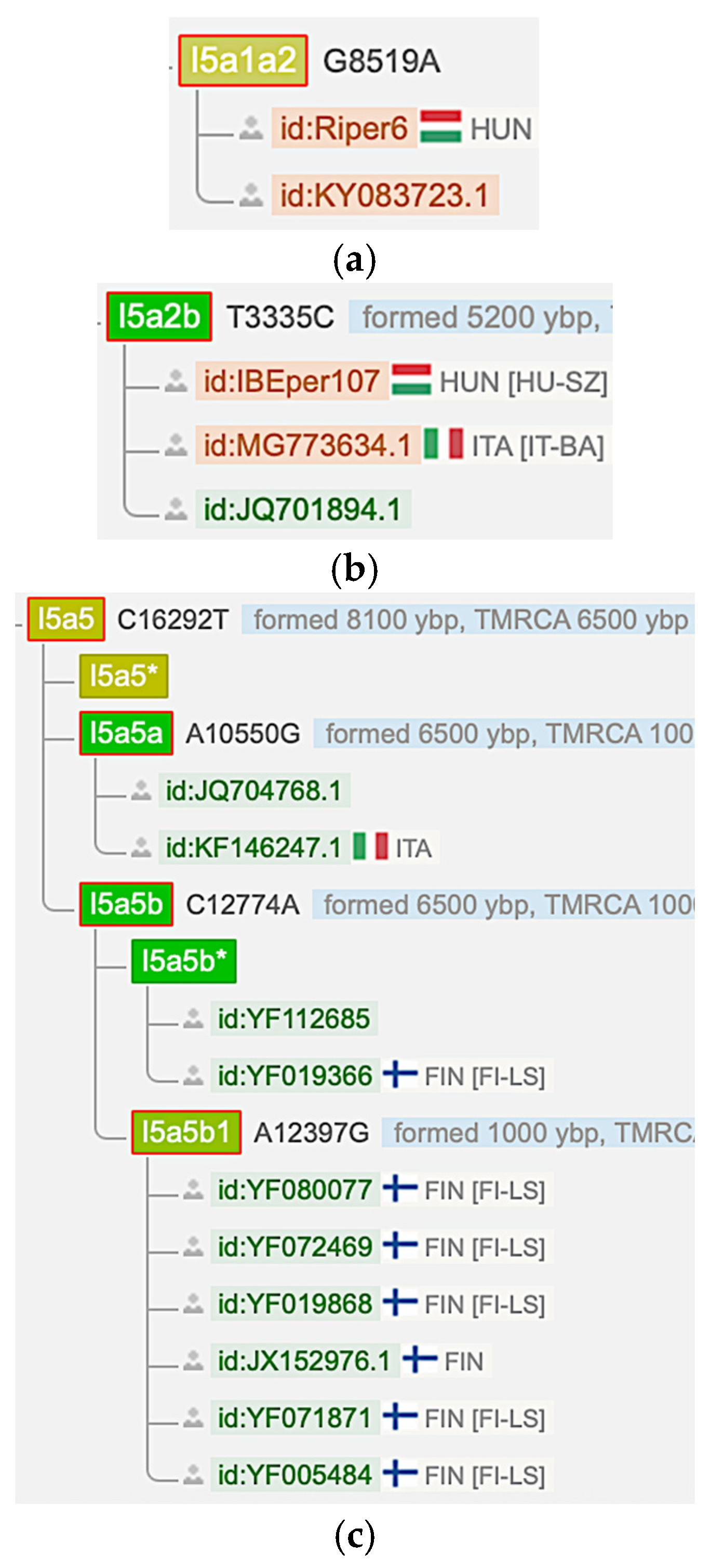
Figure 4a–c shows that an I5a1a2 Hungarian sample (Riper6) from 1000 YBP, a I5a2b Hungarian sample (IBEper107) from 975 YBP and several modern I5a5 Finnish samples were reported in the YFull database [27]. However, the YFull database did not list any Finnish or Hungarian I5c samples.
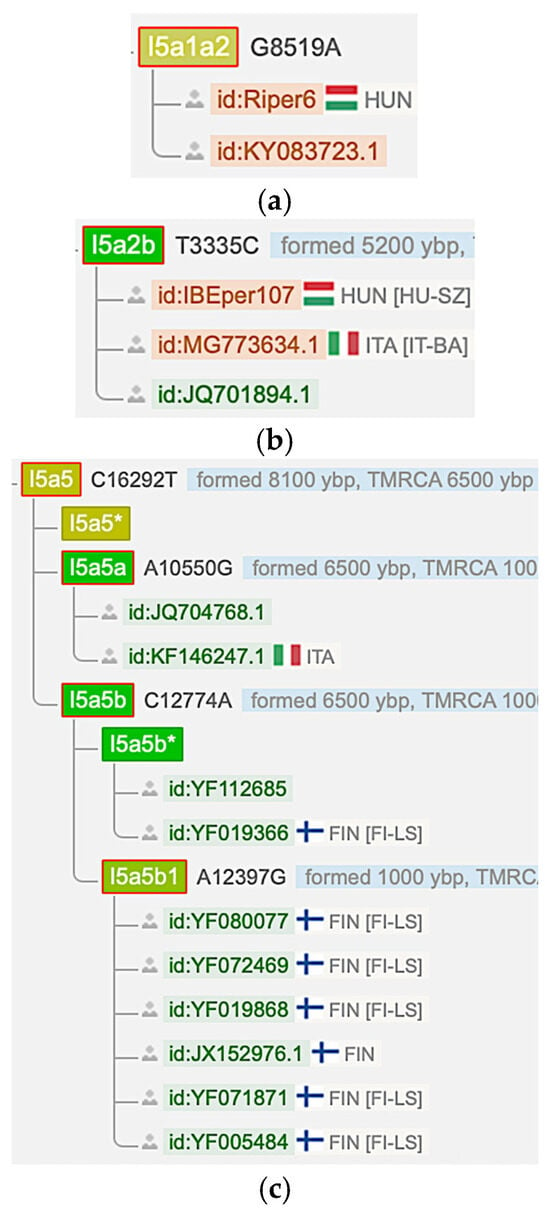
Figure 4.
Finnish and Hungarian I5a mtDNA samples from the YFull database [27]. (Data accessed: 25 April 2025). (a) An I5a1a2 mtDNA haplogroup sample (Riper6) from Hungary. (b) An I5a2b mtDNA haplogroup sample (IBEper107) from Hungary. (c) Several I5a5b1 mtDNA haplogroup samples (bottom six) from Finland.
Table 5 gives a comprehensive list of all the I5 mtDNA haplogroup samples from Eastern Europe and the Black Sea region.

Table 5.
I5, I5a, I5b and I5c samples from Eastern Europe and the Black Sea region [26,27].
3.3. The M12′G mtDNA Haplogroup
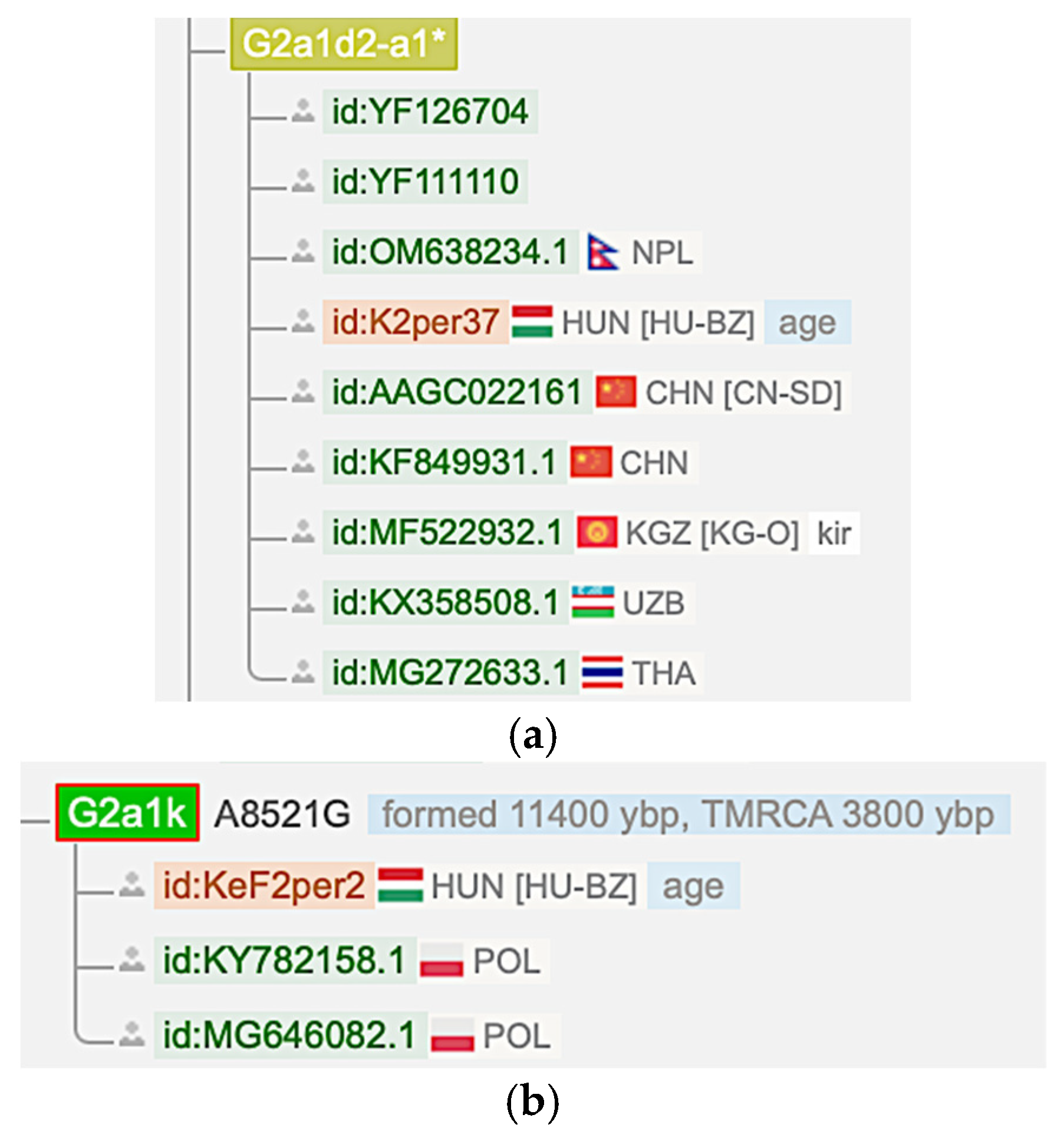
The M12′G mtDNA haplogroup has an Asian origin and is not found in Europe before 3000 YBP. The earliest known European M12′G sample is the Russia_Mezhovskaya sample (RISE523) from 3000 YBP, as shown in Table 2. Hence, it is not found in the Black Sea region and the Minoan samples before 3000 YBP. Moreover, it is also not found in Estonian and Finnish samples. Only its G2a1 sub-haplogroup can be found in some older Hungarian samples according to the YFull database [27] (Figure 5). These samples (K2per37, 1030 YBP) and (KeF2per2, 1025 YBP) can be associated with a ‘Hungarian Conqueror’ population because all Hungarian samples from the 9–10th centuries are categorized as ‘Hungarian Conqueror.’ However, they could be Avar descendants [30].
