Pharmacophore-Guided Identification of Natural Products as Potential Inhibitors of Mycobacterium ulcerans Cystathionine γ-Synthase MetB
Abstract
1. Introduction
2. Materials and Methods
2.1. Target Retrieval and Preparation
2.2. Compound Selection for Pharmacophore Generation
2.3. Ligand-Based Pharmacophore Virtual Screening
2.4. Pre-Filtering of the Library for Pharmacophore-Based Screening
2.5. Pharmacophore-Based Screening of the Library
2.6. Validation of AutoDock Vina
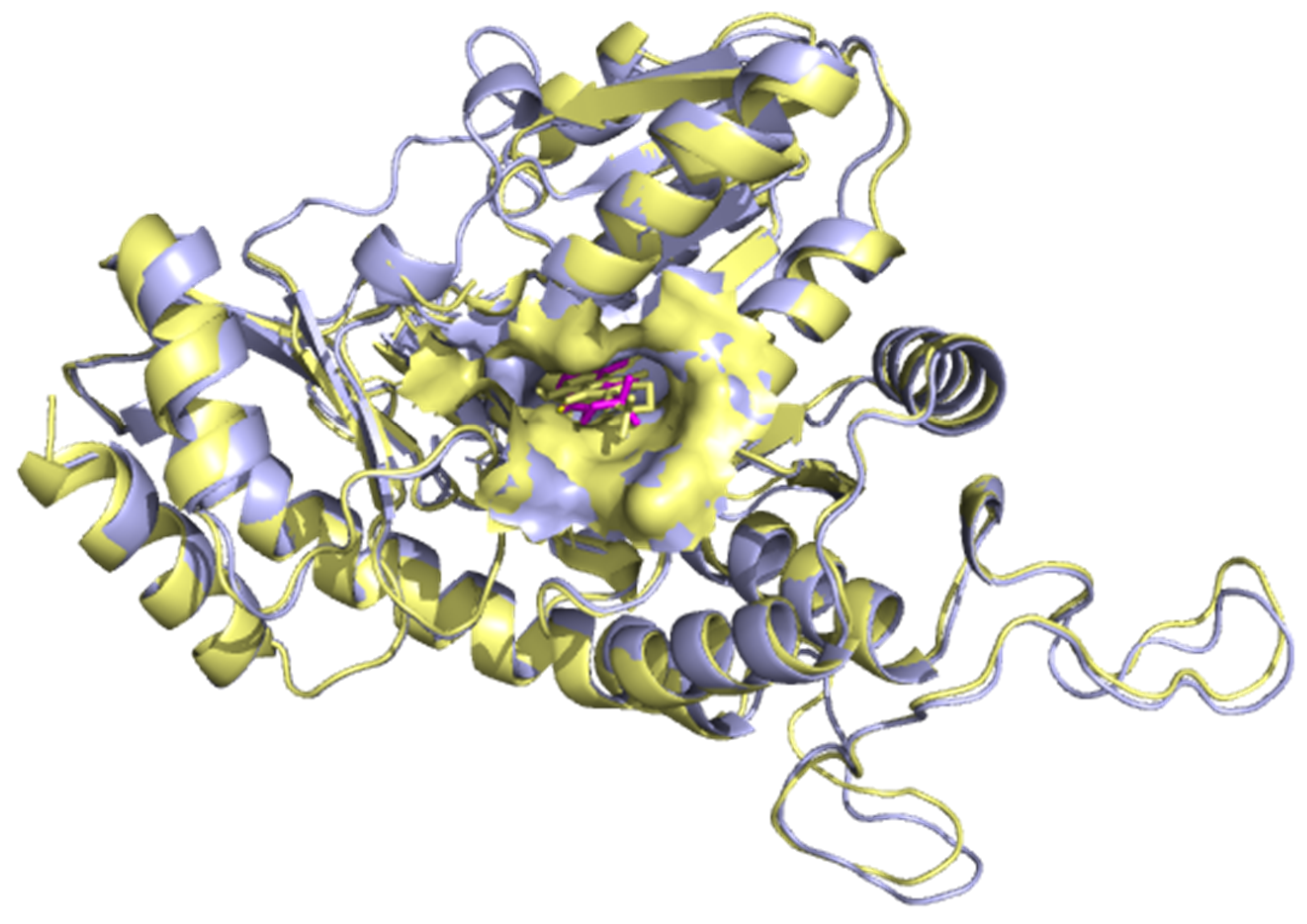
2.6.1. Superimposition of Co-Crystallized with Re-Docked Complexes
2.6.2. ROC Curve Analysis
2.7. Virtual Screening of the Library
2.8. Protein-Ligand Interaction
2.9. Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) Prediction
2.10. Prediction of Activity Spectra for Substances and Structural Similarity Analogues (PASS)
2.11. Molecular Dynamics Simulation
2.12. Molecular Mechanics Poisson-Boltzmann Surface Area Binding Free Energy Calculations
3. Results and Discussion
3.1. Target Description
3.2. Ligand-Based Pharmacophore Virtual Screening
3.2.1. Pharmacophore Generation
3.2.2. Validation of Pharmacophore Model
3.3. Validation of Generated Pharmacophore Model
3.4. Pharmacophore-Based Screening of the Library
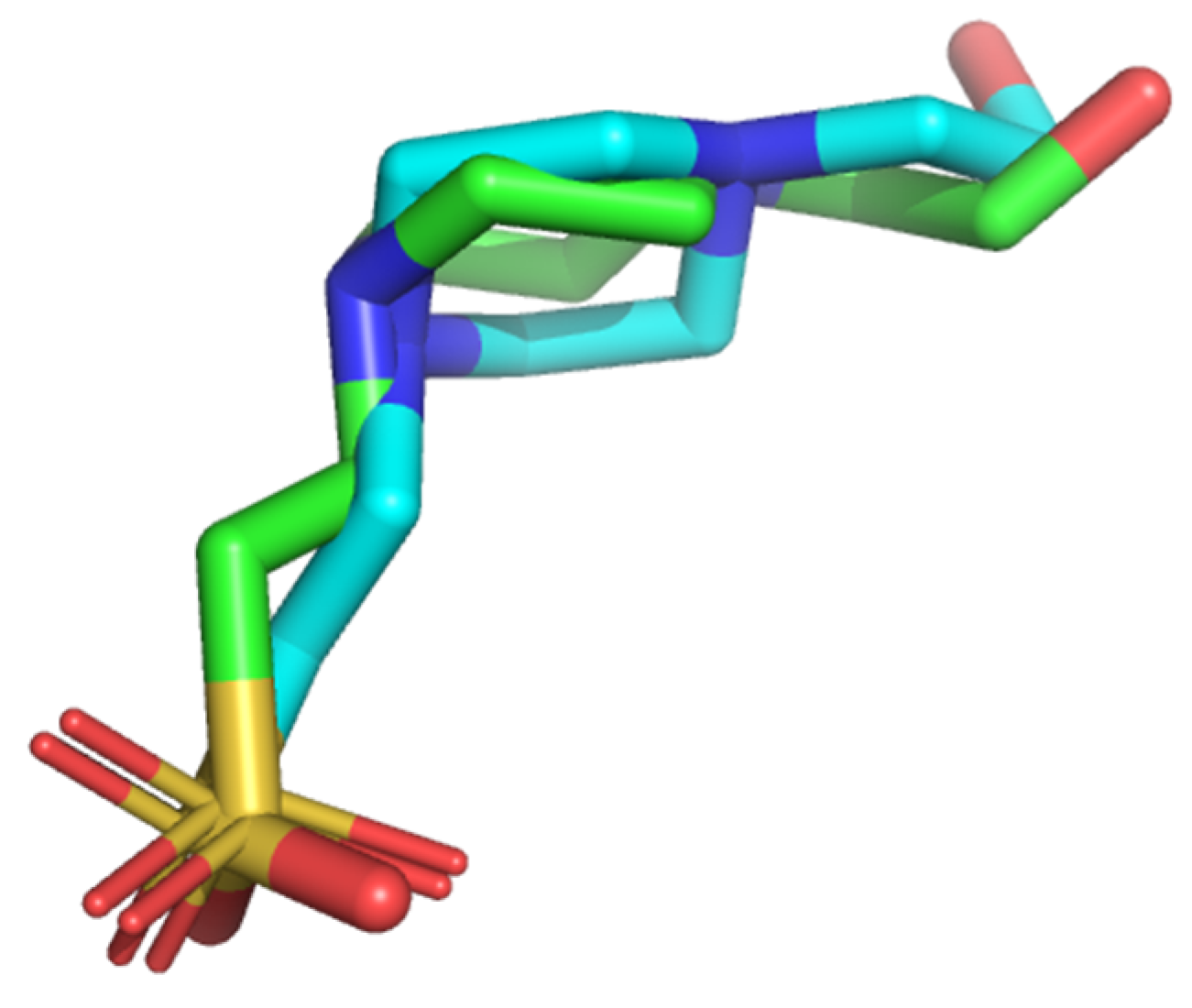
3.5. Validation of Molecular Docking Protocol
3.5.1. Superimposition of Co-Crystals with Re-Docked Complexes
3.5.2. ROC Curve Analysis of the Molecular Docking Protocol
3.6. Molecular Docking of Pharmacophore Hits
3.7. Protein-Ligand Interaction
3.8. Physicochemical Profiling
3.9. Pharmacokinetics and Toxicity Studies
3.10. Exploring Predicted Leads for Anti-Microbial and Antimycobacterial Activity.
3.11. Molecular Dynamics (MD) Simulation of Target Structure and Complexes
3.12. MM-PBSA Binding Free Energy Calculations
3.13. Leads Summary
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CGS | Cystathionine Gamma Synthase |
| PLP | Pyridoxal Phosphate |
