Quantum Dots: Synthesis, Antibody Conjugation, and HER2-Receptor Targeting for Breast Cancer Therapy
Abstract
1. Introduction
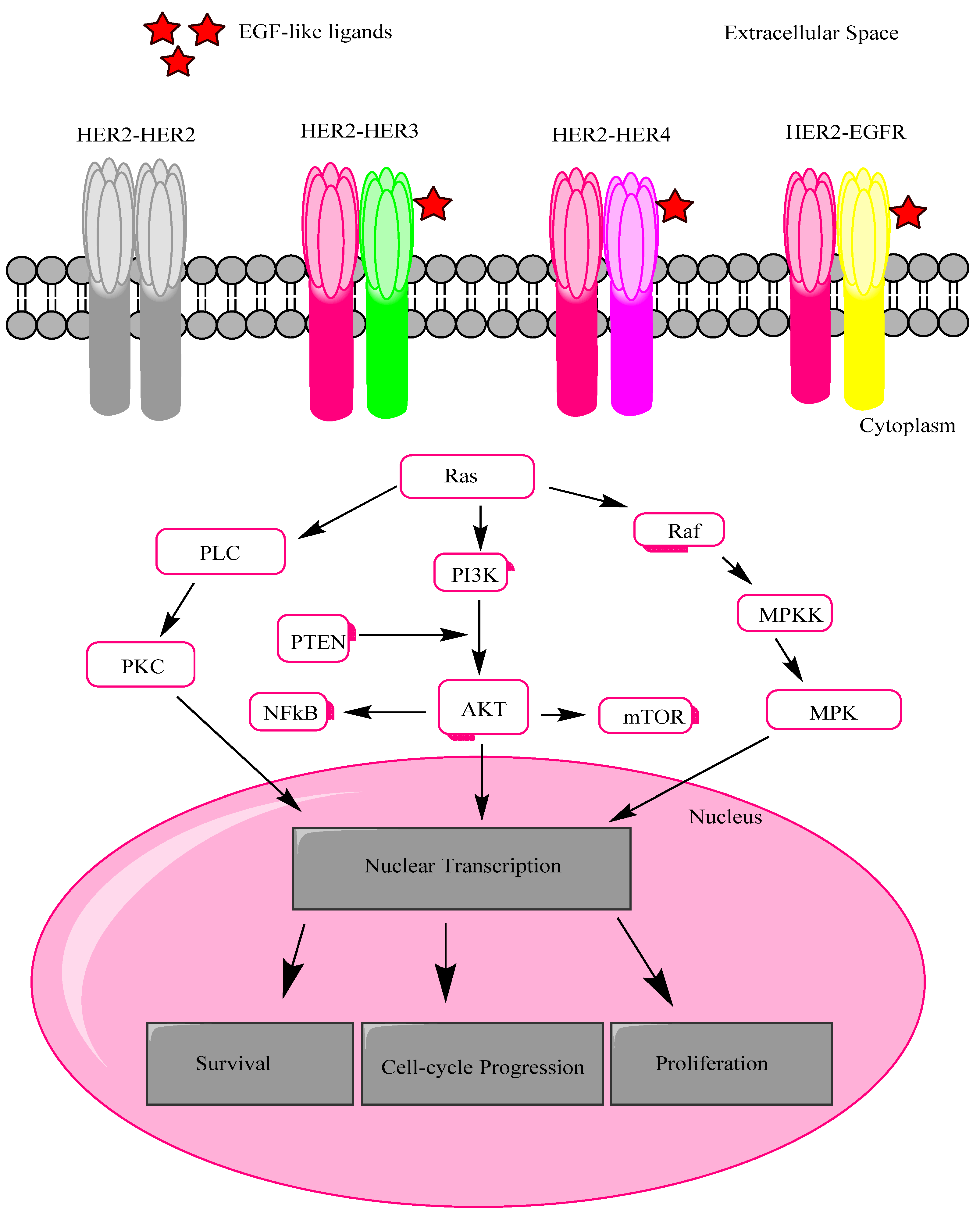
2. HER2 Receptor Mechanism
3. Methods of Synthesis of QDs
3.1. Using Ultrasonic or Microwave Irradiation
3.2. Hydrothermal Synthesis
3.3. Solvothermal Synthesis
3.4. Microorganism Biotemplate
3.5. Electrochemical Assembly
4. Synthesis of Monoclonal Antibodies
5. Polymer Coatings and Receptor Localization
6. QDs-Monoclonal Antibody Conjugation Mechanisms
6.1. Non-Covalent Linking
6.1.1. Adsorption
6.1.2. Direct Attachment to the Surface
6.1.3. Streptavidin–Biotin Binding
6.2. Covalent Linking
6.2.1. Zero-Length Coupling
6.2.2. Homobifunctional-Coupling
6.2.3. Heterobifunctional Coupling
7. HER2 Receptor Targeting by QD-mAB Conjugate
8. In-Vitro Characterization of QDS
8.1. In-Vitro Characterization of QD-mAb Conjugate
8.2. HER2 Receptor Targeting by QDs-mAb Conjugates
9. Applications of QDs
10. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ataollahi, M.; Sharifi, J.; Paknahad, M.; Paknahad, A. Breast cancer and associated factors: A review. J. Med. Life 2015, 8, 6. [Google Scholar]
- Sharma, P.C.; Sharma, D.; Sharma, A.; Bhagat, M.; Ola, M.; Thakur, V.K.; Bhardwaj, J.K.; Goyal, R.K. Recent Advances in Microbial Toxin-Related Strategies to Combat Cancer. Semin. Cancer Biol. 2021, 7, 7. [Google Scholar] [CrossRef]
- Kashyap, S.; Pal, S.; Chandan, G.; Saini, V.; Chakrabarti, S.; Saini, N.K.; Mittal, A.; Thakur, V.K.; Saini, A.K.; Saini, R.V. Understanding the Cross-Talk between Human Microbiota and Gastrointestinal Cancer for Developing Potential Diagnostic and Prognostic Biomarkers. Semin. Cancer Biol. 2021, 4, 20. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Sharma, V.; Sharma, A.; Goyal, R.; Tonk, R.K.; Thakur, V.K.; Sharma, P.C. Green Chemistry Approaches for Thiazole Containing Compounds as a Potential Scaffold for Cancer Therapy. Sustain. Chem. Pharm. 2021, 23, 100496. [Google Scholar] [CrossRef]
- Jemal, A.; Siegel, R.; Ward, E.; Hao, Y.; Xu, J.; Thun, M.J. Cancer statistics, 2009. CA A Cancer J. Clin. 2009, 59, 225–249. [Google Scholar] [CrossRef] [PubMed]
- Haghighat, M.; Naroie, A.; Rezvani, A.; Hakimi, M.; Saravani, H.; Darroudi, M.; Amini, A.; Sabaghan, M.; Khatami, M. Anticancer Property of Lanthanide Sulfate Nanostructure Against Neuroblastoma-Neuro2a Cell Line. BioNanoScience 2021, 11, 1–7. [Google Scholar] [CrossRef]
- Kardani, A.; Yaghoobi, H.; Alibakhshi, A.; Khatami, M. Inhibition of miR-155 in MCF-7 breast cancer cell line by gold nanoparticles functionalized with antagomir and AS1411 aptamer. J. Cell. Physiol. 2020, 235, 6887–6895. [Google Scholar] [CrossRef]
- Khan, A.U.; Yuan, Q.; Khan, Z.U.H.; Ahmad, A.; Khan, F.U.; Tahir, K.; Shakeel, M.; Ullah, S. An eco-benign synthesis of AgNPs using aqueous extract of Longan fruit peel: Antiproliferative response against human breast cancer cell line MCF-7, antioxidant and photocatalytic deprivation of methylene blue. J. Photochem. Photobiol. B Biol. 2018, 183, 367–373. [Google Scholar] [CrossRef]
- Khatami, M.; Kharazi, S.; Kishani Farahani, Z.; Azizi, H.; Augusto Lima Nobre, M. The anticancer effect of octagon and spherical silver nanoparticles on MCF-7 breast cancer cell line. Tehran Univ. Med. J. TUMS Publ. 2017, 75, 72–76. [Google Scholar]
- Khatami, M.; Sharifi, I.; Nobre, M.A.; Zafarnia, N.; Aflatoonian, M.R. Waste-grass-mediated green synthesis of silver nanoparticles and evaluation of their anticancer, antifungal and antibacterial activity. Green Chem. Lett. Rev. 2018, 11, 125–134. [Google Scholar] [CrossRef]
- Mahmoudvand, H.; Sepahvand, A.; Badparva, E.; Khatami, M.; Niazi, M.; Moayyedkazemi, A. Possible Association and Risk Factors of Blastocystis Infection and Colorectal Cancers in Western Iran. Arch. Clin. Infect. Dis. 2021, 16, e90861. [Google Scholar] [CrossRef]
- Groenen, L.C.; Nice, E.C.; Burgess, A.W. Structure-function relationships for the EGF/TGF-α family of mitogens. Growth Factors 1994, 11, 235–257. [Google Scholar] [CrossRef] [PubMed]
- Meric-Bernstam, F.; Hung, M.-C. Advances in targeting human epidermal growth factor receptor-2 signaling for cancer therapy. Clin. Cancer Res. 2006, 12, 6326–6330. [Google Scholar] [CrossRef] [PubMed]
- Slamon, D.J.; Clark, G.M.; Wong, S.G.; Levin, W.J.; Ullrich, A.; McGuire, W.L. Human breast cancer: Correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science 1987, 235, 177–182. [Google Scholar] [CrossRef]
- Burris III, H.A.; Tibbitts, J.; Holden, S.N.; Sliwkowski, M.X.; Phillips, G.D.L. Trastuzumab emtansine (T-DM1): A novel agent for targeting HER2+ breast cancer. Clin. Breast Cancer 2011, 11, 275–282. [Google Scholar] [CrossRef]
- Haume, K.; Rosa, S.; Grellet, S.; Śmiałek, M.A.; Butterworth, K.T.; Solov’yov, A.V.; Prise, K.M.; Golding, J.; Mason, N.J. Gold nanoparticles for cancer radiotherapy: A review. Cancer Nanotechnol. 2016, 7, 8. [Google Scholar] [CrossRef]
- Delaney, G.P.; Barton, M.B. Evidence-based estimates of the demand for radiotherapy. Clin. Oncol. 2015, 27, 70–76. [Google Scholar] [CrossRef]