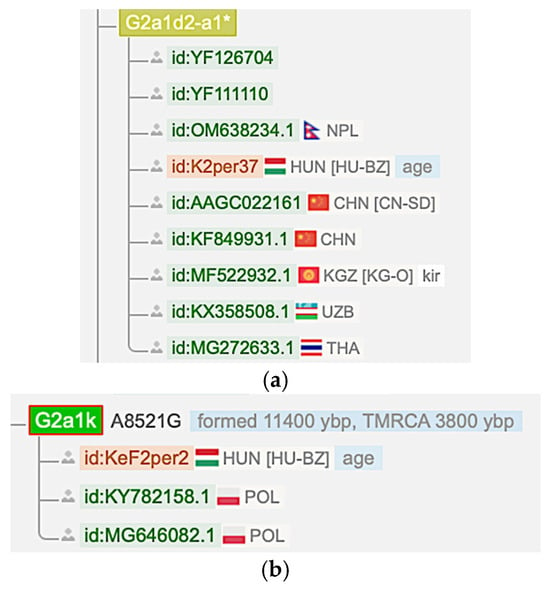
Figure 5.
Hungarian conquest period M12′G sub-haplogroup samples in the YFull database [27]. (a) A G2a1d2a mtDNA haplogroup sample (K2per37) from Hungary. (b) A G2a1k mtDNA haplogroup sample (KeF2per2) from Hungary.
3.4. Summary of the Findings
Table 6 gives a summary of the mtDNA haplogroup results.

Table 6.
The origin and TMRCA dates of the mtDNA haplogroups that were found in the Black Sea region before 3000 YBP.
4. Discussion: Alternative Hypothesis to Explain the Admixture Data
Section 4.1 presents an alternative hypothesis to explain the qpADM admixture data of Table 2. Section 4.2 gives a detailed justification of each step. Section 4.3 discusses how the new alternative hypothesis agrees with Uralic linguistic data related to the spread of agriculture and animal domestication. Finally, Section 4.4 gives a criticism of other recent proposals regarding the evolution of the Uralic language family.
4.1. An Alternative Hypothesis
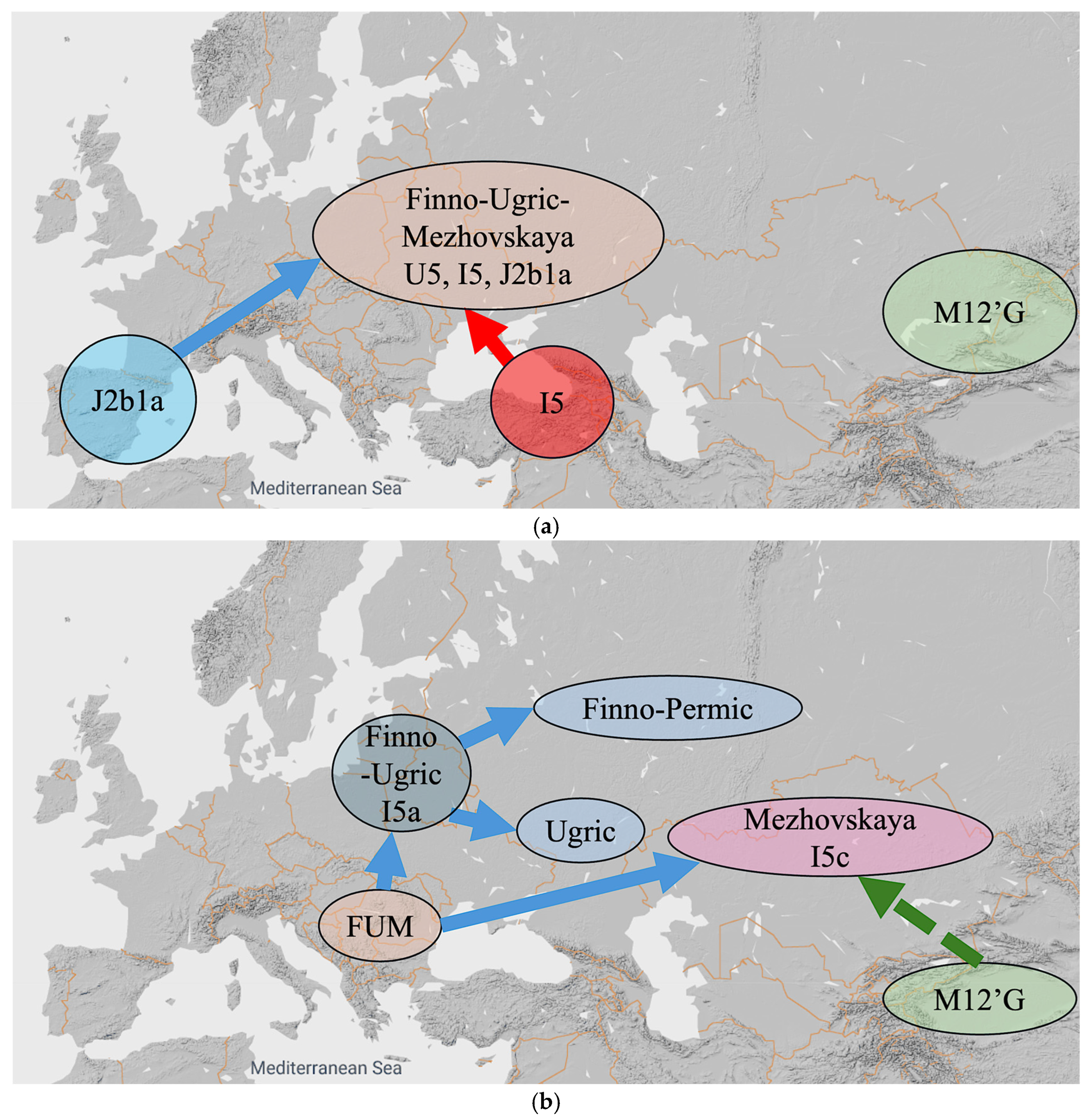
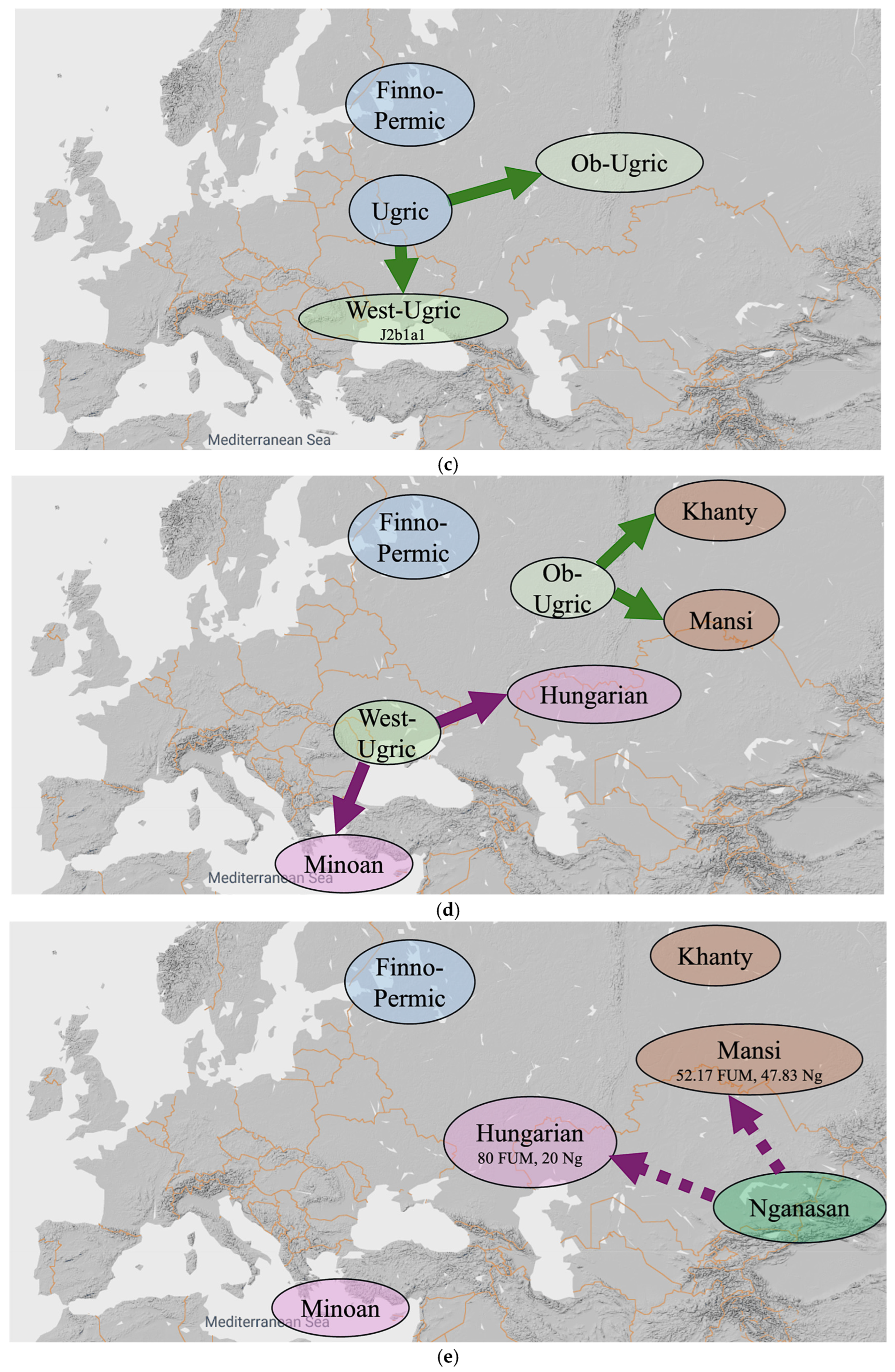
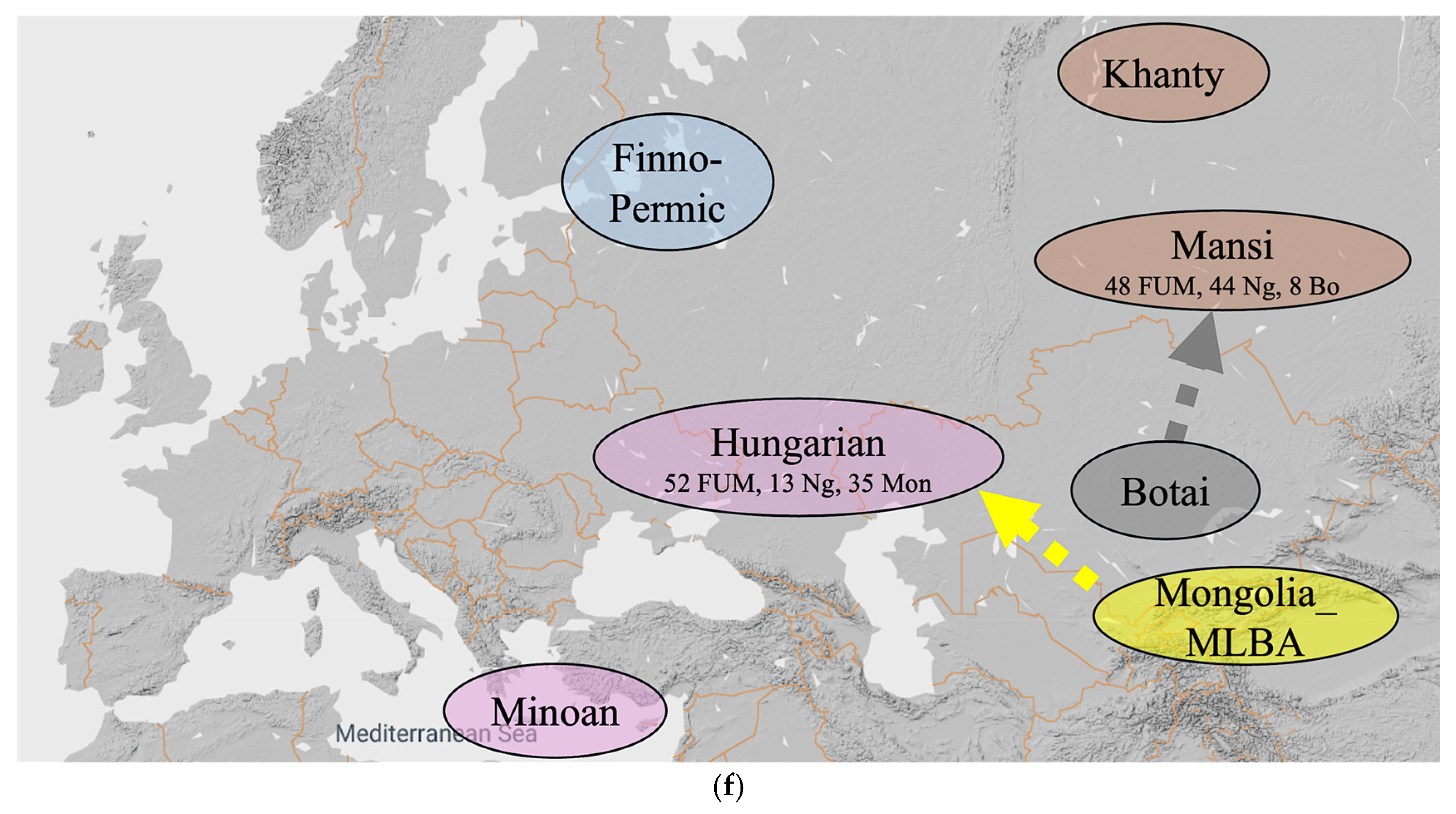
Our alternative hypothesis, which is illustrated in Figure 6, consists of ten steps listed below. Each of the ten steps is given an approximate date that keeps in mind the chronology of the origin of the various mtDNA haplogroups listed in Table 6.
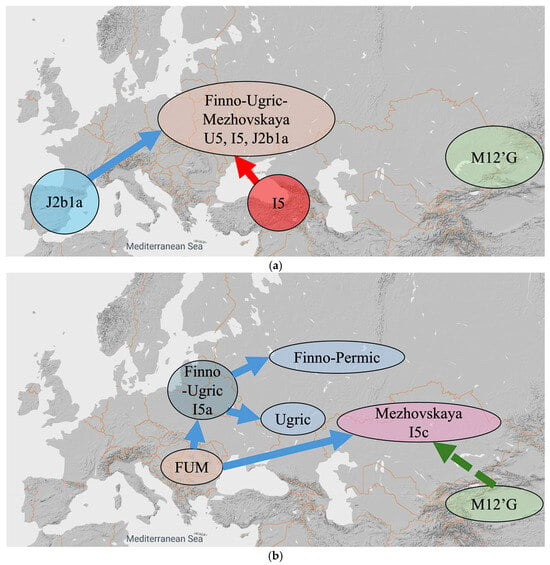
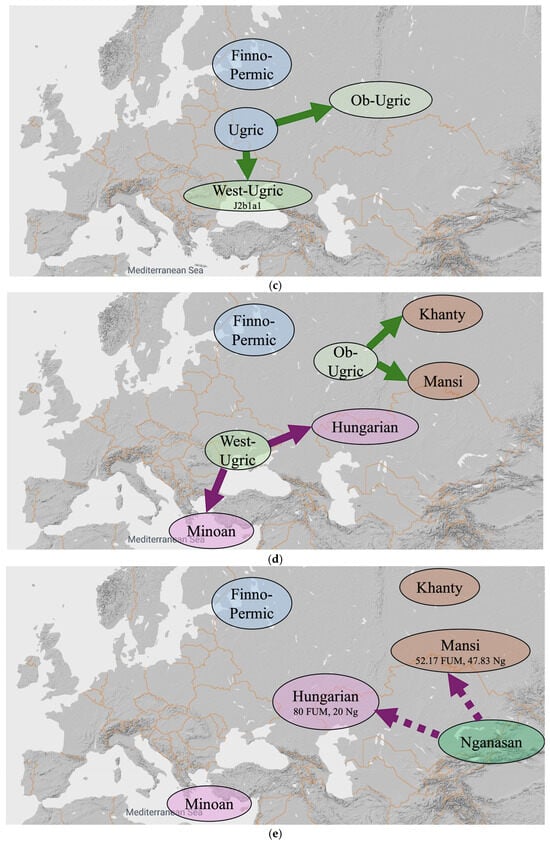