| BU | Buruli Ulcer |
| MetB | Methionine B |
| NANPDB | North African Natural Product Database |
| AfroDB | Library of natural products from African origin |
| SDF | Structure Data File |
| UFF | Universal Force Field |
| PDB | Protein Data Bank |
| MD | Molecular Dynamics |
| HPC | High-Performance Computing |
| GROMACS | GROningen MAchine for Chemical Simulations |
| TPSA | Topological Polar Surface Area |
| RMSD | Root Mean Square Deviation |
| EF | Enrichment Factor |
| ESOL | Effective Solubility |
| DUD-E | Database of Useful (Docking) Decoys—Enhanced |
| AMES | Salmonella typhimurium reverse mutation assay |
| Log P | Logarithm of the octan-1-ol/water partition coefficient |
| MW | Molecular Weight |
| ID | Identification |
| P-gp | Permeability glycoprotein |
| CYP | Cytochromes P450 |
References
- Portaels, F.; Silva, M.T.; Meyers, W.M. Buruli ulcer. Clin. Dermatol. 2009, 27, 291–305. [Google Scholar] [CrossRef]
- Evans, M.R.W.; Thangaraj, H.S.; Wansbrough-Jones, M.H. Buruli ulcer. Curr. Opin. Infect. Dis. 2000, 13, 109–112. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Buruli ulcer disease: Mycobacterium ulcerans infection: An overview of reported cases globally. Wkly. Epidemiol. Rec. 2004, 79, 194–199. [Google Scholar]
- Ampah, K.A.; Asare, P.; De-Graft Binnah, D.; Maccaulley, S.; Opare, W.; Röltgen, K.; Pluschke, G.; Yeboah-Manu, D. Burden and historical trend of Buruli ulcer prevalence in selected communities along the Offin River of Ghana. PLoS Negl. Trop. Dis. 2016, 10, e0004603. [Google Scholar] [CrossRef]
- Zhang, T.; Bishai, W.R.; Grosset, J.H.; Nuermberger, E.L. Rapid assessment of antibacterial activity against Mycobacterium ulcerans by using recombinant luminescent strains. Antimicrob. Agents Chemother. 2010, 54, 2806–2813. [Google Scholar] [CrossRef] [PubMed]