- Peydayesh, M.; Raisi, M.; Kaykavousi, K.; Gharaghani, M.A.; Abdollahpour-Alitappeh, M.; Mosazade, F.; Seifalian, A.; Khatami, M. The inhibitory effect of Tamarix hispida mediated silver nanoparticles on Cyclin D1 protein expression of human cancer cells line. Inorg. Nano-Met. Chem. 2020, 50, 1144–1149. [Google Scholar] [CrossRef]
- Poor, M.H.S.; Khatami, M.; Azizi, H.; Abazari, Y. Cytotoxic activity of biosynthesized Ag nanoparticles by Plantago major towards a human breast cancer cell line. Rend. Lincei 2017, 28, 693–699. [Google Scholar] [CrossRef]
- Sharifi, F.; Sharififar, F.; Sharifi, I.; Pournamdari, M.; Eslaminejad, T.; Khatami, M. Antioxidant, anti-proliferation and cytotoxicity activities of Gossypium hirsutum toward standard HepG2, A549, MCF-7 and U87 cancer cell lines compared to HUVEC, 3T3 normal cells. Eur. J. Med. Plants 2017, 21, 1–10. [Google Scholar] [CrossRef]
- Yaghoubi, S.; Karimi, M.H.; Lotfinia, M.; Gharibi, T.; Mahi-Birjand, M.; Kavi, E.; Hosseini, F.; Sineh Sepehr, K.; Khatami, M.; Bagheri, N. Potential drugs used in the antibody–drug conjugate (ADC) architecture for cancer therapy. J. Cell. Physiol. 2020, 235, 31–64. [Google Scholar] [CrossRef] [PubMed]
- Shakeri-Zadeh, A.; Zareyi, H.; Sheervalilou, R.; Laurent, S.; Ghaznavi, H.; Samadian, H. Gold nanoparticle-mediated bubbles in cancer nanotechnology. J. Control. Release 2020, 330, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Saini, A.K.; Kumar, N.; Tejwan, N.; Singh, T.A.; Thakur, V.K.; Das, J. Methods of Preparation of Metal-Doped and Hybrid Tungsten Oxide Nanoparticles for Anticancer, Antibacterial, and Biosensing Applications. Surf. Interfaces 2022, 28, 101641. [Google Scholar] [CrossRef]
- Irajirad, R.; Ahmadi, A.; Najafabad, B.K.; Abed, Z.; Sheervalilou, R.; Khoei, S.; Shiran, M.B.; Ghaznavi, H.; Shakeri-Zadeh, A. Combined thermo-chemotherapy of cancer using 1 MHz ultrasound waves and a cisplatin-loaded sonosensitizing nanoplatform: An in vivo study. Cancer Chemother. Pharmacol. 2019, 84, 1315–1321. [Google Scholar] [CrossRef]
- Mashayekhi, S.; Rasoulpoor, S.; Shabani, S.; Esmaeilizadeh, N.; Serati-Nouri, H.; Sheervalilou, R.; Pilehvar-Soltanahmadi, Y. Curcumin-loaded mesoporous silica nanoparticles/nanofiber composites for supporting long-term proliferation and stemness preservation of adipose-derived stem cells. Int. J. Pharm. 2020, 587, 119656. [Google Scholar] [CrossRef] [PubMed]
- Almanghadim, H.G.; Nourollahzadeh, Z.; Khademi, N.S.; Tezerjani, M.D.; Sehrig, F.Z.; Estelami, N.; Shirvaliloo, M.; Sheervalilou, R.; Sargazi, S. Application of nanoparticles in cancer therapy with an emphasis on cell cycle. Cell Biol. Int. 2021, 45, 1989–1998. [Google Scholar] [CrossRef]
- Sheervalilou, R.; Shahraki, O.; Hasanifard, L.; Shirvaliloo, M.; Mehranfar, S.; Lotfi, H.; Pilehvar-Soltanahmadi, Y.; Bahmanpour, Z.; Zadeh, S.S.; Nazarlou, Z. Electrochemical nano-biosensors as novel approach for the detection of lung cancer-related MicroRNAs. Curr. Mol. Med. 2020, 20, 13–35. [Google Scholar] [CrossRef]
- Sheervalilou, R.; Shirvaliloo, M.; Sargazi, S.; Ghaznavi, H. Recent advances in iron oxide nanoparticles for brain cancer theranostics: From in vitro to clinical applications. Expert Opin. Drug Deliv. 2021, 18, 949–977. [Google Scholar] [CrossRef]
- Liu, J.; Chen, Q.; Feng, L.; Liu, Z. Nanomedicine for tumor microenvironment modulation and cancer treatment enhancement. Nano Today 2018, 21, 55–73. [Google Scholar] [CrossRef]
- Zou, Q.; Xing, P.; Wei, L.; Liu, B. Gene2vec: Gene subsequence embedding for prediction of mammalian N6-methyladenosine sites from mRNA. RNA 2019, 25, 205–218. [Google Scholar] [CrossRef]
- Jiang, Q.; Jin, S.; Jiang, Y.; Liao, M.; Feng, R.; Zhang, L.; Liu, G.; Hao, J. Alzheimer’s disease variants with the genome-wide significance are significantly enriched in immune pathways and active in immune cells. Mol. Neurobiol. 2017, 54, 594–600. [Google Scholar] [CrossRef]
- Zou, Y.; Wu, H.; Guo, X.; Peng, L.; Ding, Y.; Tang, J.; Guo, F. MK-FSVM-SVDD: A multiple kernel-based fuzzy SVM model for predicting DNA-binding proteins via support vector data description. Curr. Bioinform. 2021, 16, 274–283. [Google Scholar] [CrossRef]
- Shirvalilou, S.; Khoei, S.; Esfahani, A.J.; Kamali, M.; Shirvaliloo, M.; Sheervalilou, R.; Mirzaghavami, P. Magnetic Hyperthermia as an adjuvant cancer therapy in combination with radiotherapy versus radiotherapy alone for recurrent/progressive glioblastoma: A systematic review. J. Neuro-Oncol. 2021, 152, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.; Xu, C.; Sun, X.; Chen, X. Nanoparticle design strategies for enhanced anticancer therapy by exploiting the tumour microenvironment. Chem. Soc. Rev. 2017, 46, 3830–3852. [Google Scholar] [CrossRef]
- Yan, S.; Yan, J.; Liu, D.; Li, X.; Kang, Q.; You, W.; Zhang, J.; Wang, L.; Tian, Z.; Lu, W. A nano-predator of pathological MDMX construct by clearable supramolecular gold (I)-thiol-peptide complexes achieves safe and potent anti-tumor activity. Theranostics 2021, 11, 6833. [Google Scholar] [CrossRef]
- Reang, J.; Sharma, P.C.; Thakur, V.K.; Majeed, J. Understanding the Therapeutic Potential of Ascorbic Acid in the Battle to Overcome Cancer. Biomolecules 2021, 11, 1130. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Alijani, H.Q.; Khatami, M.; Bagheri, F.; Iravani, S.; Sharifi, F. K-doped ZnO nanostructures: Biosynthesis and parasiticidal application. J. Mater. Res. Technol. 2021, 15, 5445–5451. [Google Scholar] [CrossRef]