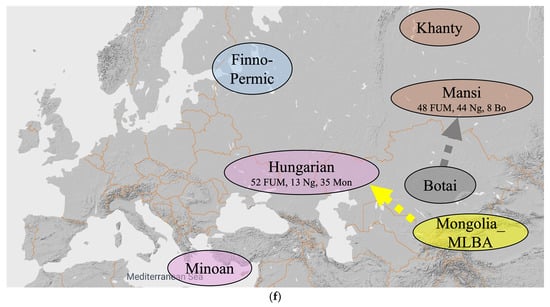
Figure 6.
An alternative hypothesis with ten steps. (a) Steps (1–4) until 11,300 YBP. (b) Steps (5–6) until 9000 YBP. (c) Step (7) until 8500 YBP. (d) Steps (8) until 5000 YBP. (e) Step (9) until 3000 YBP. (f) Step (10) until 1000 YBP.
- 30,000 YBP: The U5 mtDNA haplogroup originates in the Danube Basin, which is the homeland of the Uralic speakers.
- 20,000 YBP: The M12′G mtDNA haplogroup originates in Central Asia and slowly spreads west, reaching the Southern Urals region by 3000 YBP.
- 14,700 YBP: The Samoyedic groups move northeast from the Danube Basin following mammoth herds—which also move north with the receding ice—by 14,700 YBP. The remaining hunter-gatherers speak the common Finno-Ugric-Mezhovskaya (FUM) language. The I5 mtDNA haplogroup originates in the Black Sea region and mixes with U5 mtDNA haplogroups after 14,700 YBP. We assume that genetic mixing events did not cause major language changes. The incoming groups likely brought in some new words, but the core Proto-Finno-Ugric vocabulary and grammar were not influenced by the incomers.
- 11,300 YBP: The J2b1a mtDNA haplogroup originates in Iberia and moves east to the Black Sea region by 5000 YBP. The J2b1a also mixes with the U5 and I5 mtDNA haplogroups in Black Sea region. The Finno-Ugric-Mezhovskaya language speakers have U5, I5 and J2ba mtDNA haplogroups after this mixing (Figure 6a).