- Merritt, R.W.; Walker, E.D.; Small, P.L.; Wallace, J.R.; Johnson, P.D.; Benbow, M.E.; Boakye, D.A. Ecology and Transmission of Buruli Ulcer Disease: A systematic review. PLoS Negl. Trop. Dis. 2010, 4, e911. [Google Scholar] [CrossRef] [PubMed]
- Clifton, M.C.; Abendroth, J.; Edwards, T.E.; Leibly, D.J.; Gillespie, A.K.; Ferrell, M.; Dieterich, S.H.; Exley, I.; Staker, B.L.; Myler, P.J.; et al. Structure of the cystathionine γ-synthase MetB from Mycobacterium ulcerans. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 1154–1158. [Google Scholar] [CrossRef]
- Berney, M.; Berney-Meyer, L.; Wong, K.W.; Chen, B.; Chen, M.; Kim, J.; Wang, J.; Harris, D.; Parkhill, J.; Chan, J.; et al. Essential roles of methionine and S-adenosylmethionine in the autarkic lifestyle of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2015, 112, 10008–10013. [Google Scholar] [CrossRef]
- Walsh, D.S.; Portaels, F.; Meyers, W.M. Buruli Ulcer: Advances in understanding Mycobacterium ulcerans infection. Dermatol. Clin. 2011, 29, 1–8. [Google Scholar] [CrossRef]
- Yotsu, R.R.; Richardson, M.; Ishii, N. Drugs for treating Buruli ulcer (Mycobacterium ulcerans disease). Cochrane Database Syst. Rev. 2018, 2018, CD012118. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; MacArthur, C.; Mubila, L.; Baker, S. Control of neglected tropical diseases needs a long-term commitment. BMC Med. 2010, 8, 67. [Google Scholar] [CrossRef]
- Kwofie, S.K.; Adobor, C.; Quansah, E.; Bentil, J.; Ampadu, M.; Miller, W.A., 3rd; Wilson, M.D. Molecular docking and dynamics simulations studies of OmpATb identifies four potential novel natural product-derived anti-Mycobacterium tuberculosis compounds. Comput. Biol. Med. 2020, 122, 103811. [Google Scholar] [CrossRef]
- Siddiqui, A.A.; Iram, F.; Siddiqui, S.; Sahu, K. Role of natural products in drug discovery process. Int. J. Drug Dev. Res. 2014, 6, 172–204. Available online: https://www.ijddr.in/drug-development/role-of-natural-products-in-drug-discovery-process.php?aid=5524 (accessed on 15 February 2019).