- Zarrabi, A.; Zarepour, A.; Khosravi, A.; Alimohammadi, Z.; Thakur, V.K. Synthesis of Curcumin Loaded Smart PH-Responsive Stealth Liposome as a Novel Nanocarrier for Cancer Treatment. Fibers 2021, 9, 19. [Google Scholar] [CrossRef]
- Arshad, R.; Barani, M.; Rahdar, A.; Sargazi, S.; Cucchiarini, M.; Pandey, S.; Kang, M. Multi-Functionalized Nanomaterials and Nanoparticles for Diagnosis and Treatment of Retinoblastoma. Biosensors 2021, 11, 97. [Google Scholar] [CrossRef]
- Barani, M.; Rahdar, A.; Sargazi, S.; Amiri, M.S.; Sharma, P.K.; Bhalla, N. Nanotechnology for inflammatory bowel disease management: Detection, imaging and treatment. Sens. Bio-Sens. Res. 2021, 32, 100417. [Google Scholar] [CrossRef]
- Rahdar, A.; Sargazi, S.; Barani, M.; Shahraki, S.; Sabir, F.; Aboudzadeh, M.A. Lignin-stabilized doxorubicin microemulsions: Synthesis, physical characterization, and in vitro assessments. Polymers 2021, 13, 641. [Google Scholar] [CrossRef] [PubMed]
- Rahdar, A.; Hajinezhad, M.R.; Sargazi, S.; Zaboli, M.; Barani, M.; Baino, F.; Bilal, M.; Sanchooli, E. Biochemical, Ameliorative and Cytotoxic Effects of Newly Synthesized Curcumin Microemulsions: Evidence from In Vitro and In Vivo Studies. Nanomaterials 2021, 11, 817. [Google Scholar] [CrossRef]
- Norouzi, M.; Yasamineh, S.; Montazeri, M.; Dadashpour, M.; Sheervalilou, R.; Abasi, M.; Pilehvar-Soltanahmadi, Y. Recent advances on nanomaterials-based fluorimetric approaches for microRNAs detection. Mater. Sci. Eng. C 2019, 104, 110007. [Google Scholar] [CrossRef]
- Akbarzadeh, A.; Aval, S.F.; Sheervalilou, R.; Fekri, L.; Zarghami, N.; Mohammadian, M. Quantum dots for biomedical delivery applications. Rev. Cell Biol. Mol. Med. 2016, 2, 19. [Google Scholar] [CrossRef]
- Zhou, W.; Coleman, J.J. Semiconductor quantum dots. Curr. Opin. Solid State Mater. Sci. 2016, 20, 352–360. [Google Scholar] [CrossRef]
- Chung, S.; Revia, R.A.; Zhang, M. Graphene quantum dots and their applications in bioimaging, biosensing, and therapy. Adv. Mater. 2021, 33, 1904362. [Google Scholar] [CrossRef] [PubMed]
- Jin, T.; Tiwari, D.K.; Tanaka, S.-i.; Inouye, Y.; Yoshizawa, K.; Watanabe, T.M. Antibody–ProteinA conjugated quantum dots for multiplexed imaging of surface receptors in living cells. Mol. BioSyst. 2010, 6, 2325–2331. [Google Scholar] [CrossRef] [PubMed]
- Tol, J.; Punt, C.J. Monoclonal antibodies in the treatment of metastatic colorectal cancer: A review. Clin. Ther. 2010, 32, 437–453. [Google Scholar] [CrossRef]
- Waldmann, T.A. Monoclonal antibodies in diagnosis and therapy. Science 1991, 252, 1657–1662. [Google Scholar] [CrossRef] [PubMed]
- Vogel, C.L.; Cobleigh, M.A.; Tripathy, D.; Gutheil, J.C.; Harris, L.N.; Fehrenbacher, L.; Slamon, D.J.; Murphy, M.; Novotny, W.F.; Burchmore, M. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J. Clin. Oncol. 2002, 20, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Goldman, E.R.; Balighian, E.D.; Mattoussi, H.; Kuno, M.K.; Mauro, J.M.; Tran, P.T.; Anderson, G.P. Avidin: A natural bridge for quantum dot-antibody conjugates. J. Am. Chem. Soc. 2002, 124, 6378–6382. [Google Scholar] [CrossRef] [PubMed]
- Goldman, E.R.; Anderson, G.P.; Tran, P.T.; Mattoussi, H.; Charles, P.T.; Mauro, J.M. Conjugation of luminescent quantum dots with antibodies using an engineered adaptor protein to provide new reagents for fluoroimmunoassays. Anal. Chem. 2002, 74, 841–847. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.P.; Pinaud, F.; Antelman, J.; Weiss, S. Tracking bio-molecules in live cells using quantum dots. J. Biophotonics 2008, 1, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Iqbal, N. Human epidermal growth factor receptor 2 (HER2) in cancers: Overexpression and therapeutic implications. Mol. Biol. Int. 2014, 2014, 852748. [Google Scholar] [CrossRef] [PubMed]
- Arkhipov, A.; Shan, Y.; Kim, E.T.; Dror, R.O.; Shaw, D.E. Her2 activation mechanism reflects evolutionary preservation of asymmetric ectodomain dimers in the human EGFR family. eLife 2013, 2, e00708. [Google Scholar] [CrossRef] [PubMed]
- Moasser, M.M. The oncogene HER2: Its signaling and transforming functions and its role in human cancer pathogenesis. Oncogene 2007, 26, 6469. [Google Scholar] [CrossRef] [PubMed]
- Nahta, R.; Yuan, L.X.; Zhang, B.; Kobayashi, R.; Esteva, F.J. Insulin-like growth factor-I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer Res. 2005, 65, 11118–11128. [Google Scholar] [CrossRef]
- Karmakar, R. Quantum Dots and it method of preparations-revisited. Prajnan O Sadhona A Sci. Annu. 2015, 2, 116–142. [Google Scholar]
- Kucukyildirim, B.O.; Eker, A.A.; Eker, B. Usage of Carbon Nanotubes in Scanning Probe Microscopes as Probe. Int. J. Arts Sci. 2009, 3, 18–26. [Google Scholar]
- Tsutsui, K.; Hu, E.L.; Wilkinson, C.D. Reactive ion etched II-VI quantum dots: Dependence of etched profile on pattern geometry. Jpn. J. Appl. Phys. 1993, 32, 6233. [Google Scholar] [CrossRef]