- 11,000 YBP: The FUM broke up into a Finno-Ugric group and a Russia_Mezhovskaya group. In the Finno-Ugric group, the I5a mtDNA haplogroup forms. In the Russia_Mezhovskaya group, the I5c mtDNA haplogroup forms.
- 9000 YBP: Proto-Finno-Ugric separates into Proto-Finno-Permic and Proto-Ugric. Proto-Finno-Permic moves north and northwest all the way to Estonia and Finland, while Proto-Ugric stays around the Black Sea region. The Russia_Mezhovskaya group receives M12′G mtDNA haplogroup admixture (Figure 6b).
- 8500 YBP: Proto-Ugric broke up into Proto-West-Ugric and Proto-Ob-Ugric. Proto-Ob-Ugric moves east, while Proto-West-Ugric stays around the Black Sea region. Later, the Proto-West-Ugric acquires the J2b1a1 mtDNA haplogroup (Figure 6c).
- 5000 YBP: Proto-West-Ugric separates into Hungarian and Minoan, and later, Proto-Ob-Ugric separates into Khanty and Mansi (Figure 6d). The Proto-West-Ugric split may have been influenced by the spread of the Yamnaya culture into the northern Black Sea region c. 5050 YBP ([17], Table 33, p. 496).
- 3000 YBP: Hungarians and Mansis both receive a Nganasan admixture. The Nganasan admixture into Hungarian is weaker than the Nganasan admixture into Mansi. These admixture events result in the Hungarians having 80 percent Russia_Mezhovskaya and 20 percent Nganasan genes and the Mansis having 52.17 percent Russia_Mezhovskaya genes and 47.83 percent Nganasan genes (Figure 6e).
- 1000 YBP: The ‘Hungarian Conquerors’ receive a Mongolia_MLBA admixture that results in 52 percent Russia_Mezhovskaya genes, 13 percent Nganasan genes and 35 percent Mongolia_MLBA genes. The Mansis receive a Kazakhstan_Eneolithic_Botai admixture that results in 48 percent Russia_Mezhovskaya genes, 44 percent Nganasan genes and 8 percent Kazakhstan_Eneolithic_Botai genes (Figure 6f).
4.2. A Detailed Explanation of the Alternative Hypothesis
The explanation of the above steps is as follows.
Steps 1–4 are logical because J2b1a first appeared in Spain (Figure 1b), I5 first appeared in Anatolia (Figure 3b) and M12′G has an Asian origin.
Step 5 is logical because the Finno-Ugric peoples and the Minoans share the I5a haplogroup, which is not found in the Russia_Mezhovskaya culture. On the reverse, the Russia_Mezhovskaya culture has the I5c haplogroup, which is not found among the Finno-Ugric peoples and the Minoans according to the YFull mitochondrial DNA database [27], as shown in Figure 4.
Step 6 is logical as an explanation of how the M12′G mtDNA haplogroup was acquired by the Russia_Mezhovskaya group.
Step 7 is logical because the J2b1a1 mtDNA haplogroup is found among both Minoans and Hungarians, as shown in Table 4, but it is not found among the Mansi according to Pamjav et al. [14] and Post et al. [15].
Step 8 is logical because the Khanty and the Mansi both belong to the Ob-Ugric sub-branch of the Ugric languages [6] (pp. 523–524).
Step 9 is logical because the admixture results of Maróti et al. [25] and Török [24] imply that the Hungarian Conquerors and the Mansi populations had some Nganasan admixture at some points in their histories.
Step 10 satisfies the admixture results of Maróti et al. [25] and Török [24], which are also shown in Table 2. The addition of 35 percent Mongolia_MLBA proportionally dilutes the FUM and the Nganasan genes in the Hungarian Conqueror group. The FUM genes are inherited by both the Mansi and the Russia_Mezhovskaya populations. Hence, the Hungarian Conqueror population’s FUM genes will be displayed as Russia_Mezhovskaya genes when an admixture analysis software must choose among a limited number of possible sources that include Nganasan, Russia_Mezhovskaya, Kazakhstan_Eneolithic_Botai and Mongolia_MLBA, as shown in Table 2. Similarly, the Mansi populations’ FUM genes will be displayed as Russia_Mezhovskaya genes when an admixture analysis software is given the same possible sources. If we replace FUM with Russia_Mezhovskaya in the ovals for Hungarian and Mansi, then we get the same percentages, as shown in Table 2.
Hence, the above sequence of steps provides an alternative explanation for the admixture results obtained by Maróti et al. [25] and used by Török [24] in developing his flawed theory about the origin of Ugric peoples. The main fault of Török’s theory [24] is that the admixtures he assumes do not dilute all the already existing gene types proportionally. Appendix A gives details regarding this problem. In fact, Equations (A1)–(A4) of Appendix A explain why 80 percent Russia_Mezhovskaya and 20 percent Nganasan genes will dilute to 52 percent Russia_Mezhovskaya, 13 percent Nganasan and 35 percent Mongolia_MLBA genes after admixture with 35 percent Mongolia_MLBA. Step (9) agrees with the calculations in Equations (A1)–(A4). Similarly, Equations (A5)–(A8) of Section 3.1 explain why 52.17 percent Russia_Mezhovskaya and 43.83 percent Nganasan genes will dilute to 48 percent Russia_Mezhovskaya, 44 percent Nganasan and 8 percent Kazakhstan_Eneolithic_Botai genes after admixture of 8 percent Kazakhstan_Eneolithic_Botai genes.
On the positive side, we believe that Török’s theory is correct in assuming that the Russia_Mezhovskaya culture people may have spoken a Uralic language. That is also assumed by our alternative theory, although there are differences in the detail of how the Russia_Mezhovskaya culture fits into the Uralic language family.
According to our theory, the Russia_Mezhovskaya culture people and the Finno-Ugric people once had a common ancestor population that spoke a Proto-FUM language. This Proto-FUM split into the Finno-Ugric branch, which later branched into the Finno-Permic and the Ugric branches, and a Russia_Mezhovskaya branch. The Finno-Ugric and Russia_Mezhovskaya split apparently happened when the I5 mtDNA haplogroup already existed but the I5a and the I5c mtDNA sub-haplogroups were not yet formed, because the Proto-Finno-Ugric branch did not contain the I5c haplogroup and the Russia_Mezhovskaya branch did not contain the I5a haplogroup, according to the currently known archaeogenetic data summarized in Table 6. Step 5 assumed that the split occurred around 11,000 YBP, which is earlier than 10,900 YBP, the estimated date of the origin of the I5a and the I5c haplogroups according to the YFull database [26]. It is also much earlier than the 8100 YBP and 8900 YBP dates of the most recent common ancestors (TMRCA) of the known I5a and I5c samples, respectively [26].
4.3. The Spread of Agriculture Among the Uralic Peoples
Török’s theory [24] assumes that Proto-Ugric arose in the late Bronze Age via the admixture of the Russia_Mezhovskaya culture and the Nganasan culture. The late Bronze Age Russia_Mezhovskaya culture had to know the names of metals like bronze, agricultural crops like wheat and barley, and domesticated animals like sheep, goat and cattle. Hence, if the Ugric people were formed as Török hypothesized, then the Ugric languages would have cognate names for the above items. However, Hungarians do not have cognate words for any of those items with Finno-Ugric peoples, even the Ob-Ugric peoples (Khanty and Mansi), as shown in Table 7. Hence, Proto-Hungarian and Proto-Ob-Ugric had to separate from each other before the beginning of the Neolithic period.

Table 7.
The names for dog, horse (data rows 1–2) and other domesticated animals and Neolithic agricultural crops (other rows).
The only common domesticated animal names are for dogs, which were already domesticated by hunter-gatherer people during the Mesolithic period, and dogs, which are native to the Eurasian Steppe. There are two different names for dogs among Finno-Ugric peoples. A Finno-Permic name and an Ob-Ugric name. That means that Proto-Finno-Permic and Proto-Ob-Ugric separated before the domestication of dogs. Proto-West-Ugric (the common ancestor of Proto-Hungarian and Minoan) and Proto-Ob-Ugric (the common ancestor of Khanty and Mansi) separated after the domestication of dogs, and possibly after the domestication of horses, but before the spread of agriculture and other domestic animals. This likely happened in the late Mesolithic period.
The Mesolithic is the period that precedes the agricultural revolution of the Neolithic. The Neolithic period arrived at different regions of Eurasia at different times, as shown in Figure 7. Hence, the end of the Mesolithic period is always location dependent.
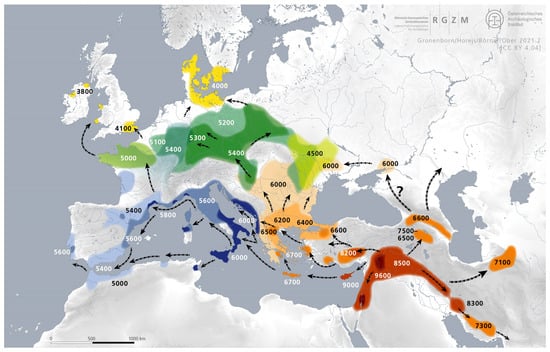
Figure 7.
The spread of agriculture reached the Danube Delta around 6000 BCE. Source: Wikipedia: https://en.wikipedia.org/wiki/Neolithic_Revolution#/media/File:Expansion_of_farming_in_western_Eurasia,_9600–4000_BCE.png (accessed on 25 April 2025). (CC by 4.04).
The Mesolithic period ended around the Danube Delta and the northern Black Sea areas with the arrival of the Neolithic period around 6000 BCE, or around 8000 YBP according to Figure 7. In our sequence of events, Step 8 placed the separation of West-Ugric and Ob-Ugric to 8500 YBP, which is about 500 years before the arrival of agriculture and animal domestication to the Danube Delta and the northern Black Sea areas. The Proto-Ugric people likely lived in these Steppe areas because of the common name for horse in Proto-Ugric, as shown in Table 7. The ancestors of the Ob-Ugric people likely migrated in an eastward direction along the northern Black Sea coast and then northward with the Don and the Volga Rivers. The West-Ugric people lived or migrated west of the Don River. The Danube Delta area was the likely place for the Minoans to start their migration to Crete [20].