- Ekor, M. The growing use of herbal medicines: Issues relating to adverse reactions and challenges in monitoring safety. Front. Pharm. 2014, 4, 177. [Google Scholar] [CrossRef]
- Dias, D.A.; Urban, S.; Roessner, U. A historical overview of natural products in drug discovery. Metabolites 2012, 2, 303–336. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Kleywegt, G.J.; Alwyn Jones, T. Model building and refinement practice. Methods Enzymol. 1997, 277, 208–230. [Google Scholar] [CrossRef]
- Lill, M.A.; Danielson, M.L. Computer-aided drug design platform using PyMOL. J. Comput. Mol. Des. 2011, 25, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Chan, H.C.S.; Hu, Z. Using PyMOL as a platform for computational drug design. Wires Comput. Mol. Sci. 2017, 7, e1298. [Google Scholar] [CrossRef]
- Bordoli, L.; Schwede, T. Automated protein structure modeling with SWISS-MODEL Workspace and the protein model portal. Methods Mol. Biol. 2012, 857, 107–136. [Google Scholar] [CrossRef] [PubMed]
- Spoel, D.V.D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- NCBI Resource Cordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2013, 41, D8–D20. [Google Scholar] [CrossRef]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bairoch, A. UniProtKB/Swiss-Prot. In Plant Bioinformatics: Methods and Protocols; Edwards, D., Totowa, E., Eds.; Humana Press: Totowa, NJ, USA, 2007; pp. 89–112. [Google Scholar] [CrossRef]
- Kong, Y.H.; Zhang, L.; Yang, Z.Y.; Han, C.; Hu, L.H.; Jiang, H.L.; Shen, X. Natural product juglone targets three key enzymes from Helicobacter pylori: Inhibition assay with crystal structure characterization. Acta Pharmacol. Sin. 2008, 29, 870–876. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Wu, D.; Bai, H.; Han, C.; Chen, J.; Chen, L.; Hu, L.; Jiang, H.; Shen, X. Enzymatic characterization and inhibitor discovery of a new Cystathionine γ-Synthase from Helicobacter pylori. J. Biochem. 2008, 143, 59–68. [Google Scholar] [CrossRef]
- Wolber, G.; Langer, T. LigandScout: 3-D Pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 2005, 45, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Shoichet, B.K. ZINC—A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005, 45, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Gidaro, M.C.; Alcaro, S.; Secci, D.; Rivanera, D.; Mollica, A.; Agamennone, M.; Giampietro, L.; Carradori, S. Identification of new anti-Candida compounds by ligand-based pharmacophore virtual screening. J. Enzym. Inhib. Med. Chem. 2016, 31, 1703–1706. [Google Scholar] [CrossRef] [PubMed]
- Lagorce, D.; Bouslama, L.; Becot, J.; Miteva, M.A.; Villoutreix, B.O. FAF-Drugs4: Free ADME-tox filtering computations for chemical biology and early stages drug discovery. Bioinformatics 2017, 33, 3658–3660. [Google Scholar] [CrossRef] [PubMed]
- Heifets, A.; Lilien, R.H. LigAlign: Flexible ligand-based active site alignment and analysis. J. Mol. Graph. Model. 2010, 29, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Goksuluk, D.; Korkmaz, S.; Zararsiz, G.; Karaagaoglu, E.A. easyROC: An interactive web-tool for ROC curve analysis using R language environment. R J. 2016, 8, 213. [Google Scholar] [CrossRef]
- Lätti, S.; Niinivehmas, S.; Pentikäinen, O.T. Rocker: Open source, easy-to-use tool for AUC and enrichment calculations and ROC visualization. J. Cheminf. 2016, 8, 45. [Google Scholar] [CrossRef]
- Mysinger, M.M.; Carchia, M.; Irwin, J.J.; Shoichet, B.K. Directory of useful decoys, enhanced (DUD-E): Better ligands and decoys for better benchmarking. J. Med. Chem. 2012, 55, 6582–6594. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Cheng, F.; Li, W.; Zhou, Y.; Shen, J.; Wu, Z.; Liu, G.; Lee, P.W.; Tang, Y. admetSAR: A comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model. 2012, 52, 3099–3105. [Google Scholar] [CrossRef] [PubMed]
- Benet, L.Z.; Hosey, C.M.; Ursu, O.; Oprea, T.I. BDDCS, the Rule of 5 and drugability. Adv. Drug Deliv. Rev. 2016, 101, 89–98. [Google Scholar] [CrossRef]
- Lagunin, A.; Stepanchikova, A.; Filimonov, D.; Poroikov, V. PASS: Prediction of activity spectra for biologically active substances. Bioinformatics 2000, 16, 747–748. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Cheng, D.; Shrivastava, S.; Tzur, D.; Gautam, B.; Hassanali, M. DrugBank: A knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008, 36, D901–D906. [Google Scholar] [CrossRef]
- Turner, P.J. XMGRACE, Version 5.1. 19. In Center for Coastal and Land-Margin Research; Oregon Graduate Institute of Science and Technology: Beaverton, OR, USA, 2005. [Google Scholar]
- Kumari, R.; Kumar, R.; Lynn, A. g_mmpbsa—A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef]
- Hevener, K.E.; Zhao, W.; Ball, D.M.; Babaoglu, K.; Qi, J.; White, S.W.; Lee, R.E. Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J. Chem. Inf. Model. 2009, 49, 444–460. [Google Scholar] [CrossRef]
- Hajian-Tilaki, K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Casp. J. Intern. Med. 2013, 4, 627–635. Available online: https://pubmed.ncbi.nlm.nih.gov/24009950/ (accessed on 2 March 2019).
