Application of VGG16 Transfer Learning for Breast Cancer Detection
Abstract
:1. Introduction
- A modified version of VGG16 tailored for breast cancer detection using transfer learning.
- The application of class weighting to address dataset imbalances and improve the sensitivity to malignant cases.
- The implementation of cyclical learning rate (CLR) scheduling to enhance the training efficiency and generalization.
- An evaluation of VGG16 against other deep learning architectures, including VGG19, InceptionV3, and AlexNet, to justify its suitability for breast cancer detection.
- An extensive BreakHis dataset evaluation using the accuracy, recall, precision, and AUC to assess benign vs. malignant classification.
2. Related Works
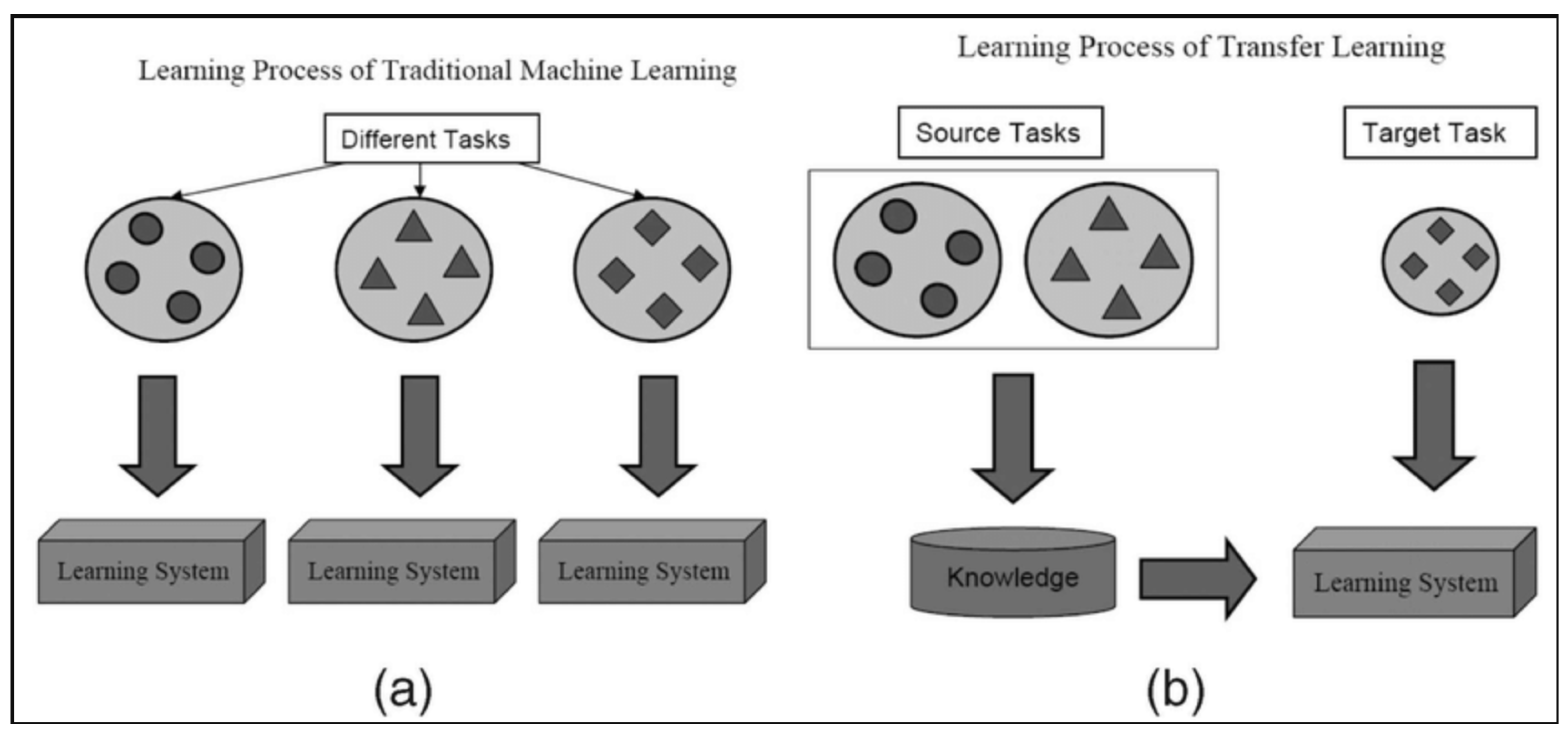
- Model Adaptability and Transfer Learning: VGG16’s sequential architecture facilitates smooth transfer learning and fine-tuning for medical imaging applications. Its structured convolutional layers enhance adaptation to histopathological datasets, ensuring reliable classification performance [13,17,22].
- Comparison with Alternative Architectures: While models such as ResNet [23], DenseNet [24], and InceptionV3 [25] achieve strong performance in medical imaging, they have more complex architectures and require substantial computational resources. Vision Transformers (ViTs) [26] and EfficientNet [27] offer promising results but demand extensive training data and are more prone to overfitting [26,27]. In contrast, VGG16, with its structured convolutional design, remains a widely used model for transfer learning and histopathological classification [22]. The architecture balances feature extraction and adaptability, making it a reliable choice for breast cancer detection [9,13].
- Study-Specific Enhancements: To further improve generalization and prevent overfitting, we integrated CLR scheduling, batch normalization, dropout, and L2 regularization. These enhancements contribute to classification stability, making VGG16 a computationally feasible and effective choice for breast cancer detection [28,29,30,31].