- Bhand, G.R.; Chaure, N.B. Synthesis of CdTe, CdSe and CdTe/CdSe core/shell QDs from wet chemical colloidal method. Mater. Sci. Semicond. Process. 2017, 68, 279–287. [Google Scholar] [CrossRef]
- Qian, H.; Li, L.; Ren, J. One-step and rapid synthesis of high quality alloyed quantum dots (CdSe–CdS) in aqueous phase by microwave irradiation with controllable temperature. Mater. Res. Bull. 2005, 40, 1726–1736. [Google Scholar] [CrossRef]
- Entezari, M.H.; Ghows, N. Micro-emulsion under ultrasound facilitates the fast synthesis of quantum dots of CdS at low temperature. Ultrason. Sonochem. 2011, 18, 127–134. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, É.; O’Riordan, A.; Doyle, H.; Moynihan, S.; Cuddihy, A.; Redmond, G. Near-infrared electroluminescent devices based on colloidal HgTe quantum dot arrays. Appl. Phys. Lett. 2005, 86, 201114. [Google Scholar] [CrossRef]
- Choucair, M.; Thordarson, P.; Stride, J.A. Gram-scale production of graphene based on solvothermal synthesis and sonication. Nat. Nanotechnol. 2009, 4, 30. [Google Scholar] [CrossRef]
- Du, C.-F.; You, T.; Jiang, L.; Yang, S.-Q.; Zou, K.; Han, K.-L.; Deng, W.-Q. Controllable synthesis of ultrasmall CuInSe 2 quantum dots for photovoltaic application. RSC Adv. 2014, 4, 33855–33860. [Google Scholar] [CrossRef]
- Lee, D.U.; Kim, D.H.; Choi, D.H.; Kim, S.W.; Lee, H.S.; Yoo, K.-H.; Kim, T.W. Microstructural and optical properties of CdSe/CdS/ZnS core-shell-shell quantum dots. Opt. Express 2016, 24, A350–A357. [Google Scholar] [CrossRef] [PubMed]
- Whaley, S.R.; English, D.; Hu, E.L.; Barbara, P.F.; Belcher, A.M. Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly. Nature 2000, 405, 665. [Google Scholar] [CrossRef] [PubMed]
- Balandin, A.; Wang, K.; Kouklin, N.; Bandyopadhyay, S. Raman spectroscopy of electrochemically self-assembled CdS quantum dots. Appl. Phys. Lett. 2000, 76, 137–139. [Google Scholar] [CrossRef]
- Bernett, M.J.; Karki, S.; Moore, G.L.; Leung, I.W.; Chen, H.; Pong, E.; Nguyen, D.-H.T.; Jacinto, J.; Zalevsky, J.; Muchhal, U.S. Engineering fully human monoclonal antibodies from murine variable regions. J. Mol. Biol. 2010, 396, 1474–1490. [Google Scholar] [CrossRef] [PubMed]
- Steinitz, M. Human Monoclonal Antibodies. Methods Mol. Biol. 2014, 111, 22. [Google Scholar]
- Medecigo, M.; Manoutcharian, K.; Vasilevko, V.; Govezensky, T.; Munguia, M.; Becerril, B.; Luz-Madrigal, A.; Vaca, L.; Cribbs, D.; Gevorkian, G. Novel amyloid-beta specific scFv and VH antibody fragments from human and mouse phage display antibody libraries. J. Neuroimmunol. 2010, 223, 104–114. [Google Scholar] [CrossRef]
- Solforosi, L.; Mancini, N.; Canducci, F.; Clementi, N.; Sautto, G.A.; Diotti, R.A.; Clementi, M.; Burioni, R. A phage display vector optimized for the generation of human antibody combinatorial libraries and the molecular cloning of monoclonal antibody fragments. New Microbiol. 2012, 35, 289–294. [Google Scholar]
- Ahmad, Z.A.; Yeap, S.K.; Ali, A.M.; Ho, W.Y.; Alitheen, N.B.M.; Hamid, M. scFv antibody: Principles and clinical application. Clin. Dev. Immunol. 2012, 2012, 980250. [Google Scholar] [CrossRef] [PubMed]
- Mahmuda, A.; Bande, F.; Al-Zihiry, K.J.K.; Abdulhaleem, N.; Majid, R.A.; Hamat, R.A.; Abdullah, W.O.; Unyah, Z. Monoclonal antibodies: A review of therapeutic applications and future prospects. Trop. J. Pharm. Res. 2017, 16, 713–722. [Google Scholar] [CrossRef]
- Carter, P.; Presta, L.; Gorman, C.M.; Ridgway, J.; Henner, D.; Wong, W.; Rowland, A.M.; Kotts, C.; Carver, M.E.; Shepard, H.M. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc. Natl. Acad. Sci. USA 1992, 89, 4285–4289. [Google Scholar] [CrossRef] [PubMed]
- Nahta, R.; Hung, M.-C.; Esteva, F.J. The HER-2-targeting antibodies trastuzumab and pertuzumab synergistically inhibit the survival of breast cancer cells. Cancer Res. 2004, 64, 2343–2346. [Google Scholar] [CrossRef] [PubMed]
- Claret, F.X.; Vu, T.T. Trastuzumab: Updated mechanisms of action and resistance in breast cancer. Front. Oncol. 2012, 2, 62. [Google Scholar]
- Hu, X.; Zhang, X.; Jin, W. Applications of electrochemiluminescence resonance energy transfer between CdSe/ZnS quantum dots and cyanine dye (Cy5) molecules in evaluating interactions and conformational changes of DNA molecules. Electrochim. Acta 2013, 94, 367–373. [Google Scholar] [CrossRef]
- Durán, G.M.; Plata, M.R.; Zougagh, M.; Contento, A.M.; Ríos, Á. Microwave-assisted synthesis of water soluble thiol capped CdSe/ZnS quantum dots and its interaction with sulfonylurea herbicides. J. Colloid. Interface Sci. 2014, 428, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Rao, J. Quantum dot bioconjugates for in vitro diagnostics & in vivo imaging. Cancer Biomark. 2008, 4, 307–319. [Google Scholar] [PubMed]
- Hong, Z.-Y.; Lv, C.; Liu, A.-A.; Liu, S.-L.; Sun, E.-Z.; Zhang, Z.-L.; Lei, A.-W.; Pang, D.-W. Clicking hydrazine and aldehyde: The way to labeling of viruses with quantum dots. ACS Nano 2015, 9, 11750–11760. [Google Scholar] [CrossRef]
- Foubert, A.; Beloglazova, N.V.; Rajkovic, A.; Sas, B.; Madder, A.; Goryacheva, I.Y.; De Saeger, S. Bioconjugation of quantum dots: Review & impact on future application. TrAC Trends Anal. Chem. 2016, 83, 31–48. [Google Scholar]