- Greiner, M.; Pfeiffer, D.; Smith, R.D. Principles and practical application of the receiver-operating characteristic analysis for diagnostic tests. Prev. Vet. Med. 2000, 45, 23–41. [Google Scholar] [CrossRef]
- Jiménez-Valverde, A. Insights into the area under the receiver operating characteristic curve (AUC) as a discrimination measure in species distribution modelling. Glob. Ecol. Biogeogr. 2011, 21, 498–507. [Google Scholar] [CrossRef]
- Lasko, T.A.; Bhagwat, J.G.; Zou, K.H.; Ohno-Machado, L. The use of receiver operating characteristic curves in biomedical informatics. J. Biomed. Inform. 2005, 38, 404–415. [Google Scholar] [CrossRef] [PubMed]
- Lewis, R.A.; Sirockin, F. 2/3D Pharmacophore definitions and their application. Ref. Modul. Chem. Mol. Sci. Chem. Eng. 2016. [Google Scholar] [CrossRef]
- Çifci, G.; Aviyente, V.; Akten, E.D. Molecular docking study based on pharmacophore modeling for novel phosphodiesteraseiv inhibitors. Mol. Inform. 2012, 31, 459–471. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar] [CrossRef]
- Lionta, E.; Spyrou, G.; Vassilatis, D.K.; Cournia, Z. Structure-based virtual screening for drug discovery: Principles, applications and recent advances. Curr. Top. Med. Chem. 2014, 14, 1923–1938. [Google Scholar] [CrossRef]
- Gimeno, A.; Ojeda-Montes, M.J.; Tomás-Hernández, S.; Cereto-Massagué, A.; Beltrán-Debón, R.; Mulero, M.; Pujadas, G.; Garcia-Vallvé, S. The Light and Dark Sides of Virtual Screening: What is there to know? Int. J. Mol. Sci. 2019, 20, 1375. [Google Scholar] [CrossRef] [PubMed]
- Kastritis, P.L.; Bonvin, A.M.J. On the binding affinity of macromolecular interactions: Daring to ask why proteins interact. J. R. Soc. Interface 2013, 10. [Google Scholar] [CrossRef]
- Du, X.; Li, Y.; Xia, Y.L.; Ai, S.M.; Liang, J.; Sang, P.; Ji, X.L.; Liu, S.Q. Insights into protein–ligand interactions: Mechanisms, models, and methods. Int. J. Mol. Sci. 2016, 17, 144. [Google Scholar] [CrossRef]
- Herschlag, D.; Pinney, M.M. Hydrogen bonds: Simple after all? Biochemistry 2018, 57, 3338–3352. [Google Scholar] [CrossRef] [PubMed]
- Bulusu, G.; Desiraju, G. Strong and weak hydrogen bonds in protein–ligand recognition. J. Indian Inst. Sci. 2019, 100, 31–41. [Google Scholar] [CrossRef]
- Hubbard, R.; Haider, M. Hydrogen bonds in proteins: Role and strength. Encycl. Life Sci. 2010, 1. [Google Scholar] [CrossRef]
- Schreiber, G.; Keating, A.E. Protein binding specificity versus promiscuity. Curr. Opin. Struct. Biol. 2011, 21, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.Y.; Zheng, H.H.; Zhang, K.Y.; Yang, F.; Kong, T.; Zhou, B.; Jiang, S.X. The roles of cytochrome P450 and P-glycoprotein in the pharmacokinetics of florfenicol in chickens. Iran. J. Vet. Res. 2018, 19, 9–14. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5960766/ (accessed on 19 June 2019).
- Devadasu, V.R.; Deb, P.K.; Maheshwari, R.; Sharma, P.; Tekade, R.K. Physicochemical, pharmaceutical, and biological considerations in GIT absorption of drugs. In Dosage Form Design Considerations; Tekade, R.K., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 149–178. [Google Scholar]
- Suenderhauf, C.; Hammann, F.; Huwyler, J. Computational prediction of blood-brain barrier permeability using decision tree induction. Molecules 2012, 17, 10429–10445. [Google Scholar] [CrossRef]
- Samiei, M.; Asgary, S.; Farajzadeh, M.; Bargahi, N.; Abdolrahimi, M.; Kananizadeh, U.; Dastmalchi, S. Investigating the mutagenic effects of three commonly used pulpotomy agents using the ames test. Adv. Pharm. Bull. 2015, 5, 121–125. [Google Scholar] [CrossRef]
- Wang, W.Q.; Duan, H.X.; Pei, Z.T.; Xu, R.R.; Qin, Z.T.; Zhu, G.C.; Sun, L.W. Evaluation by the Ames assay of the mutagenicity of UV filters using benzophenone and benzophenone-1. Int. J. Environ. Res. Public Health 2018, 15, 1907. [Google Scholar] [CrossRef]
- Yasuda, C.; Yasuda, S.; Yamashita, H.; Okada, J.; Hisada, T.; Sugiura, S. The human ether-a-go-go-related gene (hERG) current inhibition selectively prolongs action potential of midmyocardial cells to augment transmural dispersion. J. Physiol. Pharm. 2015, 66, 599–607. Available online: https://pubmed.ncbi.nlm.nih.gov/26348084/ (accessed on 21 June 2019).