3. Our Approach
3.1. Dataset and Preprocessing
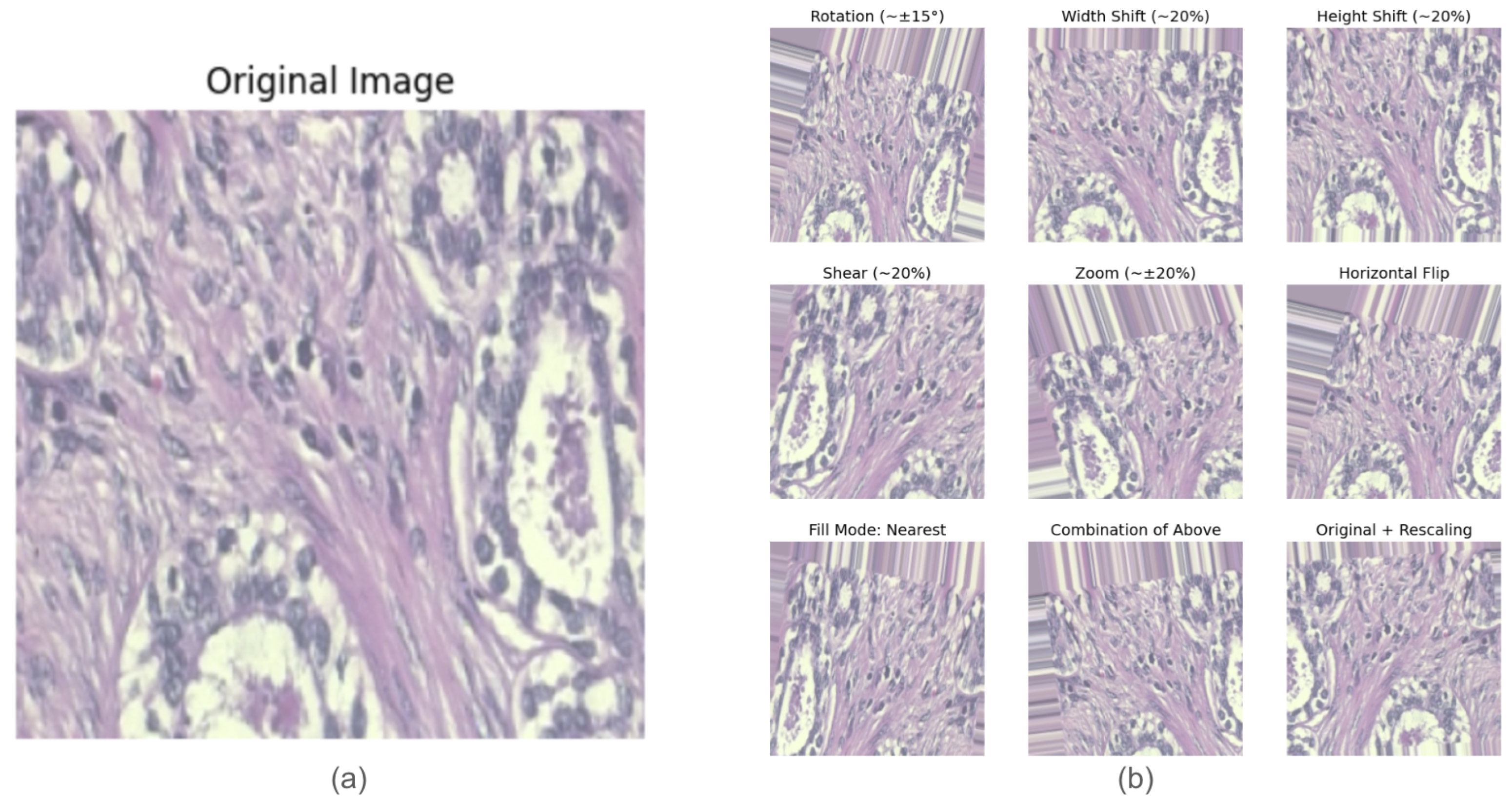
- Data Augmentation: Data augmentation techniques were applied to improve the model generalization and reduce overfitting [21]. Figure 3 shows examples of images before and after augmentation. The augmentation transformations included the following:
- –
- Rotation: Random rotations of up to 15 degrees.
- –
- Translation: Horizontal and vertical shifts of up to 20%.
- –
- Shearing: Up to 20% shearing transformation.
- –
- Zooming: Random zooms of up to 20%.
- –
- Horizontal Flipping: The creation of mirrored versions of the images.
- Handling the Class Imbalance: Given the dataset’s imbalance, where malignant samples significantly outnumber benign ones, we calculated class weights to assign higher importance to the minority class (benign) during training and counter this disparity [34]. The class weights were computed using the following formula [35]:By computing the total number of samples and dividing this value by the combined product of the number of classes and the number of samples per class, this technique assigns higher weights to the minority class (benign). For instance, if benign samples are much fewer than malignant samples, this formula will assign a higher weight to the benign class. These weights increased the penalty for misclassifying benign samples, encouraging the model to learn patterns equitably across both classes [23]. By emphasizing the minority class in this way, class weighting reduced bias toward the majority class and enhanced the model’s ability to classify both benign and malignant cases accurately.