- Hermanson, G.T. Bioconjugate Techniques; Academic Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Alivisatos, A.P.; Gu, W.; Larabell, C. Quantum dots as cellular probes. Annu. Rev. Biomed. Eng. 2005, 7, 55–76. [Google Scholar] [CrossRef] [PubMed]
- Pereira, G.; Monteiro, C.A.; Albuquerque, G.M.; Pereira, M.I.; Cabrera, M.P.; Cabral Filho, P.E.; Pereira, G.A.; Fontesa, A.; Santos, B.S. (Bio) conjugation Strategies Applied to Fluorescent Semiconductor Quantum Dots. J. Braz. Chem. Soc. 2019, 30, 2536–2561. [Google Scholar] [CrossRef]
- Sapsford, K.E.; Pons, T.; Medintz, I.L.; Mattoussi, H. Biosensing with luminescent semiconductor quantum dots. Sensors 2006, 6, 925–953. [Google Scholar] [CrossRef]
- Aldeek, F.; Safi, M.; Zhan, N.; Palui, G.; Mattoussi, H. Understanding the self-assembly of proteins onto gold nanoparticles and quantum dots driven by metal-histidine coordination. ACS Nano 2013, 7, 10197–10210. [Google Scholar] [CrossRef] [PubMed]
- Zhan, N.; Palui, G.; Mattoussi, H. Preparation of compact biocompatible quantum dots using multicoordinating molecular-scale ligands based on a zwitterionic hydrophilic motif and lipoic acid anchors. Nat. Protoc. 2015, 10, 859. [Google Scholar] [CrossRef] [PubMed]
- Tsuboi, S.; Sasaki, A.; Sakata, T.; Yasuda, H.; Jin, T. Immunoglobulin binding (B1) domain mediated antibody conjugation to quantum dots for in vitro and in vivo molecular imaging. Chem. Commun. 2017, 53, 9450–9453. [Google Scholar] [CrossRef] [PubMed]
- Tomczak, N.; Liu, R.; Vancso, J.G. Polymer-coated quantum dots. Nanoscale 2013, 5, 12018–12032. [Google Scholar] [CrossRef]
- Guo, Y.; Sakonsinsiri, C.; Nehlmeier, I.; Fascione, M.A.; Zhang, H.; Wang, W.; Pöhlmann, S.; Turnbull, W.B.; Zhou, D. Compact, polyvalent mannose quantum dots as sensitive, ratiometric FRET probes for multivalent protein–ligand interactions. Angew. Chem. Int. Ed. 2016, 55, 4738–4742. [Google Scholar] [CrossRef] [PubMed]
- Diamandis, E.P.; Christopoulos, T.K. The biotin-(strept) avidin system: Principles and applications in biotechnology. Clin. Chem. 1991, 37, 625–636. [Google Scholar] [CrossRef]
- Zhang, F.; Lees, E.; Amin, F.; Rivera_Gil, P.; Yang, F.; Mulvaney, P.; Parak, W.J. Polymer-coated nanoparticles: A universal tool for biolabelling experiments. Small 2011, 7, 3113–3127. [Google Scholar] [CrossRef] [PubMed]
- Lesch, H.P.; Kaikkonen, M.U.; Pikkarainen, J.T.; Ylä-Herttuala, S. Avidin-biotin technology in targeted therapy. Expert Opin. Drug Deliv. 2010, 7, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-y.; Wang, L.-j.; Zhang, Q.; Tang, B.; Zhang, C.-y. Single quantum dot-based nanosensor for sensitive detection of 5-methylcytosine at both CpG and non-CpG sites. Chem. Sci. 2018, 9, 1330–1338. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Liu, M.-h.; Zhang, C.-y. Integration of isothermal amplification with quantum dot-based fluorescence resonance energy transfer for simultaneous detection of multiple microRNAs. Chem. Sci. 2018, 9, 4258–4267. [Google Scholar] [CrossRef] [PubMed]
- Benoiton, N.L. Chemistry of Peptide Synthesis; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Ruan, L.; Ge, M.; Huang, X.; Ren, J. Assay of Single-Cell Apoptosis by Ensemble and Single-Molecule Fluorescence Methods: Annexin-V/Polyethylene Glycol-Functionalized Quantum Dots as Probes. Langmuir 2018, 34, 10040–10047. [Google Scholar] [CrossRef]
- Pati, M.L.; Fanizza, E.; Hager, S.; Groza, D.; Heffeter, P.; Laurenza, A.G.; Laquintana, V.; Curri, M.L.; Depalo, N.; Abate, C. Quantum dot based luminescent nanoprobes for sigma-2 receptor imaging. Mol. Pharm. 2017, 15, 458–471. [Google Scholar] [CrossRef]
- Raichlin, S.; Sharon, E.; Freeman, R.; Tzfati, Y.; Willner, I. Electron-transfer quenching of nucleic acid-functionalized CdSe/ZnS quantum dots by doxorubicin: A versatile system for the optical detection of DNA, aptamer–substrate complexes and telomerase activity. Biosens. Bioelectron. 2011, 26, 4681–4689. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-H.; Li, Y.-Q.; Zhang, H.-L.; Wang, H.-Q.; Lin, S.; Chen, J.; Zhao, Y.-D.; Luo, Q.-M. Bioconjugation of concanavalin and CdTe quantum dots and the detection of glucose. Colloids Surf. A: Physicochem. Eng. Asp. 2010, 364, 82–86. [Google Scholar] [CrossRef]
- Chen, Z.; Lu, M. Novel electrochemical immunoassay for human IgG1 using metal sulfide quantum dot-doped bovine serum albumin microspheres on antibody-functionalized magnetic beads. Anal. Chim. Acta 2017, 979, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Chen, K.; Miao, Z.; Ren, G.; Chen, X.; Gambhir, S.S.; Cheng, Z. Affibody-based nanoprobes for HER2-expressing cell and tumor imaging. Biomaterials 2011, 32, 2141–2148. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, A.Q.; Torchynska, T.; Espinola, J.C.; Gómez, J.J.; Douda, J. Electronic effects in emission of core/shell CdSe/ZnS quantum dots conjugated to anti-Interleukin 10 antibodies. J. Lumin. 2013, 143, 38–42. [Google Scholar] [CrossRef]
- Kobirumaki, F.; Yumoto, M.; Watanabe, M. Real-time tracking of single quantum dots conjugated with monoclonal anti-smooth muscle myosin antibody in intact and skinned taenia caeci from guinea pig. In Proceedings of the Proceedings of Annual Meeting of the Physiological Society of Japan, Tokyo, Japan, 25–27 March 2008; p. 66. [Google Scholar]