- Danker, T.; Möller, C. Early identification of hERG liability in drug discovery programs by automated patch clamp. Front. Pharm. 2014, 5, 203. [Google Scholar] [CrossRef] [PubMed]
- Guha, R. On exploring structure activity relationships. Methods Mol. Biol. 2013, 993, 81–94. [Google Scholar] [CrossRef]
- Benchabane, Y.; Di Giorgio, C.; Boyer, G.; Sabatier, A.S.; Allegro, D.; Peyrot, V.; De Méo, M. Photo-inducible cytotoxic and clastogenic activities of 3,6-di-substituted acridines obtained by acylation of proflavine. Eur. J. Med. Chem. 2009, 44, 2459–2467. [Google Scholar] [CrossRef] [PubMed]
- Stepanchikova, A.V.; Lagunin, A.A.; Filimonov, D.A.; Poroikov, V.V. Prediction of Biological Activity Spectra for Substances: Evaluation on the Diverse Sets of Drug-Like Structures. 2003. Available online: https://www.ingentaconnect.com/content/ben/cmc/2003/00000010/00000003/art00003 (accessed on 8 May 2019).
- Tsouh Fokou, P.V.; Nyarko, A.K.; Appiah-Opong, R.; Tchokouaha Yamthe, L.R.; Ofosuhene, M.; Boyom, F.F. Update on medicinal plants with potency on Mycobacterium ulcerans. Biomed. Res. Int. 2015, 2015, 1–16. [Google Scholar] [CrossRef]
- Kwofie, S.K.; Dankwa, B.; Odame, E.A.; Agamah, F.E.; Doe, L.; Teye, J.; Agyapong, O.; Miller, W.A., 3rd; Mosi, L.; Wilson, M.D. In Silico screening of isocitrate lyase for novel anti-buruli ulcer natural products originating from Africa. Molecules 2018, 23, 1550. [Google Scholar] [CrossRef]
- Dunn, M.F.; Ramírez-Trujillo, J.A.; Hernández-Lucas, I. Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology 2009, 155, 3166–3175. [Google Scholar] [CrossRef] [PubMed]
- Lirio, S.B.; Macabeo, A.P.; Paragas, E.M.; Knorn, M.; Kohls, P.; Franzblau, S.G.; Wang, Y.; Aguinaldo, M.A. Antitubercular constituents from Premna odorata Blanco. J. Ethnopharmacol. 2014, 154, 471–474. [Google Scholar] [CrossRef]
- Sasikumar, K.; Ghosh, A.R.; Dusthackeer, A. Antimycobacterial potentials of quercetin and rutin against Mycobacterium tuberculosis H37Rv 3. Biotech 2018, 8, 427. [Google Scholar] [CrossRef]
- Butova, T.; Zaitseva, S.; Butov, D.; Stepanenko, G. Morphological changes in experimental tuberculosis resulting from treatment with quercetin and polyvinylpyrrolidone. Int. J. Mycobacteriol. 2016, 5, S103–S104. [Google Scholar] [CrossRef] [PubMed]
- Hughes, W.T.; Dankner, W.M.; Yogev, R.; Huang, S.; Paul, M.E.; Flores, M.A.; Kline, M.W.; Wei, L.J. Pediatric AIDS Clinical Trials Group 254 Team. Comparison of atovaquone and azithromycin with trimethoprim-sulfamethoxazole for the prevention of serious bacterial infections in children with hiv infection. Clin. Infect. Dis. 2005, 40, 136–145. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Molecular Docking, Estimating Free Energies of Binding, and AutoDock’s Semi-Empirical Force Field. Dr. Sebastian Raschka. 2014. Available online: https://sebastianraschka.com/Articles/2014_autodock_energycomps.html (accessed on 10 February 2020).
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Kwofie, S.K.; Broni, E.; Teye, J.; Quansah, E.; Issah, I.; Wilson, M.D.; Miller, W.A., 3rd; Tiburu, E.K.; Bonney, J. Pharmacoinformatics-based identification of potential bioactive compounds against Ebola virus protein VP24. Comput. Biol. Med. 2019, 113, 103414. [Google Scholar] [CrossRef] [PubMed]











| Compound | IC50 (µM) | Structure |
|---|---|---|
| Juglone | 7 ± 0.7 |  |
| α-Lapachone | 11 ± 3 |  |
| 9-Hydroxy-α-Lapachone | 9 ± 1 |  |
| Paulownin | 19 ± 2 |  |
| Yangambin | 27 ± 6 |  |
| Name | Pharmacophore-Fit Score |