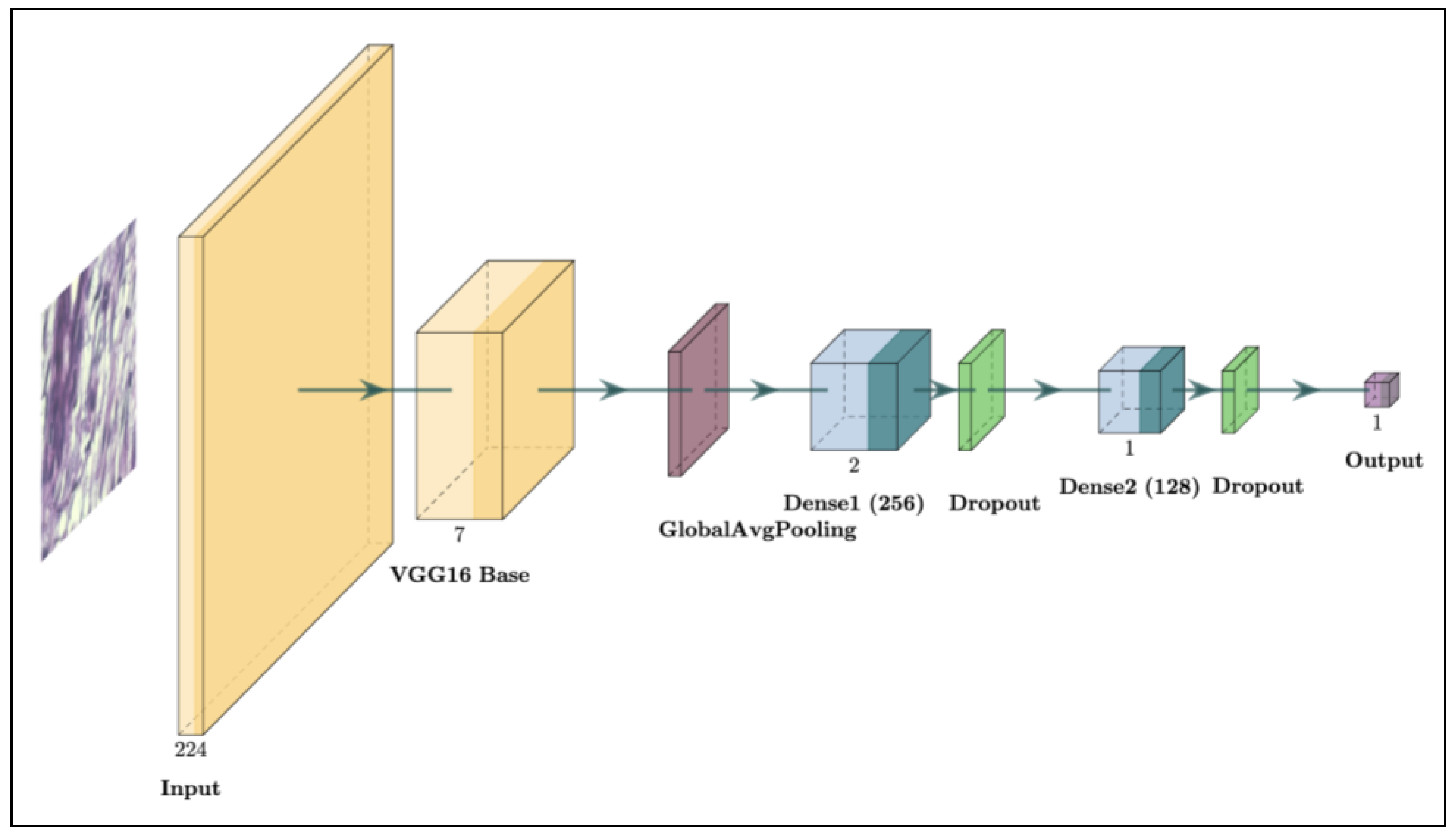
3.2. Model Architecture
3.2.1. Base Model: VGG16
- Global Average Pooling: A specialized layer, called GlobalAveragePooling2D, was introduced after the convolutional layers to replace the traditional Flatten layer. This pooling technique reduces the spatial dimensions of the feature maps, creating a more compact and generalized feature vector while reducing the risk of overfitting [38]. The Global Average Pooling layer computes the average output of each feature map, condensing it into a single value per map. This approach is effective in image classification tasks as it encourages the model to capture the global context rather than focusing on specific spatial locations, which can be advantageous in tasks like histopathological analysis where spatial features vary widely across samples.
- Dense Layers: Two Dense layers with 256 and 128 units, respectively, were added after the pooling layer. Each dense layer uses the ReLU (Rectified Linear Unit) activation function to introduce non-linearity and enable the model to learn complex patterns within the data [39]. Dense layers are essential in transforming the compacted features into higher-level representations for binary classification. L2 regularization, with a penalty coefficient of , was applied to both dense layers to help prevent overfitting by penalizing large weight values [31]. This regularization technique improved the model’s generalization by preventing overly complex fits to the training data, a key factor in medical imaging tasks where training datasets are often limited.
- Batch Normalization and Dropout: Batch normalization layers were incorporated after each dense layer to standardize the outputs, stabilizing and accelerating the training process [29]. Batch normalization mitigates internal covariate shifts by normalizing the layer inputs, leading to faster convergence and reduced sensitivity to the initialization parameters. Following each batch normalization layer, a Dropout layer with a rate of 0.25 was added. To mitigate overfitting, dropout randomly deactivates parts of the network during each training phase, supporting more generalized learning outcomes [30]. This combination of normalization and dropout has been shown to improve both a model’s stability and robustness, especially in TL scenarios where overfitting can be common due to limited data.
- Output Layer: A final fully connected layer with one output neuron and a sigmoid activation function was added to generate a probability score for binary classification. The sigmoid function outputs a value between 0 and 1, which can be interpreted as the probability of a sample belonging to the positive class [40]. If the probability is below 0.5, it indicates a higher likelihood of the tumor being benign; if the probability is 0.5 or higher, it indicates a higher likelihood of malignancy. This simple yet effective design makes it well suited for binary classification, ensuring clinically meaningful predictions [16,18].