- Jaiswal, J.K.; Mattoussi, H.; Mauro, J.M.; Simon, S.M. Long-term multiple color imaging of live cells using quantum dot bioconjugates. Nat. Biotechnol. 2003, 21, 47. [Google Scholar] [CrossRef]
- Medintz, I.L.; Uyeda, H.T.; Goldman, E.R.; Mattoussi, H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat. Mater. 2005, 4, 435. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Gerion, D. Fluorescent CdSe/ZnS nanocrystal− peptide conjugates for long-term, nontoxic imaging and nuclear targeting in living cells. Nano Lett. 2004, 4, 1827–1832. [Google Scholar] [CrossRef]
- Han, S.-J.; Rathinaraj, P.; Park, S.-Y.; Kim, Y.K.; Lee, J.H.; Kang, I.-K.; Moon, J.-S.; Winiarz, J.G. Specific intracellular uptake of herceptin-conjugated CdSe/ZnS quantum dots into breast cancer cells. BioMed Res. Int. 2014, 2014, 954307. [Google Scholar] [CrossRef] [PubMed]
- Tada, H.; Higuchi, H.; Wanatabe, T.M.; Ohuchi, N. In vivo real-time tracking of single quantum dots conjugated with monoclonal anti-HER2 antibody in tumors of mice. Cancer Res. 2007, 67, 1138–1144. [Google Scholar] [CrossRef] [PubMed]
- Nurunnabi, M.; Cho, K.J.; Choi, J.S.; Huh, K.M.; Lee, Y.-k. Targeted near-IR QDs-loaded micelles for cancer therapy and imaging. Biomaterials 2010, 31, 5436–5444. [Google Scholar] [CrossRef] [PubMed]
- Núñez, C.; Estévez, S.V.; del Pilar Chantada, M. Inorganic nanoparticles in diagnosis and treatment of breast cancer. JBIC J. Biol. Inorg. Chem. 2018, 23, 331–345. [Google Scholar] [CrossRef] [PubMed]
- Yue, D.; Wang, M.; Deng, F.; Yin, W.; Zhao, H.; Zhao, X.; Xu, Z. Biomarker-targeted fluorescent probes for breast cancer imaging. Chin. Chem. Lett. 2018, 29, 648–656. [Google Scholar] [CrossRef]
- Colombo, M.; Corsi, F.; Foschi, D.; Mazzantini, E.; Mazzucchelli, S.; Morasso, C.; Occhipinti, E.; Polito, L.; Prosperi, D.; Ronchi, S. HER2 targeting as a two-sided strategy for breast cancer diagnosis and treatment: Outlook and recent implications in nanomedical approaches. Pharmacol. Res. 2010, 62, 150–165. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Long, J.; Zhang, B.; Liu, C.; Ji, S.; Xu, J.; Yu, X.; Ni, Q. Quantum dots in cancer therapy. Expert Opin. Drug Deliv. 2012, 9, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Kuskovsky, I.L.; Fung, J.; Robinson, R.; Herman, I.; Neumark, G.; Zhou, X.; Guo, S.; Tamargo, M. Determination of size and composition of optically active CdZnSe/ZnBeSe quantum dots. Appl. Phys. Lett. 2003, 83, 3779–3781. [Google Scholar] [CrossRef]
- Rameshwar, T.; Samal, S.; Lee, S.; Kim, S.; Cho, J.; Kim, I.S. Determination of the size of water-soluble nanoparticles and quantum dots by field-flow fractionation. J. Nanosci. Nanotechnol. 2006, 6, 2461–2467. [Google Scholar] [CrossRef] [PubMed]
- Hapke-Wurst, I.; Zeitler, U.; Schumacher, H.; Haug, R.; Pierz, K.; Ahlers, F. Size determination of InAs quantum dots using magneto-tunnelling experiments. Semicond. Sci. Technol. 1999, 14, L41. [Google Scholar] [CrossRef]
- Yamauchi, T.; Matsuba, Y.; Ohyama, Y.; Tabuchi, M.; Nakamura, A.; Akahane, K.; Lan, S.; Kawamura, T.; Takamasu, T.; Kido, G. Quantum Size Effects of InAs-and InGaAs-Quantum Dots Studied by Scanning Tunneling Microscopy/Spectroscopy. Appl. Phys. Lett. 1997, 71, 1083. [Google Scholar] [CrossRef]
- Drbohlavova, J.; Adam, V.; Kizek, R.; Hubalek, J. Quantum dots—characterization, preparation and usage in biological systems. Int. J. Mol. Sci. 2009, 10, 656–673. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-O.; So, H.-M.; Jeon, E.-K.; Chang, H.; Won, K.; Kim, Y.H. Aptamers as molecular recognition elements for electrical nanobiosensors. Anal. Bioanal. Chem. 2008, 390, 1023–1032. [Google Scholar] [CrossRef]
- Zhang, P.; Lu, H.; Chen, J.; Han, H.; Ma, W. Simple and sensitive detection of HBsAg by using a quantum dots nanobeads based dot-blot immunoassay. Theranostics 2014, 4, 307. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, Q.; Meng, Q.; Wu, F.; Zhang, L.; Tang, Y.; Guan, Y.; An, L. Quantum dots-based lateral flow immunoassay combined with image analysis for semiquantitative detection of IgE antibody to mite. Int. J. Nanomed. 2017, 12, 4805. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vo, N.; Ngo, H.; Vu, D.; Duong, A.; Lam, Q. Conjugation of E. coli O157: H7 antibody to CdSe/ZnS quantum dots. J. Nanomater. 2015, 2015, 8. [Google Scholar] [CrossRef]
- Kotagiri, N.; Li, Z.; Xu, X.; Mondal, S.; Nehorai, A.; Achilefu, S. Antibody quantum dot conjugates developed via copper-free click chemistry for rapid analysis of biological samples using a microfluidic microsphere array system. Bioconjugate Chem. 2014, 25, 1272–1281. [Google Scholar] [CrossRef]
- Ahmad, S.A.A.; Zaini, M.S.; Kamarudin, M.A. An Electrochemical Sandwich Immunosensor for the Detection of HER2 using Antibody-Conjugated PbS Quantum Dot as a label. J. Pharm. Biomed. Anal. 2019, 174, 608–617. [Google Scholar]
- Rizvi, S.B.; Rouhi, S.; Taniguchi, S.; Yang, S.Y.; Green, M.; Keshtgar, M.; Seifalian, A.M. Near-infrared quantum dots for HER2 localization and imaging of cancer cells. Int. J. Nanomed. 2014, 9, 1323. [Google Scholar]