|---|---|
| Pyrogallol | 67.16 |
| Chrysophanol | 67.04 |
| ZINC00058187 | 67.02 |
| 3-methoxy-4-hydroxyphenol 1-O-beta-D-glucopyranoside | 66.97 |
| Gossypetin 3,7,8-trimethyl ether | 66.88 |
| 3′-hydroxyflindulatin | 66.88 |
| Vanillin | 66.85 |
| 4′-methyl gossypetin | 66.82 |
| Isoscutellarein | 66.81 |
| Corniculatusin | 66.75 |
| Sexangularetin | 66.75 |
| Bucegin | 66.74 |
| Isoscutellarein 8-methyl ether | 66.74 |
| Onopordin | 66.74 |
| Gentisic acid | 66.74 |
| 1,8-dihydroxy-3,5-dimethoxyxanthone | 66.74 |
| Herbacetin | 66.74 |
| ZINC14490611 | 66.72 |
| Vanillic acid | 66.70 |
| Betavulgarin | 66.56 |
| Epitaxifolin | 66.42 |
| P-hydroxybenzoic acid | 66.40 |
| 2,5-dihydroxybenzaldehyde | 66.36 |
| 2,4′-dihydroxy-3′-methoxyacetophenone | 66.35 |
| Omega-hydroxypropioguaiacone | 66.34 |
| 5-(hydroxymethyl)-2-furancarboxylic acid | 66.34 |
| Aloe-emodin | 66.30 |
| Catechin | 66.30 |
| ZINC05854400 | 66.19 |
| 2,5-dihydroxybenzyl alcohol | 66.17 |
| ZINC00013245 | 66.12 |
| Acetovanillone | 66.11 |
| 4-(2-formyl-5-hydroxymethylpyrrol-1-yl) butyric acid | 66.07 |
| Gossypetin 3,8-dimethyl ether | 65.97 |
| 3,7-dihydroxy-8-methoxy-3-(3′,4′-methylenedioxybenzyl)chroman-4-one | 65.89 |
| 2,3-dihydroxy-1-(4-hydroxy-3-methoxyphenyl)-1-propanone | 65.88 |
| Shikimic acid-4-O-gallate | 65.85 |
| ZINC13328057 | 65.77 |
| Gentisic acid 5-O-glucoside | 65.44 |
| No. | ZINC ID/Compound Name | Binding Energy/ kcal/mol |
|---|---|---|
| Known Inhibitors | ||
| 1 | 9-hydroxy-alpha-lapachone | −8.8 |
| 2 | Alpha-Lapachone | −8.7 |
| 3 | Paulownin | −8.5 |
| 4 | Juglone | −7.3 |
| 5 | Yangambin | −7.2 |
| Pharmacophore Hits | ||
| 6 | Chrysophanol | −8.9 |
| 7 | Aloe-emodin | −8.6 |
| 8 | Herbacetin | −8.5 |
| 9 | Isoscutellarein | −8.4 |
| 10 | Onopordin | −8.4 |
| 11 | Betavulgarin | −8.4 |
| 12 | ZINC05854400 | −8.4 |
| 13 | ZINC14490611 | −8.3 |
| 14 | Bucegin | −8.3 |
| 15 | Isoscutellarein 8-methyl ether | −8.2 |
| 16 | Sexangularetin | −8.2 |
| 17 | Corniculatusin | −8.2 |
| 18 | 4′-methyl gossypetin | −8.2 |
| 19 | 1,8-dihydroxy-3,5-dimethoxyxanthone | −8.1 |
| 20 | Epitaxifolin | −8.1 |
| 21 | ZINC13328057 | −7.9 |
| 22 | Catechin | −7.7 |
| 23 | Gossypetin 3,8-dimethyl ether | −7.7 |
| 24 | Gossypetin 3,7,8-trimethyl ether | −7.7 |
| 25 | ZINC00058187 | −7.6 |
| 26 | Shikimic acid−4-O-gallate | −7.6 |
| 27 | 3,7-dihydroxy-8-methoxy-3-(3′,4′-methylenedioxybenzyl)chroman-4-one | −7.6 |
| 28 | 3′-hydroxyflindulatin | −7.6 |
| 29 | Gentisic acid 5-O-glucoside | −7.1 |
| Compounds | Pa | Pi |
|---|---|---|
| Gentisic acid 5-O-glucoside | 0.618 | 0.008 |
| ZINC05854400 | 0.489 | 0.017 |
| ZINC00058187 | 0.411 | 0.027 |
| Sexangularetin | 0.405 | 0.029 |
| Isoscutellarein 8-methyl ether | 0.404 | 0.029 |
| Isoscutellarein | 0.403 | 0.029 |
| Herbacetin | 0.399 | 0.030 |
| Onopordin | 0.396 | 0.031 |
| Bucegin | 0.394 | 0.031 |
| Gossypetin 3,8-dimethyl ether | 0.393 | 0.032 |
| Gossypetin 3,7,8-trimethyl ether | 0.384 | 0.034 |
| 3′-hydroxyflindulatin | 0.384 | 0.034 |
| Epitaxifolin | 0.381 | 0.035 |
| ZINC14490611 | 0.379 | 0.035 |
| Corniculatusin | 0.379 | 0.035 |
| 4′-methyl gossypetin | 0.373 | 0.037 |
| Chrysophanol | 0.371 | 0.038 |
| 1,8-dihydroxy-3,5-dimethoxyxanthone | 0.368 | 0.038 |
| Aloe-emodin | 0.360 | 0.040 |
| ZINC13328057 | 0.358 | 0.041 |
| Betavulgarin | 0.354 | 0.042 |
| Catechin | 0.350 | 0.043 |
| Shikimic acid-4-O-gallate | 0.327 | 0.065 |
| Antimycobacterial | Anti-Ulcerative | ||||
|---|---|---|---|---|---|
| Compounds | Pa | Pi | Compounds | Pa | Pi |