3.2.2. Fine-Tuning the Base Model
3.2.3. Compilation and Hyperparameters
3.3. Comparative Analysis with Other Pre-Trained Models
4. Result Analysis
4.1. Evaluation Metrics
- Accuracy: The proportion of the total correct predictions, calculated asThe accuracy provides a general measure of the model’s performance across both classes.
- Precision: The ratio of correctly predicted positive samples to the total predicted positive samples, given byThe precision evaluates the model’s accuracy in predicting malignant cases, which is crucial to minimize false alarms.
- Recall: The ratio of correctly identified positive samples to all actual positive samples, defined asThe recall is especially important in identifying malignant cases accurately to avoid missed detections.
- F1-Score: A harmonic mean of the precision and recall, calculated asThe F1-score balances precision and recall, making it effective in handling class imbalances.
- Area Under the ROC Curve (AUC): The AUC evaluates the model’s proficiency in distinguishing between classes by calculating the area beneath the Receiver Operating Characteristic (ROC) curve, which illustrates the relationship between the True Positive Rate (TPR) and the False Positive Rate (FPR). A higher AUC value signifies improved classification performance, showcasing the model’s ability to accurately distinguish positive instances and reduce the number of false positives. This metric provides a comprehensive evaluation of the model’s classification capabilities, which is essential for achieving accurate and reliable diagnostic results.
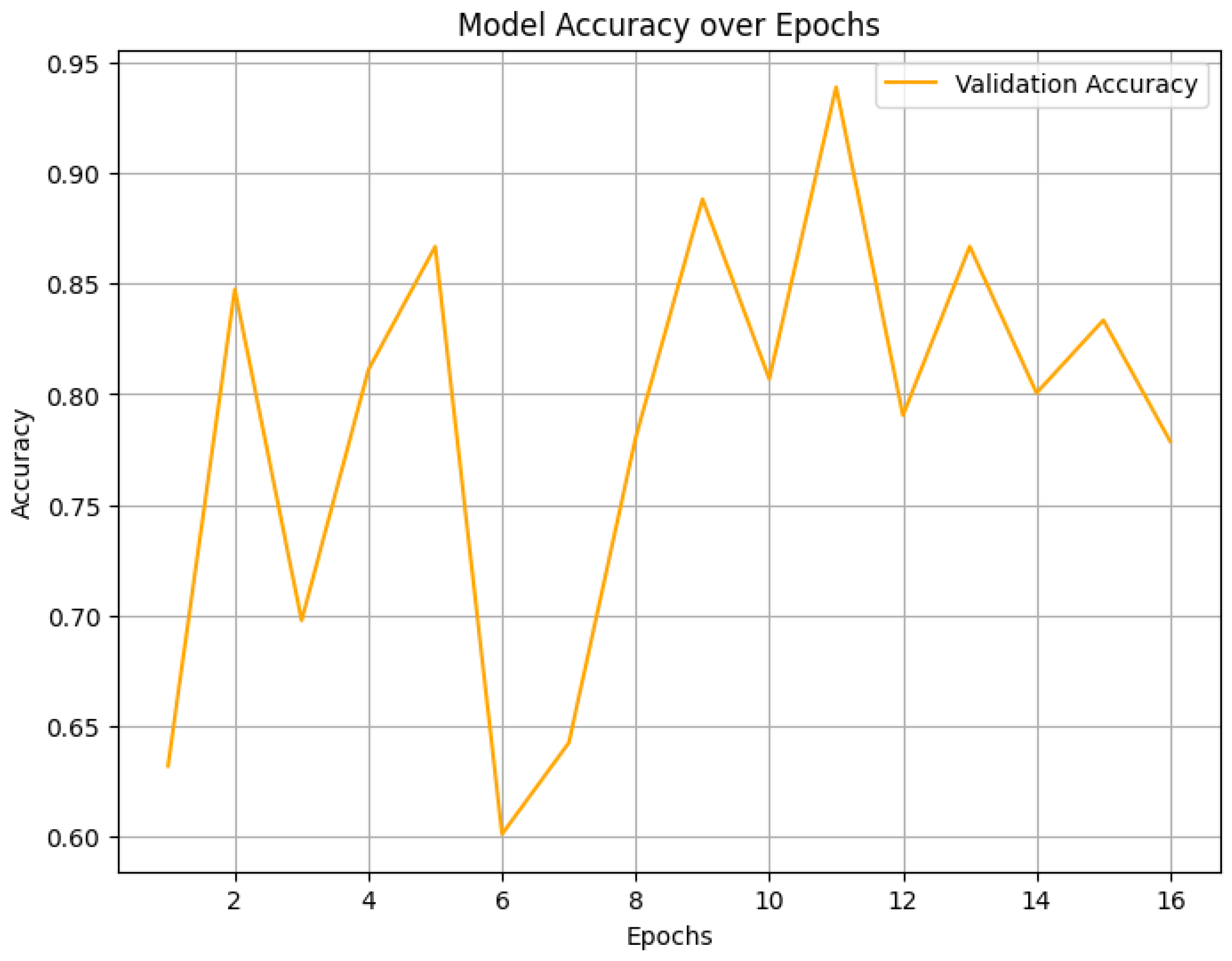
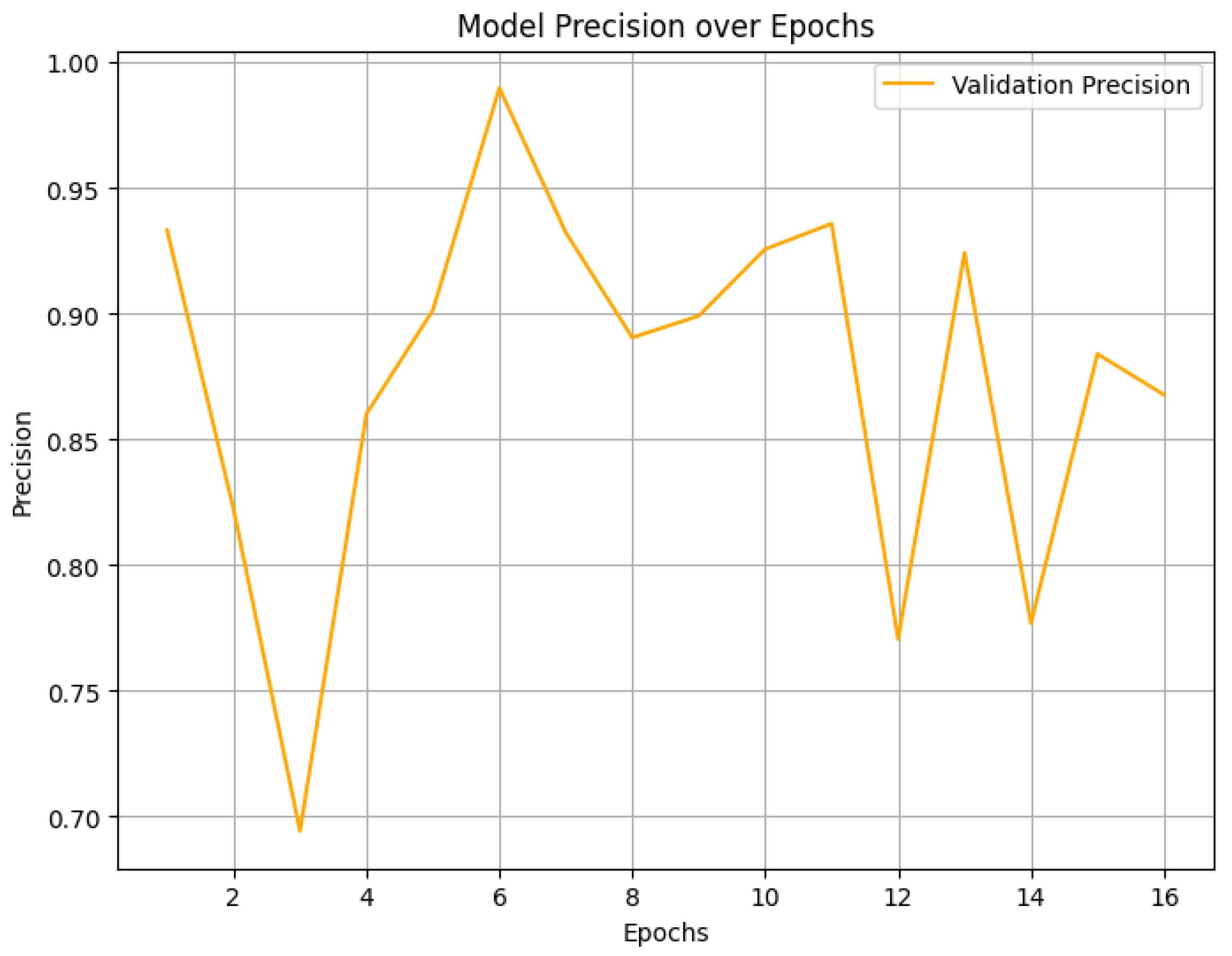
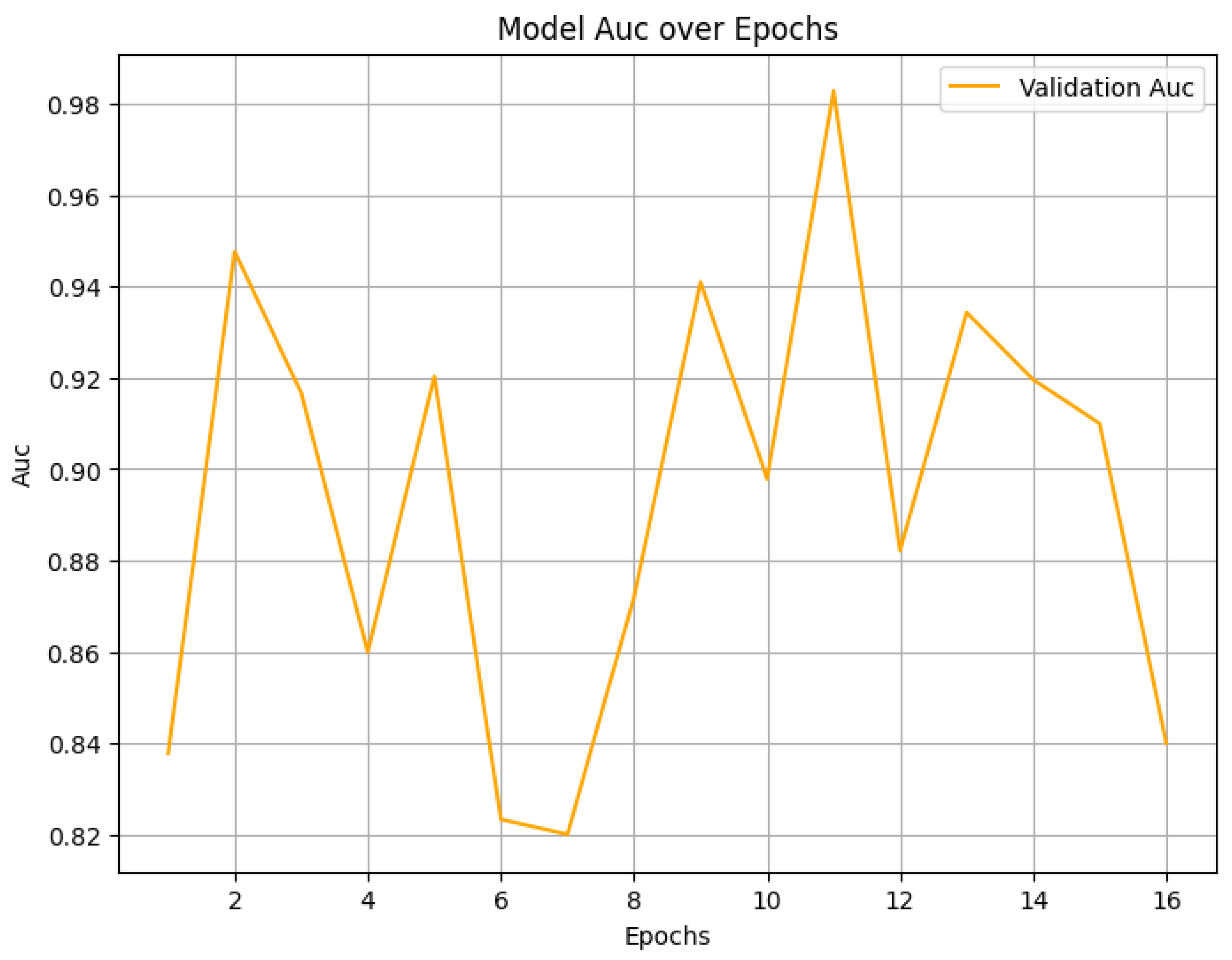
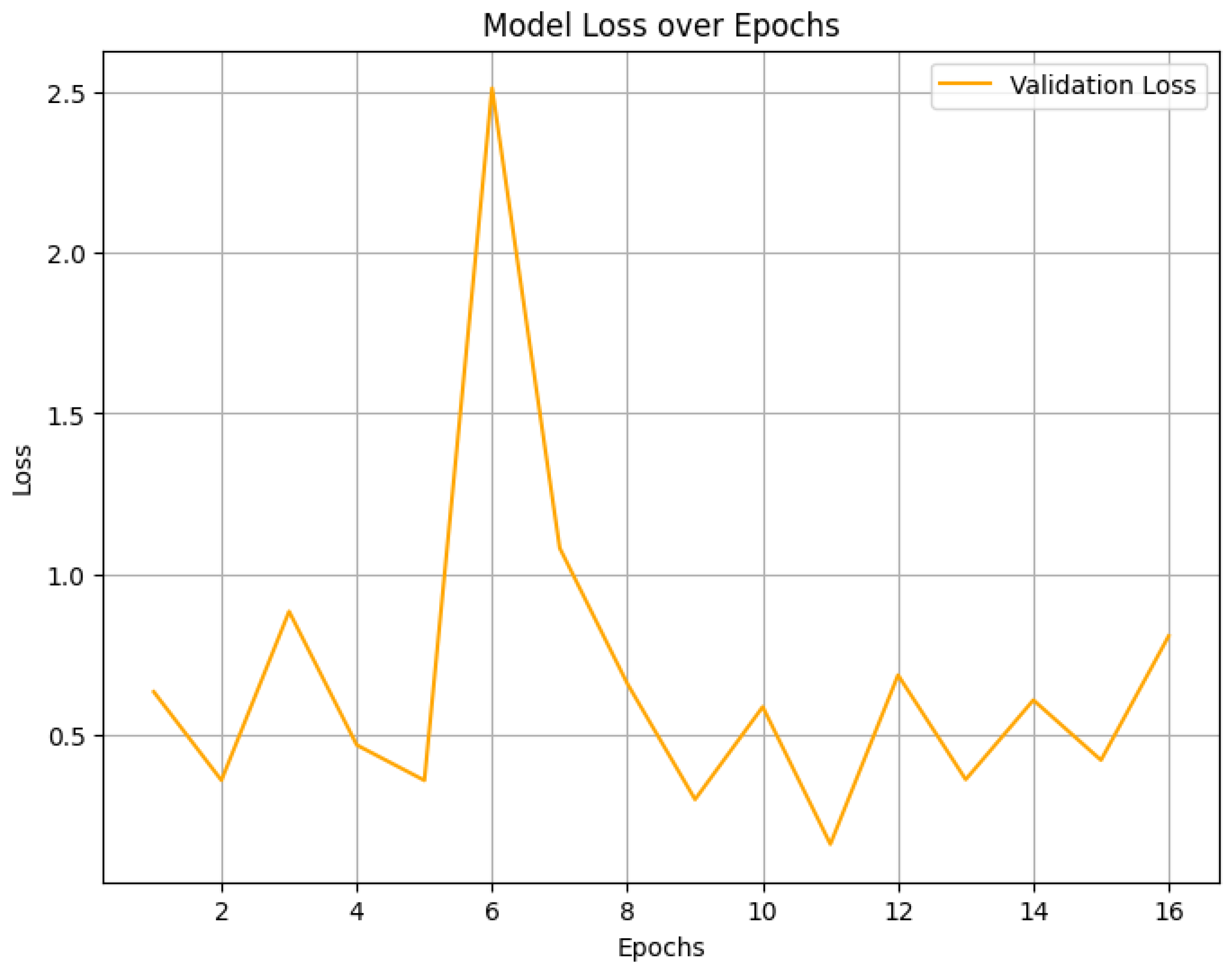
4.2. Final Performance Metrics
4.3. Different Pre-Trained Models’ Performance Comparison
4.4. Comparative Analysis with Previous Studies
4.5. Discussion
5. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. (Eds.) SEER Cancer Statistics Review, 1975–2017. 2020. Available online: https://seer.cancer.gov/csr/1975_2017/ (accessed on 13 January 2025).