- Pinaud, F.; King, D.; Moore, H.-P.; Weiss, S. Bioactivation and cell targeting of semiconductor CdSe/ZnS nanocrystals with phytochelatin-related peptides. J. Am. Chem. Soc. 2004, 126, 6115–6123. [Google Scholar] [CrossRef] [PubMed]
- Gerion, D.; Pinaud, F.; Williams, S.C.; Parak, W.J.; Zanchet, D.; Weiss, S.; Alivisatos, A.P. Synthesis and properties of biocompatible water-soluble silica-coated CdSe/ZnS semiconductor quantum dots. J. Phys. Chem. B 2001, 105, 8861–8871. [Google Scholar] [CrossRef]
- Parce, J.W.; Nikiforov, T.T.; Mehta, T.B.; Kopf-Sill, A.R.; Chow, A.W.; Knapp, M.R. Sequencing by Incorporation. U.S. Patent 6613513B1, 2 September 2003. [Google Scholar]
- Nehilla, B.J.; Vu, T.Q.; Desai, T.A. Stoichiometry-Dependent Formation of Quantum Dot− Antibody Bioconjugates: A Complementary Atomic Force Microscopy and Agarose Gel Electrophoresis Study. J. Phys. Chem. B 2005, 109, 20724–20730. [Google Scholar] [CrossRef] [PubMed]
- Rakovich, T.Y.; Mahfoud, O.K.; Mohamed, B.M.; Prina-Mello, A.; Crosbie-Staunton, K.; Van Den Broeck, T.; De Kimpe, L.; Sukhanova, A.; Baty, D.; Rakovich, A. Highly sensitive single domain antibody–quantum dot conjugates for detection of HER2 biomarker in lung and breast cancer cells. ACS Nano 2014, 8, 5682–5695. [Google Scholar] [CrossRef]
- Kairdolf, B.A.; Smith, A.M.; Stokes, T.H.; Wang, M.D.; Young, A.N.; Nie, S. Semiconductor quantum dots for bioimaging and biodiagnostic applications. Annu. Rev. Anal. Chem. 2013, 6, 143–162. [Google Scholar] [CrossRef]
- Azzazy, H.M.; Mansour, M.M.; Kazmierczak, S.C. From diagnostics to therapy: Prospects of quantum dots. Clin. Biochem. 2007, 40, 917–927. [Google Scholar] [CrossRef]
- Schroeder, J.; Shweky, I.; Shmeeda, H.; Banin, U.; Gabizon, A. Folate-mediated tumor cell uptake of quantum dots entrapped in lipid nanoparticles. J. Control. Release 2007, 124, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Walling, M.A.; Novak, J.A.; Shepard, J.R. Quantum dots for live cell and in vivo imaging. Int. J. Mol. Sci. 2009, 10, 441–491. [Google Scholar] [CrossRef] [PubMed]
- Maysinger, D.; Behrendt, M.; Lalancette-Hébert, M.; Kriz, J. Real-time imaging of astrocyte response to quantum dots: In vivo screening model system for biocompatibility of nanoparticles. Nano Lett. 2007, 7, 2513–2520. [Google Scholar] [CrossRef] [PubMed]
- Bakalova, R.; Zhelev, Z.; Aoki, I.; Masamoto, K.; Mileva, M.; Obata, T.; Higuchi, M.; Gadjeva, V.; Kanno, I. Multimodal silica-shelled quantum dots: Direct intracellular delivery, photosensitization, toxic, and microcirculation effects. Bioconjugate Chem. 2008, 19, 1135–1142. [Google Scholar] [CrossRef]
- Chang, J.-C.; Su, H.-L.; Hsu, S.-h. The use of peptide-delivery to protect human adipose-derived adult stem cells from damage caused by the internalization of quantum dots. Biomaterials 2008, 29, 925–936. [Google Scholar] [CrossRef]
- Hoshino, A.; Hanaki, K.-I.; Suzuki, K.; Yamamoto, K. Applications of T-lymphoma labeled with fluorescent quantum dots to cell tracing markers in mouse body. Biochem. Biophys. Res. Commun. 2004, 314, 46–53. [Google Scholar] [CrossRef]
- Beer, A.J.; Schwaiger, M. Imaging of integrin αvβ3 expression. Cancer Metastasis Rev. 2008, 27, 631–644. [Google Scholar] [CrossRef]
- Molaei, M.J. Carbon quantum dots and their biomedical and therapeutic applications: A review. RSC Adv. 2019, 9, 6460–6481. [Google Scholar] [CrossRef]
- Bhuckory, S.; Mattera, L.; Wegner, K.; Qiu, X.; Wu, Y.-T.; Charbonniere, L.; Reiss, P.; Hildebrandt, N. Direct conjugation of antibodies to the ZnS shell of quantum dots for FRET immunoassays with low picomolar detection limits. Chem. Commun. 2016, 52, 14423–14425. [Google Scholar] [CrossRef]
- Chakraborty, I.; Pradeep, T. Atomically precise clusters of noble metals: Emerging link between atoms and nanoparticles. Chem. Rev. 2017, 117, 8208–8271. [Google Scholar] [CrossRef] [PubMed]
- Ghazani, A.A.; Lee, J.A.; Klostranec, J.; Xiang, Q.; Dacosta, R.S.; Wilson, B.C.; Tsao, M.S.; Chan, W.C. High throughput quantification of protein expression of cancer antigens in tissue microarray using quantum dot nanocrystals. Nano Lett. 2006, 6, 2881–2886. [Google Scholar] [CrossRef]
- Chen, J.; Pan, J.; Zhao, J.; Qiu, X.; Zheng, J.; Wang, Z.; Huang, Y.; Chu, H. Quantum dot imaging for HSP70 and HSF-1 kinetics in SCC-25 cells with or without leucine deprivation following heat shock. Oncol. Rep. 2013, 29, 2255–2260. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.-Z.; Chen, C.; Jiang, G.; Tian, W.-Q.; Li, Y.; Sun, S.-R. Quantum dot-based immunofluorescent imaging of Ki67 and identification of prognostic value in HER2-positive (non-luminal) breast cancer. Int. J. Nanomed. 2014, 9, 1339. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, J.J.; Yazan, L.S.; Abdullah, C.A.C. A review on current nanomaterials and their drug conjugate for targeted breast cancer treatment. Int. J. Nanomed. 2017, 12, 2373. [Google Scholar] [CrossRef] [PubMed]
- Kievit, F.M.; Zhang, M. Cancer nanotheranostics: Improving imaging and therapy by targeted delivery across biological barriers. Adv. Mater. 2011, 23, H217–H247. [Google Scholar] [CrossRef]