The Ob-Ugric migration occurred much later than the Proto-Samoyedic migration, although they went in the same direction. The Proto-Samoyedic people may have migrated northeast with the receding ice and in the pursuit of mammoth herds, as suggested by Krantz [21] and Wiik [22]. The early age of this migration and the mixing with Paleo-Siberian peoples would explain the great genetic distance between Samoyedic peoples like the Nganasans and the Selkups and the rest of the Finno-Ugric peoples. The Nganasans do not have the I5 or J2b1a haplogroups. Since the YFull database estimate for the formation of I5 is 14,700 YBP and the formation of J2b1a is 11,300 YBP, the separation of the Samoyedic branch likely took place before 14,700 YBP, which is Step 3 in our alternative hypothesis. That roughly coincides with the end of the last Ice Age.
Figure 8 shows the most likely Uralic family tree as a summary of our alternative hypothesis and the above discussion. This Uralic family tree deviates significantly from several earlier proposals, such as that of Honkola et al. [32], which put the split of Proto-Ugric into Proto-Hungarian and Proto-Ob-Ugric at 3300 YBP. More recently, Bakró-Nagy et al. [6], p. 54, avoided giving a date for the split of Proto-Ugric, and in fact talked only about an Ugric ‘sprachbund’, that is, a group of languages that had areal contacts and borrowed words from each other. This later proposal is more compatible with our Figure 8 if we assume that the areal contacts involved the Minoan language as well as the other Ugric languages.
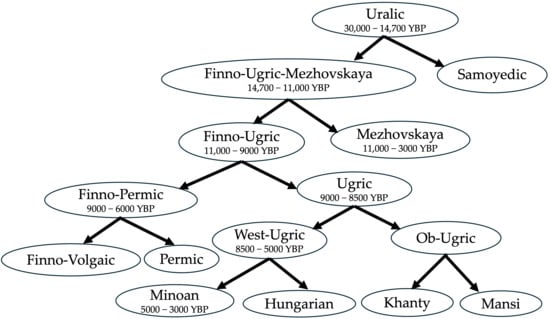
The Proto-Finno-Permic homeland was likely in the forest zone in the middle Volga River region because there is no common name for horse in Proto-Finno-Permic [8]. However, the common Proto-Finno-Permic names for some agricultural crops and domestic animals (see Table 7) implies that the split into Proto-Finno-Volgaic and Proto-Permic occurred after the Neolithization of the middle Volga River region, which occurred around 7300 YBP according to Zeng et al. ([33], Extended Data Figure 2). The Neolithization had to take some time to affect the Proto-Finno-Permic language. Hence, we have given an estimated date of 6000 YBP for the split into Proto-Finno-Volgaic and Proto-Permic.
There are many lexical and morphosyntactic parallels between the Finnic and the Volgaic languages ([6], p. 32) to assume a spread of the Finnic languages along the Volga River to the northwest. This spread was slow, with Proto-Saami having a homeland in what is now Southern Finland and Karelia and some areas south-east of it ([6], p. 35). The Proto-Saami homeland broke up only sometime between 0 and 500 ([6], p. 36). Similarly, the Permic languages spread slowly north following the Kama and the Pechora Rivers.
4.4. A Criticism of Other Uralic Language Family Origin Theories
The discussion in Section 4.3 implies that other archaeogenetics-based proposals that hypothesize any Bronze Age culture, such as the Seima-Turbino culture (Grünthal et al. [34]), to have been a Proto-Finno-Ugric culture, also cannot be valid because of the same reasons, i.e., the lack of Proto-Finno-Ugric cognate words for agricultural crops and domestic animals, as shown in Table 7.
The proposal of Zeng et al. [33] is an extension of that of Grünthal et al. [34]. Zeng et al. [33] associates the Yakutia Late Neolithic–Bronze Age culture (Yakutia_LNBA) with the Proto-Uralic people. According to [33], the Proto-Uralic people expanded to the Altai-Sayan and then to West Siberian regions associated with the Seima-Turbino culture.
Zeng et al. [33] provides a valuable collection of archaeogenetic data. The problem lies only in the incorrect interpretation of the data. For example, Zeng et al. [33], Supplementary Information 13.4, mentions that at Tatarka (in the Alai-Sayan region), only Y-chromosome N subclades N-F1419 and N-M2126 were found, which are “closely related to those found at today’s Uralic speakers.” Furthermore, at Rostovka (in Western Siberia, on the banks of the middle Irtysh River), a specific subclade of N-M2126, namely N-Z1936, was found, which “is found at high frequency only among Uralic-speaking populations and is most prevalent among Finnic-speaking populations around the Baltic Sea.” Based on such data, Zeng et al. [33] propose the following sequence of transitions:
Proto-Uralic = Yakutia_LNBA > Seima-Turbino culture (Tatarka, Rostovka etc.) > present Uralic speakers
Unfortunately, the above proposal of Zeng et al. [33] has the same linguistic problem as [34] had. The Yakutia_LNBA culture cannot be Proto-Uralic because then there would be Proto-Uralic protowords for Neolithic agricultural crop names and domesticated animals, as well as some metals. However, Table 7 shows that it is not true. Originating the Uralic language family from any Neolithic or Bronze Age culture is a stillborn idea.
If Zeng et al. [33] would have proposed a purely genetic migration as follows:
then there would be no problem, because the above-mentioned Y-chromosome haplogroups and other genes associated with them could have spread in the Bronze Age from east to west without changing the basic language of any of the Uralic peoples, although a few word borrowings may have occurred. Word borrowings are typically associated with the cultural innovations of the period. The Seima-Turbino culture is well-known for its metal works, with some nice examples shown in [33], Extended Data Figure 13. Hence, some metal names may have been adopted from the Seima-Turbino culture by some Finno-Ugric peoples. For example, Hungarian ezüst ‘silver’ has apparent cognates in the Permic languages (Komi eziś ‘silver’ and Udmurt azveś ‘silver’) [31] and in some Iranian languages (Ossetic ævzist ‘silver’ [35]). All these words may go back to some Seimo-Turbino word for silver. However, that would be only the borrowing of a wanderword and not a proof that the Seimo-Turbino culture spoke a Uralic language.
Yakutia_LNBA > Seima-Turbino culture (Tatarka, Rostovka etc.) > derived genes in some Uralic populations
Gyuris et al. [36] gave further support to Zeng et al. [33] by showing a genetic connection between the Hungarian Conquerors and the early medieval, i.e., 750–1000 CE, Karayakupovo archaeological horizon, which includes the Uyelgi cemetery near Chelyabinsk in the middle Irtysh River area. According to Gyuris et al. [36], those Hungarian Conquerors who showed the closest relationship to the Karayakupovo archaeological horizon also carried Yakutia_LNBA genes but no Hun/Xiongnu genes. Therefore, Gyuris et al. [36] argue that there was no mixing of Uralic-origin and Hun/Xiongnu genes in the Southern Urals region, in contradiction to the theory of Török [24].
The Carpathian Basin received many waves of migration from the Ural Mountains, Western Siberia and even Yakutia. While these archaeogenetic connections are interesting, it must be kept in mind that the languages of most migrant groups, including the Hungarian Conquerors, are unknown for the lack of written records. It cannot be excluded that Hungarian language speakers participated in several migrations. Some Hungarian Conquerors may have spoken a dialect of Hungarian, but they were not the first to bring the Hungarian language to the Carpathian Basin, because written Hungarian language documents already appear there in the 3rd century [10].
5. Conclusions
Incorrect hypotheses about the development of language families based on incorrect interpretations of the available archaeogenetic data can have negative consequences for linguistics. Any hypothesis about the Uralic language family that seems to disallow the inclusion of the Minoan language must be suspect given the linguistic results [5,9,10,11,12,13] that point to the Minoan language being an extinct member of the Ugric branch of the Uralic languages. As future work, any new archaeogenetics-based proposal will be carefully analyzed, especially those that are suspected to be incorrect from a linguistic point of view.
Although there are some relevant Finno-Volgaic cognates [8], they only mean that agriculture and animal domestication were introduced sometime before Proto-Finno-Volgaic separated into different branches. However, that happened thousands of years later than the dissolution of the Proto-Finno-Ugric unity. Hopefully, this paper called attention to this commonly misunderstood problem, thereby helping to prevent some stillborn proposals. However, there remain many questions about Uralic prehistory. One limitation of the current study is that it also could not precisely identify the earliest arrival of the Hungarian language to the Carpathian Basin. It remains an open problem to investigate whether it happened in the 3rd century or can be traced further back in time.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The author declares no conflicts of interest.
Appendix A
Why Is a New Theory Needed?
We show, using a mathematical analysis, that Török’s hypothesis [24] is impossible. We reason by contradiction. Let us assume that there was a common ancestor population of the Hungarian Conquerors and the Mansis. Furthermore, we assume that the common ancestor population was composed of Nganasan and Russia_Mezhovskaya populations.
We will reason by contradiction. Assume that in the common ancestor population there was an x percent Nganasan and a y percent Russia_Mezhovskaya genetic admixture. We can talk about a proto-Hungarian Conqueror group and a proto-Mansi group after the separation from the common ancestor.
The proto-Hungarian Conqueror group received a 35 percent admixture from the Mongolia_MLBA population. This genetic admixture diluted the Nganasan and the Russia_Mezhovskaya genes in equal proportions until the earlier x percent Nganasan proportion was reduced to 13 percent. Since the Mongolia_MLBA population does not contain Nganasan genes, 13 out of 100 − 35 = 65, the non-Mongolia_MLBA portion before the admixture of Mongolia_MLBA, must have the same concentration as the proto-Hungarian Conqueror group before the admixture, which is x out of 100. Hence, it must be the case that
which gives
This means that the 20 percent Nganasan genes in the proto-Hungarian Conqueror group was reduced to 13 percent Nganasan genes after the addition of 35 percent Mongolia_MLBA.