- Bashar, M.A.; Begam, N. Breast cancer surpasses lung cancer as the most commonly diagnosed cancer worldwide. Indian J. Cancer 2022, 59, 438–439. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Morgan, E.; Rumgay, H.; Mafra, A.; Singh, D.; Laversanne, M.; Vignat, J.; Gralow, J.R.; Cardoso, F.; Siesling, S.; et al. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast 2022, 66, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Nover, A.B.; Jagtap, S.; Anjum, W.; Yegingil, H.; Shih, W.Y.; Shih, W.H.; Brooks, A.D. Modern breast cancer detection: A technological review. J. Biomed. Imaging 2009, 2009, 902326. [Google Scholar] [CrossRef] [PubMed]
- Nelson, H.D.; Tyne, K.; Naik, A.; Bougatsos, C.; Chan, B.K.; Humphrey, L. Screening for breast cancer: An update for the US Preventive Services Task Force. Ann. Intern. Med. 2009, 151, 727–737. [Google Scholar] [CrossRef]
- Yedjou, C.G.; Tchounwou, S.S.; Aló, R.A.; Elhag, R.; Mochona, B.; Latinwo, L. Application of machine learning algorithms in breast cancer diagnosis and classification. Int. J. Sci. Acad. Res. 2021, 2, 3081. [Google Scholar]
- Rodriguez, L.G.; Caballero, J.M.; Niguidula, J.D.; Calibo, D.I.; Rodriguez, C.A. eCommerce Sales Attrition: A Business Intelligence Visualization. In Proceedings of the Big Data Technologies and Applications: 8th International Conference, BDTA 2017, Gwangju, Republic of Korea, 23–24 November 2017; Proceedings 8. Springer: Cham, Switzerland, 2018; pp. 107–112. [Google Scholar]
- Pan, S.J.; Yang, Q. A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 2010, 22, 1345–1359. [Google Scholar] [CrossRef]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef]
- Raghu, M.; Zhang, C.; Kleinberg, J.; Bengio, S. Transfusion: Understanding transfer learning for medical imaging. Adv. Neural Inf. Process. Syst. 2019, 32, 1–22. [Google Scholar]
- Weiss, K.; Khoshgoftaar, T.M.; Wang, D. A survey of transfer learning. J. Big Data 2016, 3, 9. [Google Scholar] [CrossRef]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? Adv. Neural Inf. Process. Syst. 2014, 27, 1–9. [Google Scholar]
- Shallu; Mehra, R. Breast cancer histology images classification: Training from scratch or transfer learning? Ict Express 2018, 4, 247–254. [Google Scholar] [CrossRef]
- Ghafoorian, M.; Karssemeijer, N.; Heskes, T.; van Uden, I.W.; Sanchez, C.I.; Litjens, G.; de Leeuw, F.E.; van Ginneken, B.; Marchiori, E.; Platel, B. Location sensitive deep convolutional neural networks for segmentation of white matter hyperintensities. Sci. Rep. 2017, 7, 5110. [Google Scholar] [CrossRef]
- Li, F.; Liu, M.; The Alzheimer’s Disease Neuroimaging Initiative. A hybrid convolutional and recurrent neural network for hippocampus analysis in Alzheimer’s disease. J. Neurosci. Methods 2019, 323, 108–118. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Rana, M.; Bhushan, M. Classifying breast cancer using transfer learning models based on histopathological images. Neural Comput. Appl. 2023, 35, 14243–14257. [Google Scholar] [CrossRef]
- Hossain, A.A.; Nisha, J.K.; Johora, F. Breast cancer classification from ultrasound images using VGG16 model based transfer learning. Int. J. Image Graph. Signal Process. 2023, 13, 12. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Prusty, S.; Dash, S.K.; Patnaik, S. A novel transfer learning technique for detecting breast cancer mammograms using VGG16 bottleneck feature. ECS Trans. 2022, 107, 733. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- He, H.; Garcia, E.A. Learning from imbalanced data. IEEE Trans. Knowl. Data Eng. 2009, 21, 1263–1284. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Tan, M.; Le, Q. EfficientNet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning (ICML), Long Beach, CA, USA, 10–15 June 2019; pp. 6105–6114. [Google Scholar]
- Smith, L.N. Cyclical learning rates for training neural networks. In Proceedings of the 2017 IEEE Winter Conference on Applications of Computer Vision (WACV), Santa Rosa, CA, USA, 24–31 March 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 464–472. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2015; pp. 448–456. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Krogh, A.; Hertz, J.A. A simple weight decay can improve generalization. Adv. Neural Inf. Process. Syst. 1992, 4, 950–957. [Google Scholar]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. A dataset for breast cancer histopathological image classification. IEEE Trans. Biomed. Eng. 2015, 63, 1455–1462. [Google Scholar]
- Bayramoglu, N.; Kannala, J.; Heikkila, J. Deep Learning for Magnification Independent Breast Cancer Histopathology Image Classification. In Proceedings of the 23rd International Conference on Pattern Recognition (ICPR), Cancun, Mexico, 4–8 December 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 2440–2445. [Google Scholar]
- Cui, Y.; Jia, M.; Lin, T.Y.; Song, Y.; Belongie, S. Class-balanced loss based on effective number of samples. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 9268–9277. [Google Scholar]