| Top-Down Approach | ||||
|---|---|---|---|---|
| Method | Procedure | Advantages | Particle Size (nm) | Ref |
| Electron beam lithography | Beam of electron strikes with electron-sensitive film | A high degree of flexibility | ≈30 | [58] |
| Focused ion beam (FIB) techniques | FIB functioned at high or low beam currents for imaging and can be operated at site-specific sputtering | Ultra-small radius particles | 8–20 | [58,59] |
| Etching techniques | Following the trapping of reactive gas species in an etching system, the formed plasma breaks down the gas molecules into more reactive fragments with the assistance of a controlled radiofrequency voltage | Close-packed arrays formed | 40 | [58,60] |
| Bottom–Up Approach | ||
|---|---|---|
| Method | Description | Ref |
| Wet-chemical methods | Precipitation technique using one or more solvents. Different process parameters are carefully controlled, and QDs of specific size and shape are prepared by a controlling ratio of cationic and anionic species, temperature, and double-layer thickness. Bhand et al. synthesized CdTe QDs using the wet-chemical method and employed characterization techniques to investigate their optical, compositional, and morphological properties. The resulting QDs were in size range of 2.5–4 nm from the absorption peak position. | [61] |
| Physical vapor deposition (PVD) | Condensation is the phenomenon involved in this case. The most commonly used PVD technique is sputtering. In PVD, a solid or liquid source is used to vaporize the material and transported to a vacuum, and then condensation occurs. Melvin et al. synthesized CdSe/CdTe colloidal QDs by a physical vapor deposition method. The PVD method has previously been used to generate niobium oxide (Nb2O5), CdSe/CdTeQDs, and other nanocrystals. | [58] |
| Targeted Deliveries | Details | Ref |
|---|---|---|
| Near-IR QDs-loaded micelles | The high absorption of the near-IR QDs-loaded micelles by HER2 receptor-overexpressing carcinoma cells, which are distributed fast in-vivo. | [112] |
| Inorganic nanoparticles with a core/shell configuration. Metals (e.g., iron oxide, gold, and QDs) or organic fluorescent dyes encapsulated in silica form the core, while metals or organic polymers form the shell | The HER2 receptor is exploited to target magnetic NPs in breast cancer tissue for imaging and diagnostic purposes. | [113] |
| Biomarker-targeted fluorescent probes | Breast cancer cells and associated animal models are scanned using fluorescent probes. Fluorescent probes are superior to other diagnostic methods in terms of high selectivity, sensitivity, specificity, signal intensity, and early diagnosis. | [114] |
| QDS-based inorganic NPs for HER2 receptor targeting | The application of targeted multimodal NPs as vectors in therapeutic purposes, sources of signals in diagnostic purposes, and monitor endogenous response simultaneously. | [115] |
| QDs in breast cancer therapy | As a result of their ultra-sensitivity, bio-conjugated QDs were used to detect metastasis, especially micro-metastasis. QDs may also be employed to track circulating tumor cells or even circulating stem cells, which are essential prognosis factors and sources of cancer recurrence. | [116] |
| Advantages | Disadvantages | Ref |
|---|---|---|
| Biological imaging, diagnosis, and therapeutic applications | High toxicity | [149] |
| High photostability and no photobleaching | In high breast cancer cell lines, QDs are epigenetic and genotoxic carcinogens | [149] |
| Versatility for bio-conjugation with various biomolecules | Detection or diagnosis of cancer is limited to superficial sites | [150] |
| In comparison with organic fluorophores, the QDs amplify signal brightness up to 10–100 times more | As a result of the oxidative breakdown of its heavy metal core, metal ions will be released into the environment, where they will attach to the thiol functional group of intracellular proteins and cause the function of subcellular organelles to be impaired | [150] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fatima, I.; Rahdar, A.; Sargazi, S.; Barani, M.; Hassanisaadi, M.; Thakur, V.K. Quantum Dots: Synthesis, Antibody Conjugation, and HER2-Receptor Targeting for Breast Cancer Therapy. J. Funct. Biomater. 2021, 12, 75. https://doi.org/10.3390/jfb12040075
Fatima I, Rahdar A, Sargazi S, Barani M, Hassanisaadi M, Thakur VK. Quantum Dots: Synthesis, Antibody Conjugation, and HER2-Receptor Targeting for Breast Cancer Therapy. Journal of Functional Biomaterials. 2021; 12(4):75. https://doi.org/10.3390/jfb12040075
Chicago/Turabian StyleFatima, Iqra, Abbas Rahdar, Saman Sargazi, Mahmood Barani, Mohadeseh Hassanisaadi, and Vijay Kumar Thakur. 2021. "Quantum Dots: Synthesis, Antibody Conjugation, and HER2-Receptor Targeting for Breast Cancer Therapy" Journal of Functional Biomaterials 12, no. 4: 75. https://doi.org/10.3390/jfb12040075
APA StyleFatima, I., Rahdar, A., Sargazi, S., Barani, M., Hassanisaadi, M., & Thakur, V. K. (2021). Quantum Dots: Synthesis, Antibody Conjugation, and HER2-Receptor Targeting for Breast Cancer Therapy. Journal of Functional Biomaterials, 12(4), 75. https://doi.org/10.3390/jfb12040075