Similarly, the earlier y percent Russia_Mezhovskaya proportion was reduced to 52 percent. Hence, we can calculate that
which gives
The proto-Mansi group received an 8 percent admixture from the Kazakhstan_Eneolithic_Botai population. This genetic admixture diluted the Nganasan and the Russia_Mezhovskaya genes in equal proportions until the earlier x percent Nganasan proportion was reduced to 44 percent and the earlier y percent Russia_Mezhovskaya proportion was reduced to 48 percent. Hence, it must be the case that
which gives
Similarly, we can calculate that
which gives
Hence, the qpADM admixture data for the Hungarian Conqueror population imply that x = 20 and y = 80, while the qpADM admixture data for the Mansi population imply that x = 47.83 and y = 52.17. This is a contradiction. Hence, there could not have been a common ancestor population between the Hungarian Conquerors and the Mansis that was composed of Nganasan and Russia_Mezhovskaya populations.
Appendix B
Minoan–Uralic Language Links
Greek language etymologists have identified a Pre-Greek layer of about 1100 words within the ancient Greek language [7]. Those words that are borrowed from well-known Mediterranean languages are indicated as such in their etymological dictionaries. The Pre-Greek layer consists of borrowings from a different, single unknown language according to Beekes [37], p. 44. This language, which is non-Indo-European ([37], p. 45), is most likely the Minoan language.
Independently, Uralic language etymologists reconstructed a set of protowords from the hypothetical Proto-Ugric language, which was spoken by the common ancestor of the Hungarians, Khanty and Mansi [8]. The Proto-Ugric words include also words that are inherited from the still older Proto-Finno-Ugric language [8]. The etymological dictionary of Rédei [8] lists 625 Hungarian words that have a Proto-Ugric origin.
While the Greek and the Uralic etymologists worked independently on their etymological dictionaries, I pointed out [10,11] that there are many parallels between the set of 1100 Pre-Greek and 625 Hungarian words with Proto-Ugric origins. Table A1, which is updated from [10], shows 110 parallels. These word parallels suggest that the Minoan language was an Ugric language from which the Greek language borrowed some words.

Table A1.
Pre-Greek and Proto-Ugric word parallels. The corresponding consonant phonemes are highlighted in red. For some words we used the International Phonetic Alphabet notation to make the pronunciation clear.
Table A1.
Pre-Greek and Proto-Ugric word parallels. The corresponding consonant phonemes are highlighted in red. For some words we used the International Phonetic Alphabet notation to make the pronunciation clear.
| Pre-Greek [7,37] | Pre-Greek Meaning [7] | Hungarian [8] | Hungarian Meaning [8] | PFU, PU [8] | ID [8] |
|---|---|---|---|---|---|
| ἄγ-ν-ος (#91) | withy tree | ág | bough, branch, tree | ϑaŋɜ | 1745 |
| αἴθου-σα (#105) | portico | aj-tó | door | ȣjɜ | 1873 |
| άκιδ-ν-ός (#91) | thin | kes-hed | lean | käńćɜ | 1773 |
| ἀκκώ | bogey, vain woman | ük, ik | great-grandmother | ewkkɜ | 139 |
| ἄλοξ, ἄλοκ-ος | furrow | lyuk | hole, crevice | lowkkɜ | 493 |
| Ἄρη-ς | war god | ara-t, ir-t | cut, divide, eradicate | šurɜ | 1014 |
| Ἀχιλλε-ύς | Achilles | hal > haló | to die > mortal | kola | 339 |
| βαλ-μ-ός (#90) | breast | váll | shoulder | wolka | 1161 |
| βασκά-ς | duck | vöcsök | duck | wajće | 1106 |
| βῆkα | vine on trees | vék-ony | thin | wekkɜ | 1136 |
| Βλ-άβη (#53) | damage | vál | to separate, fall apart | walka | 1110 |
| γαῖα < * γή-aῖa | earth goddess | anya | mother | ańa | 15 |
| γή | earth | kő | stone | kiwe | 322 |
| Εἰλείθι-υ-α (#91) | goddess of birth | él > él-et | to live > life | elä | 132 |
| Ελέ-νη 1 | mother of life | ne | woman | niŋä | 598 |
| Ἐρεχθε-ύς | Athenians’ ancestor | ér > ere-get | to reach, let go, originate | šärɜ | 1000 |
| ἕψω /epsɔː/ | boil | fő > fő-z | to heat, simmer cook | peje | 735 |
| ϝάνα-κ-ες 2 (69b) | the Dioskouroi | vén > vén-ek | old > the old people | wȣ̈nɜ | 1180 |
| ἥρω-ς | lord, hero | ër | man | ir-kä | 152 |
| θάτα-ς | sacrificer | táltos | magician, diviner | tultɜ | 1862 |
| θρόμβο-ς | clot, curd | töm > * tereb | stocky > spreading | temɜ | 1046 |
| θύσ-θλα 3 | Bacchic ritual tool | tűz | fire | tüwɜtɜ | 1864 |
| /thɔrak-s/ 4 (69b) | cuirass, trunk, chest | toro-k | throat | turɜ | 1863 |
| ἰσχίο-ν (#91) | hip-joint | segg | buttocks | śäŋkɜ | 951 |
| Καδ-μῖλος 5 | younger of two boys | * kat > két | two | kakta | 227 |
| καλλα-ρ-ίας (#101a) | a kind of cod fish | hal | fish | kala | 228 |
| κάλ-ύβη (#53) | sleeping tent | hál | stay for the night | kalɜ | 231 |
| καραβί-ς | kind of a sea crab | hara-p | to bite | karɜ | 249 |
| καρκί-ν-ος (#91) | crab | hara-g | anger, bicker | kurɜ | 426 |
| κέλῦ-φ-ος (#141) | husk/peel, eggshell | ki > kívül > kül | out > outside | ki | 1776 |
| κιθάρα | lyre | tár > ki-tár | to open, outstretch (arms) | tara | 1026 |
| κίρ-βα (#53) | leather pouch | here-zacskó | scrotum | koj-ra | 333 |
| κισσ-ύβι-ον (#53, #91) | rustic cup | köcsö-g | vessel | kičɜ | 300 |
| κολ οβό-ς (#53) | curtailed, maimed | halo-k | crack, gap to cut tree | kolɜ | 342 |
| κομμόο-μ-αι (#90) | embellish/adorn self | hom-lok | forehead | kuma | 393 |
| κόττ αβο-ς (#53) | game with two vases | kettő | two | kakta | 227 |
| κρω-σσ-ός (#108a) | pail, pitcher, bottle | hor-d > hor-dó | to drag, draw; barrel | kurɜ | 1784 |
| κυδοί-μ-ός (#90) | din of battle | had | army | kunta | 400 |
| κύμ-βη (#53) | head | hom-lok | forehead | kuma | 393 |
| λαίθα-ργ-ος (#102) | guileful, treacherous | lát | to see | lȣttɜ | 505 |
| λἀξ /lak-s/ | with the heal or foot | lök | push, shove | likkä | 485 |
| λἀπα-θ-ος (#65) | pitfall for animals | láp | drift objects > mud | lȣppɜ | 504 |
| λάτ-ας | cf. Ἀσγε-λάτας | lát | to see | lȣttɜ | 505 |
| λἀττα | Cretan fly | légy | a fly | lȣńćɜ | 501 |
| λέπω | peel off | lep | cover | leppɜ | 479 |
| λίβα-ς | three-legged stand | láb | leg | luwe | 498 |
| λογγά-ζω | to linger, abide < abode | lak | cover, roof, abode | lakka | 451 |
| /lɔbɛːk-s/ 6 (69c) | vulture | lebë-g | to fly | lempɜ | 475 |
| μακε-δν-ός (#38) | tall | mag-as | tall | muŋkɜ | 563 |
| μαλ-θακός | weak, tender, soft | mál | peel off, wash off | mȣlɜ | 569 |
| μάλκη | numbness from cold | mele-g | warm | mälɜ | 1803 |
| μἀρ-πτω (#100) | catch, seize | mar | bite, break | mura | 566 |
| μαρἀ-σσ-αι (#108a) | dogs, swine | mar > mar-ós | bite, break > biter | mura | 566 |
| μάργο-ς | mad, furious | mérëg > mérges | poison, anger > angry | mirkkɜ | 547 |
| μέγα-ρ-ον (#101b, 91) | temple inner space | mag-as | tall | muŋkɜ | 563 |
| μέρμερο-ς 7 | difficult, awesome | mer | to dare, risk | märɜ | 1806 |
| μήρ-ινθ-ος (#81) | cord, thread | mér | to measure | merɜ | 538 |
| μίτη-ς | substance of bees | méz | honey | mete | 539 |
| μῦθο-ς | word, discourse, tale | mese | tale | mańćɜ | 1800 |
| μύραι-να (#91) | eel | már-t | to dip | marɜ | 1801 |
| Νηρε-ύς | Gaia and Pontus’ son | nyiro-k | moist place, swamp | ńorɜ | 639 |
| νύ-μ-φ-η (#90, #141) | nymph | nő | woman | niŋä | 598 |
| νῶκα-ρ (#101a) | slumbering | nyug-szik | rest | ńuŋɜ | 648 |
| Ὀδυσσεύ-ς | Odysseus | út > uta-zó | road, path > traveler | utka | 1096 |
| ὄλ-βο-ς (#53) | prosperity, bliss | ál-d | bless, spell magic | alɜ | 9 |
| ὄνο-ς | cod-like fish family | őn | fish type | šäwnä* | 886 |
| ὄρυμ-ος | altar, sanctuary | orr > orom | nose > mountain | wōre | 1144 |
| πανό-ς | torch | fejér > fény | light | päjɜ | 717 |
| πελε-μ-ίζω (#90) | to shake, tremble | fél > félelem 8 | to fear > fear, worry (n.) | pele | 739 |
| πήλη-κ-ος 9 (#69c) | helmet | fő > föl | head, top > up | päŋe | 729 |
| πήρα | leather bag | fér | to fit into something | pȣ̈rɜ | 823 |