- King, G.; Zeng, L. Logistic regression in rare events data. Political Anal. 2001, 9, 137–163. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Liu, S.; Deng, W. Very deep convolutional neural network based image classification using small training sample size. In Proceedings of the 2015 3rd IAPR Asian Conference on Pattern Recognition (ACPR), Kuala Lumpur, Malaysia, 3–6 November 2015; IEEE: Piscataway, NJ, USA, 2015; pp. 730–734. [Google Scholar]
- Lin, M.; Chen, Q.; Yan, S. Network in network. arXiv 2013, arXiv:1312.4400. [Google Scholar]
- Nair, V.; Hinton, G.E. Rectified linear units improve restricted Boltzmann machines. In Proceedings of the 27th International Conference on Machine Learning (ICML-10), Haifa, Israel, 21–24 June 2010; pp. 807–814. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Murphy, K.P. Machine Learning: A Probabilistic Perspective; MIT Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Zhang, J.; He, T.; Sra, S.; Jadbabaie, A. Why gradient clipping accelerates training: A theoretical justification for adaptivity. In Proceedings of the International Conference on Learning Representations, Addis Ababa, Ethiopia, 26–30 April 2020. [Google Scholar]
- Prechelt, L. Early stopping-but when. In Neural Networks: Tricks of the Trade; Springer: Berlin/Heidelberg, Germany, 2002; pp. 55–69. [Google Scholar]
- Chollet, F. Deep Learning with Python; Simon and Schuster: New York, NY, USA, 2021. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 25, 1–9. [Google Scholar] [CrossRef]
- Otten, J.D.; Karssemeijer, N.; Hendriks, J.H.; Groenewoud, J.H.; Fracheboud, J.; Verbeek, A.L.M.; de Koning, H.J.; Holland, R. Effect of recall rate on earlier screen detection of breast cancers based on the Dutch performance indicators. J. Natl. Cancer Inst. 2005, 97, 748–754. [Google Scholar] [CrossRef]
- Agarwal, P.; Yadav, A.; Mathur, P. Breast cancer prediction on breakhis dataset using deep cnn and transfer learning model. In Proceedings of the Data Engineering for Smart Systems: Proceedings of SSIC 2021, Jaipur, India, 22–23 January 2022; Springer: Singapore, 2022; pp. 77–88. [Google Scholar]
- Singh, O.; Adnan, M.H.; Tabassum, T.; Rahman, A. A VGG16-Based Deep Learning System for Accurate Detection of Breast Cancer in Histopathology Images. J. Adv. Res. Artif. Intell. Its Appl. 2024, 1, 57–64. [Google Scholar]



| Magnification | Benign | Malignant | Total |
|---|---|---|---|
| 40× | 652 | 1370 | 1995 |
| 100× | 644 | 1437 | 2081 |
| 200× | 623 | 1390 | 2013 |
| 400× | 588 | 1232 | 1820 |
| Total | 2480 | 5429 | 7909 |
| Metric | Random (Before Augmentation) | Random (After Augmentation) | Stratified (After Augmentation) |
|---|---|---|---|
| Validation Accuracy | 83.09% | 93.68% | 93.09% |
| Precision | 83.58% | 93.22% | 92.22% |
| Recall | 93.80% | 97.91% | 98.22% |
| AUC | 0.9065 | 0.9838 | 0.9768 |
| Loss | 0.3742 | 0.1442 | 0.2043 |
| F1-Score | 88.43% | 95.52% | 95.13% |
| Model | Acc. (%) | Prec. (%) | Rec. (%) | AUC | Time (s/Epoch) |
|---|---|---|---|---|---|
| M-VGG16 | 93.68 | 93.22 | 97.91 | 0.9838 | 112.5 |
| M-VGG19 | 92.30 | 91.85 | 96.55 | 0.9810 | 130.8 |
| M-InceptionV3 | 91.45 | 92.00 | 95.12 | 0.9756 | 98.7 |
| M-AlexNet | 85.68 | 88.00 | 83.95 | 0.9403 | 57.2 |
| Model | Memory (MB) | Inference Time (ms) | Approx. GFLOPs |
|---|---|---|---|
| M-VGG16 | 56.76 | 41.56 | 1476.04 |
| M-VGG19 | 77.02 | 44.00 | 2008.68 |
| M-InceptionV3 | 85.30 | 58.20 | 31.44 |
| M-AlexNet | 14.68 | 25.78 | 21.84 |
| Recent Articles on Detecting Breast Cancer with TL | Dataset | Model | Accuracy (%) | Recall (%) | Precision (%) | F1-Score (%) |
|---|---|---|---|---|---|---|
| Mehra et al. [13] | BreakHis | Standard VGG16 | 92.60 | 93.00 | 93.00 | 93.00 |
| Rana et al. [17] | BreakHis | VGG16 with smaller kernel sizes | 67.51 | 95.24 | 36.86 | 55.28 |
| Agarwal et al. [49] | BreakHis | VGG16 with SMOTE preprocessing | 94.67 | 80.52 | 92.60 | 85.21 |
| Singh et al. [50] | BreakHis | VGG16-based CNN | 93.00 | 91.00 | 94.00 | 92.47 |
| Proposed M-VGG16 Model | BreakHis | Modified VGG16 with CLR scheduler | 93.68 | 97.91 | 93.22 | 95.52 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fatima, T.; Soliman, H. Application of VGG16 Transfer Learning for Breast Cancer Detection. Information 2025, 16, 227. https://doi.org/10.3390/info16030227
Fatima T, Soliman H. Application of VGG16 Transfer Learning for Breast Cancer Detection. Information. 2025; 16(3):227. https://doi.org/10.3390/info16030227
Chicago/Turabian StyleFatima, Tanjim, and Hamdy Soliman. 2025. "Application of VGG16 Transfer Learning for Breast Cancer Detection" Information 16, no. 3: 227. https://doi.org/10.3390/info16030227
APA StyleFatima, T., & Soliman, H. (2025). Application of VGG16 Transfer Learning for Breast Cancer Detection. Information, 16(3), 227. https://doi.org/10.3390/info16030227