| Gentisic acid 5-O-glucoside | 0.623 | 0.009 | ZINC05854400 | 0.637 | 0.008 |
| Isoscutellarein 8-methyl ether | 0.568 | 0.012 | Gentisic acid 5-O-glucoside | 0.550 | 0.016 |
| ZINC00058187 | 0.51 | 0.018 | Isoscutellarein | 0.536 | 0.017 |
| Sexangularetin | 0.47 | 0.024 | Isoscutellarein 8-methyl ether | 0.521 | 0.02 |
| Isoscutellarein | 0.465 | 0.025 | Sexangularetin | 0.486 | 0.026 |
| ZINC05854400 | 0.446 | 0.029 | ZINC00058187 | 0.333 | 0.076 |
| Antituberculosis | Antioxidant | ||||
| Compounds | Pa | Pi | Compounds | Pa | Pi |
| Gentisic acid 5-O-glucoside | 0.548 | 0.008 | Isoscutellarein | 0.876 | 0.003 |
| Isoscutellarein 8-methyl ether | 0.502 | 0.02 | ZINC05854400 | 0.837 | 0.003 |
| ZINC00058187 | 0.492 | 0.013 | Sexangularetin | 0.825 | 0.003 |
| Sexangularetin | 0.443 | 0.021 | Isoscutellarein 8-methyl ether | 0.777 | 0.004 |
| Isoscutellarein | 0.438 | 0.022 | Gentisic acid 5-O-glucoside | 0.637 | 0.004 |
| Dermatological | ZINC00058187 | 0.343 | 0.017 | ||
| Compounds | Pa | Pi | |||
| ZINC05854400 | 0.501 | 0.032 | |||
| Gentisic acid 5-O-glucoside | 0.457 | 0.041 | |||
| ZINC00058187 | 0.356 | 0.065 | |||
| Compound | van der Waal Energy (KJ/mol) | Electrostatic Energy (KJ/mol) | Polar Solvation Energy (KJ/mol) | SASA Energy (KJ/mol) | Binding Energy (KJ/mol) |
|---|---|---|---|---|---|
| Gentisic 5-O glucoside | 1.793 +/− 6.832 kJ/mol | −633.686 +/− 68.712 kJ/mol | 398.347 +/− 34.000 kJ/mol | −6.320 +/− 2.460 kJ/mol | −239.865 +/− 68.428 kJ/mol |
| Isoscutellarein | −101.940 +/− 79.145 kJ/mol | −12.961+/− 12.615 kJ/mol | 54.056 +/− 51.199 kJ/mol | −9.944 +/− 8.391 kJ/mol | −70.790 +/− 60.419 kJ/mol |
| ZINC05854400 | −117.075 +/− 30.579 kJ/mol | 356.619 +/− 91.497 kJ/mol | 95.183 +/− 77.435 kJ/mol | −13.820 +/− 3.671 kJ/mol | 320.907 +/− 54.474 kJ/mol |
| Compound Name | IUPAC Name Other Names | 2D Structure |
|---|---|---|
| Gentisic acid 5-O glucoside | 2-hydroxy-5-[3,4,5-trihydroxy-6- (hydroxymethyl)oxan-2-yl]oxybenzoic acid |  |
| Isoscutellarein | 5,7,8-trihydroxy-2- (4-hydroxyphenyl)chromen-4-one |  |
| ZINC05854400 | Gartanin |  |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwofie, S.K.; Dolling, N.N.O.; Donkoh, E.; Laryea, G.M.; Mosi, L.; Miller, W.A., III; Adinortey, M.B.; Wilson, M.D. Pharmacophore-Guided Identification of Natural Products as Potential Inhibitors of Mycobacterium ulcerans Cystathionine γ-Synthase MetB. Computation 2021, 9, 32. https://doi.org/10.3390/computation9030032
Kwofie SK, Dolling NNO, Donkoh E, Laryea GM, Mosi L, Miller WA III, Adinortey MB, Wilson MD. Pharmacophore-Guided Identification of Natural Products as Potential Inhibitors of Mycobacterium ulcerans Cystathionine γ-Synthase MetB. Computation. 2021; 9(3):32. https://doi.org/10.3390/computation9030032
Chicago/Turabian StyleKwofie, Samuel K., Nigel N. O. Dolling, Emmanuel Donkoh, Godwin M. Laryea, Lydia Mosi, Whelton A. Miller, III, Michael B. Adinortey, and Michael D. Wilson. 2021. "Pharmacophore-Guided Identification of Natural Products as Potential Inhibitors of Mycobacterium ulcerans Cystathionine γ-Synthase MetB" Computation 9, no. 3: 32. https://doi.org/10.3390/computation9030032
APA StyleKwofie, S. K., Dolling, N. N. O., Donkoh, E., Laryea, G. M., Mosi, L., Miller, W. A., III, Adinortey, M. B., & Wilson, M. D. (2021). Pharmacophore-Guided Identification of Natural Products as Potential Inhibitors of Mycobacterium ulcerans Cystathionine γ-Synthase MetB. Computation, 9(3), 32. https://doi.org/10.3390/computation9030032