| πίθο-ς | wine jar | fazé-k | pot, kettle | pata | 710 |
| πλα-στή (#109) | (clay) wall | fal | wall | paδɜ | 687 |
| πύελό-ς | bathtub | foly-ik | to flow | pȣlɜ | 1832 |
| πύλη | wing of double gate | fél > ajtófél | jamb, doorpost | pele | 738 |
| ῥί-ς | nose | orr | nose | wōre | 1144 |
| ῥίπτω | to throw | rëp-ít | to fly, throw, shake | rȣ̈ppɜ | 868 |
| ῥώδ-ιγγ-ες (#69a) | bruise | rút | ugly | rȣtɜ | 866 |
| σαυκό-ν (#91) | dry | szík | salty, dry | ćȣ̈kkɜ | 1737 |
| σαῠλο-ς | (animal) walking | szala-d | rust > run | ćaδa | 49 |
| σι-σύρα | goat’s fur cloak | szőr | fur | säkrɜ | 1844 |
| σίγραι | wild swine | csokor | herd, group, bouquet | ćukkɜ-rɜ | 76 |
| σίττ-υβο-ς (#53) | cauldron | süt | to bake, shine (sun) | čittɜ | 1744 |
| σοφία | clever | szép | beautiful, old | śeppä | 956 |
| σπυρί-ς | basket | csupo-r | (birch bark) vessel | ćuppɜ | 80 |
| σύρ-ιγξ-ις 10 (#69a) | pipe-like objects | szár | leg, stem, stalk | sȣrɜ | 1854 |
| σφά-ζω | to slay, slaughter | csap | to hit | ćappɜ | 51 |
| τέραμ-να (#91) | house, residence | tér > terem | space, room, square | tärɜ | 1860 |
| τέρχ-ν-ος (#91) | sprout, twig, fruit | törk-öly * | pressed grape | tȣ̈rkkɜ | 1085 |
| τόξο-ν (#91) | quiver | tegez | quiver | täŋɜtɜ | 1859 |
| τρέφω | to cause to curdle | töm > törp-ül | to shrink | temɜ | 1046 |
| τύλη | bulge, callosity, nail | toll | feather | tulka | 1075 |
| τύλλο-ς | box, chest | tál | serving dish | talɜ | 1857 |
| τύμβο-ς | burial mound | domb | hill | tȣmpɜ | 1865 |
| τύρ-ανν-ος (#24) | tyrant | tőr | dagger, sword | terä | 1049 |
| ὑστρι-χ-ίς (#69b) | hedgehog, whip | ostor | whip | oćtɜrɜ | 658 |
| φάσσα | wood-pigeon | fecs-ke | swallow (bird) | päč-kɜ | 711 |
| φαῦλο-ς | bad, unfit, ill, mean | bal | left | palɜ | 698 |
| φήλη-κ-ος 11 (69b) | wild fig | *boló-k > bogyók | berries | pola | 789 |
| φιάλη | flat vessel | fël | high, long | piδe | 759 |
| φλε-ν (#91) | to burn | fül | to burn | pilɜ | 1826 |
| φορί-ν-η (#91) | hide | bőr | skin, birch tree bark | perɜ | 751 |
| φορκό-ν (#91) | white/grey, wrinkled | far > far-k-as | behind > tail > wolf | purɜ | 821 |
| φωρια-μ-ός (#90) | chest, trunk | fara-g | to carve, hollow out | parɜ | 708 |
| χαρά-σσω (#108b) | to carve, engrave | hor-zso-l | to scrape | korɜ | 367 |
| Χάρω-ν (#91) | doom, death demon | hara-g | anger, bicker | kurɜ | 426 |
| χιτών | apron, tunic | köt > köté-ny | tie, knit | kit-ke | 320 |
| χλεμε-ρ-ός (#101) | warm, verdant | këll > kelle-m | necessary > pleasant | kel-ke | 281 |
| ψάλλω /psallɔ/ | pluck | foszli-k | pluck off feather | puśɜ | 826 |
| ψόθο-ς /psothos/ | ashes | füs-t | smoke | pičɜ | 1825 |
1 This a compound word with the first part related to Hungarian élet ‘life’ from Proto-Finno-Ugric * elä ‘life.’ 2 Also occurs as ϝάναξ. 3 This a compound word with the second part related to Hungarian tál ‘plate’ from Proto-Ugric * talɜ ‘plate.’ 4 We give here the pronunciation instead of the original form θώραξ, that hides the suffix. 5 This a compound word with the second part related to Pre-Greek μέλλαξ ‘young boy’ [7]. 6 We give here the pronunciation instead of the original form λώβηξ that hides the suffix. 7 Root duplication. 8 Easily contractable into félem, which also means ‘I fear (something).’ 9 Also written as πήληξ in some texts. 10 Here the original suffix seems to be -ιγγ- but with the addition of an -s the second γ became a devoiced /k/. 11 Also written as φήληξ in some texts.
Etymologists consider a set of word parallels more likely to be borrowings if they show regular sound changes. Word initial regular sound changes are shown in Table A2.

Table A2.
Word initial regular sound changes among Pre-Greek, Hungarian and PFU or PU.
Table A2.
Word initial regular sound changes among Pre-Greek, Hungarian and PFU or PU.
| Pre-Greek | Hungarian | PFU, PU |
|---|---|---|
| σ | cs, s | č |
| σ | cs, s, sz | ć |
| κ, χ | h | k, before back vowel |
| κ, χ | k | k, before front vowel |
| γ | h, k | k, before any (vowel-)semivowel |
| λ | l, ly | l |
| μ | m | m |
| ν | n | n |
| ν | ny | ń |
| π, φ | b, f | p |
| ρ | r | r |
| σ | sz | s |
| σ | s, sz | ś |
| - | - | š |
| - | - | ϑ |
| θ, τ | d, t | t |
| β, F, - | v, - | w |
The word medial regular sound changes are shown in Table A3. In both Table A2 and Table A3, the regular sound changes in the Hungarian column with respect to the Proto-Finno-Ugric and the Proto-Ugric column follow the usual regular sound change rules of Uralic linguistics, as described for example in Csúcs [38].

Table A3.
Word medial regular sound changes among Pre-Greek, Hungarian and PFU or PU.
Table A3.
Word medial regular sound changes among Pre-Greek, Hungarian and PFU or PU.
| Pre-Greek | Hungarian | PFU, PU |
|---|---|---|
| σσ | cs | č |
| σ | cs, s | ć |
| λ | l | δ |
| ι | j, - | j |
| κ | - | k |
| γ, γγ, κ, κκ, χ | k | kk |
| ρ | r | kr |
| δ, ττ | t, tt | kt |
| λ, λλ | l, ly | l, ll |
| λ | l, ll | lk |
| τ | lt | lt |
| μ, μμ | m | m |
| β, μβ | b, mb | mp |
| ν | n, ny | n |
| γ, κ | g, - | ŋ |
| γ, κ, χ | g, gg | ŋk |
| δ | d | nt |
| ι | ny | ń |
| δ, θ, ττ | gy | ńć |
| β, π, φ | p | pp |
| ρ | r | r |
| σσ | s | s |
| σ, τ | sz | ś |
| σ | sz | š |
| δ, θ, σ, τ | t, z | t |
| δ, θ, τ, ττ | t | tk, tt |
| β, - | v, - | w |
Table A1, Table A2 and Table A3 imply that the Minoan language is an extinct Ugric language. The Pre-Greek word suffixes that were analyzed and numbered by Beekes [36], pp. 29–42, also hint at the Minoan grammar and have recognizable Hungarian parallels (see Table A4).
Beekes [37] merged different suffixes into a single group in some cases. For example, we break up #69-ιγγ/κ/χ into #69a-ιγγ, #69b -κ/χ and #69c-κ in Table A4. Beekes [37] also combined two suffixes into a single suffix. For example, Beekes’ suffixes #38-δν, #102-ργ and #109-στ can be explained as a combination of two suffixes, as shown in Table A4.

Table A4.
Cognate Pre-Greek, Hungarian and PFU or PU suffixes and their meanings.
Table A4.
Cognate Pre-Greek, Hungarian and PFU or PU suffixes and their meanings.
| Id [37] | Pre-Greek | Hungarian | Hungarian Meaning [31] | Hungarian Origin [31] |
|---|---|---|---|---|
| 24 | -ανν | -nyél 1 | related to H. nyél ‘handle’ and Mansi äń ‘tooth, hook’ [39]. Only occurs in τύραννος ‘dagger-toothed’ > ‘tyrant.’ | Finno-Ugric, #595 [8] |
| 38 | -δν | -tn | combination of 65 and 91 | |
| 53 | -ηβ-α/ο | -p | noun and adjective former. Same as #141 but with p > b voicing between vowels. Vowels can change or be dropped | Uralic *-pp |
| 65 | -θ | -t | verb and noun former | Uralic *-tt |
| 69a | -ιγγ | -g | noun and adjective former | Finno-Ugric *-ŋk |
| 69b | -κ/χ | -k | noun plural | Finno-Ugric *-k |
| 69c | -κ 2 | -g | frequentative verb or noun former | Finno-Ugric *-ŋk |
| 81 | -ινθ | -d | noun former, common in place names | Finno-Ugric *-nt |
| 90 | -μ | -m | noun and adjective former | Uralic *-m |
| 91 | -ν | -n | noun and adjective former | Uralic *-n |
| 101a | -ρ | -r | diminutive noun former | Finno-Ugric *-r |
| 101b | -ρ | -r | frequentative | Finno-Ugric *-r |
| 102 | -ργ | -rg | combination of 101b and 69c | |
| 105 | -σα | -sz/s/ | noun and adjective former | Uralic *-ś |
| 108a | -σσ | -s/∫/ | noun and adjective former | Uralic *-ć |
| 108b | -σσ | -s/∫/ | verb former | Uralic *-ć |
| 109 | -στ | -st 3 | combination of 108a and 65 | |
| 141 | -φ | -p | noun and adjective former | Uralic *-pp |
1 This word can occur in itself, but it is used frequently as the second element of compound words, as in, for example, Hungarian ásónyél ‘the handle of an axe.’ 2 The original form was likely -γ that was devoiced to -κ after the addition of an -(o)s suffix. 3 This occurs in Hungarian ezüst ‘silver’ (see Section 4.4) and in Hungarian füst ‘smoke’ (last row of Table A1).
The morphological analysis of Table A4 also suggests that Minoan was a Uralic language. Further, morphological analysis is beyond the scope of this paper. However, the proposed translations of twenty-eight Linear A inscriptions of the Minoan Civilization can be found in Revesz [11], a translation of the Arkalochori Axe and the Malia Altar Stone can be found in [12], and a translation of the Phaistos Disk can be found in [13]. Unfortunately, these translations and decipherments, which use mostly hypothetical Proto-Ugric words and grammar, are harder to explain, and even the reading direction of the Phaistos Disk was subject to considerable controversy for many years [40]. Hence, the reader is encouraged to consult the original publications [11,12,13].
References
- Ventris, M.; Chadwick, J. Documents in Mycenaean Greek, 2nd ed.; Cambridge University Press: Cambridge, UK, 2013. [Google Scholar]
- Lazaridis, I.; Mittnik, A.; Patterson, N.; Mallick, S.; Rohland, N.; Pfrengle, S.; Furtwängler, A.; Peltzer, A.; Posth, C.; Vasilakis, A.; et al. Genetic origins of the Minoans and Mycenaeans. Nature 2017, 548, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Godart, L.; Olivier, J.-P. Recueil des Inscriptions en Lineaire A; École Française d’Athènes: Athina, Greece, 1976; Volume 1–5, p. 1985. [Google Scholar]
- Manning, S. Chronology and Terminology. In The Oxford Handbook of the Bronze Age Aegean; Cline, E., Ed.; Oxford University Press: Oxford, UK, 2012; pp. 11–28. [Google Scholar]
- Revesz, P.Z. A vowel harmony testing algorithm to aid in ancient script decipherment. In Proceedings of the 24th International Conference on Circuits, Systems, Communications and Computers, Chania, Greece, 19–22 July 2020; IEEE Press: Piscataway, NJ, USA, 2020; pp. 35–38. [Google Scholar]
- Bakró-Nagy, M.; Laakso, J.; Skribnik, E. (Eds.) The Oxford Guide to the Uralic Languages; Oxford University Press: Oxford, UK, 2022. [Google Scholar]
- Beekes, R.S.P. Etymological Dictionary of Greek; Brill NV: Leiden, The Netherlands, 2009. [Google Scholar]
- Rédei, K. (Ed.) Uralisches Etymologisches Wörterbuch; Akadémiai Kiadó: Budapest, Hungary, 1988. [Google Scholar]
- Revesz, P.Z. Minoan and Finno-Ugric regular sound changes discovered by data mining. In Proceedings of the 24th International Conference on Circuits, Systems, Communications and Computers, Chania, Greece, 19–22 July 2020; IEEE Press: Piscataway, NJ, USA, 2020; pp. 241–246. [Google Scholar]
- Revesz, P.Z. A tale of two sphinxes: Proof that the Potaissa Sphinx is authentic and other Aegean influences on early Hungarian inscriptions. Mediterr. Archaeol. Archaeom. 2024, 24, 191–216. [Google Scholar]
- Revesz, P.Z. Establishing the West-Ugric language family with Minoan, Hattic and Hungarian by a decipherment of Linear A. WSEAS Trans. Inf. Sci. Appl. 2017, 14, 306–335. [Google Scholar]
- Revesz, P.Z. A translation of the Arkalochori Axe and the Malia Altar Stone. WSEAS Trans. Inf. Sci. Appl. 2017, 14, 124–133. [Google Scholar]
- Revesz, P.Z. A computer-aided translation of the Phaistos Disk. Int. J. Comput. 2016, 10, 94–100. [Google Scholar]
- Pamjav, A.; Dudás, E.; Krizsán, K.; Galambos, A. A Y-chromosomal study of Mansi population from Konda River Basin in Ural. Forensic Sci. Int. Genet. Suppl. Ser. 2019, 7, 602–603. [Google Scholar] [CrossRef]
- Post, H.; Németh, E.; Klima, L.; Flores, R.; Fehér, T.; Türk, A.; Székely, G.; Sahakyan, H.; Mondal, M.; Montinaro, F.; et al. Y-chromosomal connection between Hungarians and geographically distant populations of the Ural Mountain region and West Siberia. Sci. Rep. 2019, 9, 7786. [Google Scholar] [CrossRef] [PubMed]
- Evans, A. The Palace of Minos; Macmillan and Company: London, UK, 1921; Volume 1. [Google Scholar]
- Gimbutas, M. The Civilization of the Goddess: The World of Old Europe; HarperCollins: San Francisco, CA, USA, 1991. [Google Scholar]
- Haarmann, H. The Mystery of the Danube Civilisation: The Discovery of Europe’s Oldest Civilisation; Marix: Wiesbaden, Germany, 2020. [Google Scholar]
- Revesz, P.Z. Minoan archaeogenetic data mining reveals Danube Basin and western Black Sea littoral origin. Int. J. Biol. Biomed. Eng. 2019, 13, 108–120. [Google Scholar]
- Revesz, P.Z. Archaeogenetic data mining supports a Uralic-Minoan homeland in the Danube Basin. Information 2024, 15, 646. [Google Scholar] [CrossRef]
- Krantz, G. Geographical Development of European Languages; P. Lang: New York, NY, USA, 1988. [Google Scholar]
- Wiik, K. The Uralic and Finno-Ugric phonetic substratum in Proto-Germanic. Linguist. Ural. 1997, 33, 258–280. [Google Scholar] [CrossRef]
- Hajdú, P. Über die alten Siedlungsraume der uralischen Sprachfamilie. Acta Linguist. Hung. 1964, 14, 47–83. [Google Scholar]
- Török, T. Integrating linguistic, archaeological and genetic perspectives unfold the origin of Ugrians. Genes 2013, 14, 1345. [Google Scholar] [CrossRef] [PubMed]
- Maróti, Z.; Neparáczki, E.; Schütz, O.; Maár, K.; Varga, G.I.; Kovács, B.; Kalmár, T.; Nyerki, E.; Nagy, I.; Latinovics, D.; et al. The genetic origin of Huns, Avars, and conquering Hungarians. Curr. Biol. 2022, 32, 2858–2870. [Google Scholar] [CrossRef] [PubMed]
- Ancient DNA Database. Available online: https://haplotree.info/maps/ancient_dna (accessed on 20 April 2025).
- Yfull Database. Available online: https://www.yfull.com (accessed on 20 April 2025).
- Family Tree DNA (FTDNA) Database. Available online: https://www.familytreedna.com (accessed on 20 April 2025).
- Skourtanioti, E.; Ringbauer, H.; Gnecchi Ruscone, G.A.; Bianco, R.A.; Burri, M.; Freund, C.; Furtwängler, A.; Gomes Martins, N.F.; Knolle, F.; Neumann, G.U.; et al. Ancient DNA reveals admixture history and endogamy in the prehistoric Aegean. Nat. Ecol. Evol. 2023, 7, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Gerber, D.; Csáky, V.; Szeifert, B.; Borbély, N.; Jakab, K.; Mező, G.; Petkes, Z.; Szücsi, F.; Évinger, S.; Líbor, C.; et al. Ancient genomes reveal Avar-Hungarian transformations in the 9th-10th centuries CE Carpathian Basin. Sci. Adv. 2024, 10, eadq5864. [Google Scholar] [CrossRef] [PubMed]
- Zaicz, G. (Ed.) Etimológiai Szótár: Magyar Szavak és Toldalékok Eredete, (Etymological Dictionary: Origin of Hungarian Words and Affixes); Tinta Publishing: Budapest, Hungary, 2006. [Google Scholar]
- Honkola, T.; Vesakoski, O.; Korhonen, K.; Lehtinen, J.; Syrjänen, K.; Wahlberg, N. Cultural and climatic changes shape the evolutionary history of the Uralic languages. J. Evol. Biol. 2013, 26, 1244–1253. [Google Scholar] [CrossRef] [PubMed]
- Zeng, T.C.; Vyazov, L.A.; Kim, A.; Flegontov, P.; Sirak, K.; Maier, R.; Lazaridis, I.; Akbari, A.; Frachetti, M.; Tishkin, A.A.; et al. Ancient DNA reveals the prehistory of the Uralic and Yeniseian peoples. Nature 2025, 644, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Grünthal, R.; Heyd, V.; Holopainen, S.; Janhunen, J.A.; Khanina, O.; Miestamo, M.; Nichols, J.; Saarikivi, J.; Sinnemäki, K. Drastic demographic events triggered the Uralic spread. Diachronica 2022, 39, 490–524. [Google Scholar] [CrossRef]
- English-Ossetic Dictionary, Glosbe. Available online: https://glosbe.com/en/os (accessed on 30 September 2025).
- Gyuris, B.; Vyazov, L.; Türk, A.; Flegontov, P.; Szeifert, B.; Langó, P.; Mende, B.G.; Csáky, V.; Chizhevskiy, A.A.; Gazimzyanov, I.R.; et al. Long shared haplotypes identify the southern Urals as a primary source for the 10th-century Hungarians. Cell 2025, 188, 6064–6078. [Google Scholar] [CrossRef] [PubMed]
- Beekes, R.S.P. Pre-Greek Phonology, Morphology, Lexicon; Brill NV: Leiden, The Netherlands, 2014. [Google Scholar]
- Csúcs, S. Miért Finnugor Nyelv a Magyar? Tinta Könyvkiadó: Budapest, Hungary, 2019. [Google Scholar]
- Mansi Dictionary of Munkácsi and Kálmán. Available online: https://www.babel.gwi.uni-muenchen.de/munka/ (accessed on 20 September 2025).
- Revesz, P.Z. Experimental evidence for a left-to-right reading direction of the Phaistos Disk. Mediterr. Archaeol. Archaeom. 2022, 22, 79–96. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).